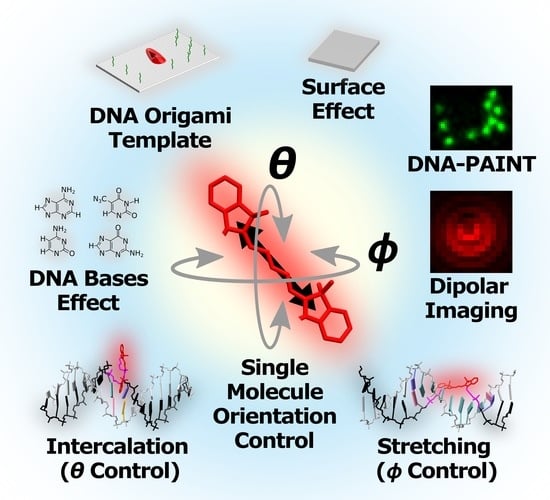
Strategies for Controlling the Spatial Orientation of Single Molecules Tethered on DNA Origami Templates Physisorbed on Glass Substrates: Intercalation and Stretching
Abstract
:1. Introduction
2. Results and Discussion
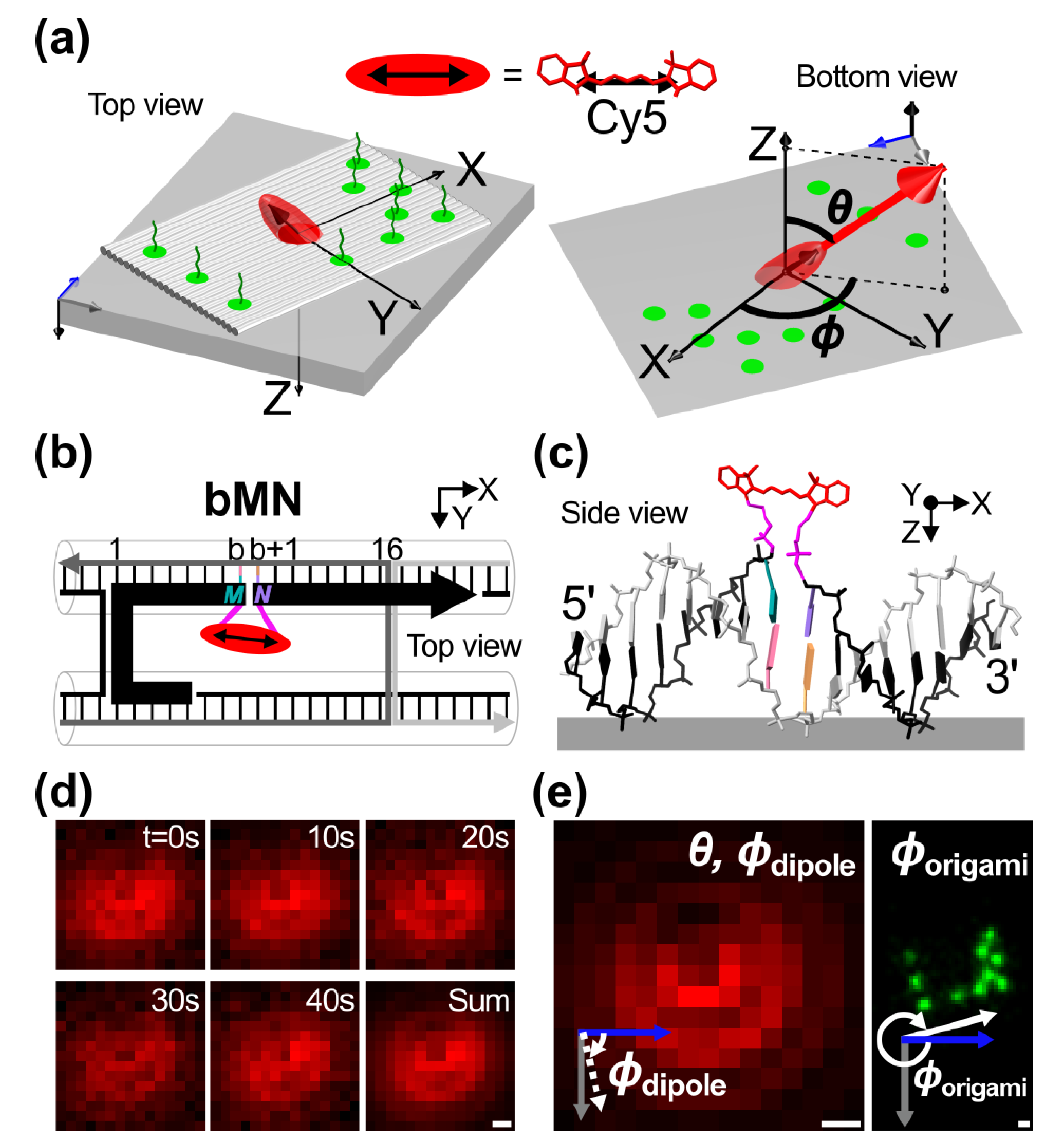
2.1. Template Design and Orientation Measurements
2.2. θ Orientation Control via Intercalation and Attachment Position
2.2.1. Effect of the Neighboring Bases
2.2.2. Effect of the Surface
2.3. ϕ Orientation Control via Mechanical Stretching
3. Materials and Methods
3.1. DNA Origami Fabrication
3.2. Glass Substrate Preparation
3.3. Chamber Preparation and DNA Origami Physisorption onto Glass Substrate
3.4. Fluorescence Microscopy for Single Dipole Imaging
3.5. Fluorescence Microscopy for Super-Resolution
3.6. Data Processing
3.7. Density Functional Theory (DFT)
3.8. Molecular Dynamics (MD)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seeman, N.C.; Sleiman, H.F. DNA Nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar] [CrossRef]
- Zhong, Q.; Ihle, A.; Ahles, S.; Wegner, H.A.; Schirmeisen, A.; Ebeling, D. Constructing Covalent Organic Nanoarchitectures Molecule by Molecule via Scanning Probe Manipulation. Nat. Chem. 2021, 13, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kashida, H.; Kishida, H.; Yada, H.; Okamoto, H.; Asanuma, H. Development of a Robust Model System of FRET Using Base Surrogates Tethering Fluorophores for Strict Control of Their Position and Orientation within DNA Duplex. J. Am. Chem. Soc. 2013, 135, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, A.; Thachuk, C.; Mitskovets, A.; Atwater, H.A.; Kirkpatrick, D.; Rothemund, P.W.K. Absolute and Arbitrary Orientation of Single-Molecule Shapes. Science 2021, 371, eabd6179. [Google Scholar] [CrossRef]
- Wang, X.; Deshmukh, R.; Sha, R.; Birktoft, J.J.; Menon, V.; Seeman, N.C.; Canary, J.W. Orienting an Organic Semiconductor into DNA 3D Arrays by Covalent Bonds. Angew. Chem. Int. Ed. 2022, 61, e202115155. [Google Scholar] [CrossRef]
- Lin, K.; Tutunnikov, I.; Qiang, J.; Ma, J.; Song, Q.; Ji, Q.; Zhang, W.; Li, H.; Sun, F.; Gong, X.; et al. All-Optical Field-Free Three-Dimensional Orientation of Asymmetric-Top Molecules. Nat. Commun. 2018, 9, 5134. [Google Scholar] [CrossRef] [Green Version]
- Simpson, G.J.; García-López, V.; Daniel Boese, A.; Tour, J.M.; Grill, L. How to Control Single-Molecule Rotation. Nat. Commun. 2019, 10, 4631. [Google Scholar] [CrossRef] [Green Version]
- Backlund, M.P.; Lew, M.D.; Backer, A.S.; Sahl, S.J.; Moerner, W.E. The Role of Molecular Dipole Orientation in Single-Molecule Fluorescence Microscopy and Implications for Super-Resolution Imaging. ChemPhysChem 2014, 15, 587–599. [Google Scholar] [CrossRef] [Green Version]
- Ishii, T.; Bencheikh, F.; Forget, S.; Chénais, S.; Heinrich, B.; Kreher, D.; Sosa Vargas, L.; Miyata, K.; Onda, K.; Fujihara, T.; et al. Enhanced Light–Matter Interaction and Polariton Relaxation by the Control of Molecular Orientation. Adv. Opt. Mater. 2021, 9, 2101048. [Google Scholar] [CrossRef]
- Hofmann, A.; Schmid, M.; Brütting, W. The Many Facets of Molecular Orientation in Organic Optoelectronics. Adv. Opt. Mater. 2021, 9, 2101004. [Google Scholar] [CrossRef]
- Wasielewski, M.R.; Forbes, M.D.E.; Frank, N.L.; Kowalski, K.; Scholes, G.D.; Yuen-Zhou, J.; Baldo, M.A.; Freedman, D.E.; Goldsmith, R.H.; Goodson, T.; et al. Exploiting Chemistry and Molecular Systems for Quantum Information Science. Nat. Rev. Chem. 2020, 4, 490–504. [Google Scholar] [CrossRef]
- Feynman, R.P. There’s Plenty of Room at the Bottom. Eng. Sci. 1960, 23, 22–36. [Google Scholar]
- Mirkovic, T.; Ostroumov, E.E.; Anna, J.M.; van Grondelle, R.; Govindjee; Scholes, G.D. Light Absorption and Energy Transfer in the Antenna Complexes of Photosynthetic Organisms. Chem. Rev. 2016, 117, 249–293. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhan, Y.; Zhang, Y.; Mao, C.; Xie, X.; Lin, Y. The Biological Applications of DNA Nanomaterials: Current Challenges and Future Directions. Signal Transduct. Target. Ther. 2021, 6, 351. [Google Scholar] [CrossRef]
- Lacroix, A.; Sleiman, H.F. DNA Nanostructures: Current Challenges and Opportunities for Cellular Delivery. ACS Nano 2021, 15, 3631–3645. [Google Scholar] [CrossRef]
- Cervantes-Salguero, K.; Freeley, M.; Chávez, J.L.; Palma, M. Single-Molecule DNA Origami Aptasensors for Real-Time Biomarker Detection. J. Mater. Chem. B 2020, 8, 6352–6356. [Google Scholar] [CrossRef]
- Ochmann, S.E.; Joshi, H.; Büber, E.; Franquelim, H.G.; Stegemann, P.; Saccà, B.; Keyser, U.F.; Aksimentiev, A.; Tinnefeld, P. DNA Origami Voltage Sensors for Transmembrane Potentials with Single-Molecule Sensitivity. Nano Lett. 2021, 21, 8634–8641. [Google Scholar] [CrossRef]
- Dickinson, G.D.; Mortuza, G.M.; Clay, W.; Piantanida, L.; Green, C.M.; Watson, C.; Hayden, E.J.; Andersen, T.; Kuang, W.; Graugnard, E.; et al. An Alternative Approach to Nucleic Acid Memory. Nat. Commun. 2021, 12, 2371. [Google Scholar] [CrossRef]
- Kuzyk, A.; Jungmann, R.; Acuna, G.P.; Liu, N. DNA Origami Route for Nanophotonics. ACS Photonics 2018, 5, 1151–1163. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Li, H.; Tang, P.; Lan, X. Progress and Perspective on Chiral Plasmonic Nanostructures Enabled by DNA Programming Methodology. Mater. Adv. 2021, 2, 7336–7349. [Google Scholar] [CrossRef]
- Piantanida, L.; Naumenko, D.; Torelli, E.; Marini, M.; Bauer, D.M.; Fruk, L.; Firrao, G.; Lazzarino, M. Plasmon Resonance Tuning Using DNA Origami Actuation. Chem. Commun. 2015, 51, 4789–4792. [Google Scholar] [CrossRef] [PubMed]
- Kellis, D.L.; Sarter, C.; Cannon, B.L.; Davis, P.H.; Graugnard, E.; Lee, J.; Pensack, R.D.; Kolmar, T.; Jäschke, A.; Yurke, B.; et al. An All-Optical Excitonic Switch Operated in the Liquid and Solid Phases. ACS Nano 2019, 13, 2986–2994. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.; Bakke, M.R.; Gudnason, D.A.; Sandahl, A.F.; Hansen, R.A.; Knudsen, J.B.; Kodal, A.L.B.; Birkedal, V.; Gothelf, K.V. A Single Molecule Polyphenylene-Vinylene Photonic Wire. ACS Nano 2021, 15, 9404–9411. [Google Scholar] [CrossRef]
- Boulais, E.; Sawaya, N.P.D.; Veneziano, R.; Andreoni, A.; Banal, J.L.; Kondo, T.; Mandal, S.; Lin, S.; Schlau-Cohen, G.S.; Woodbury, N.W.; et al. Programmed Coherent Coupling in a Synthetic DNA-Based Excitonic Circuit. Nat. Mater. 2017, 17, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA Origami. Nat. Rev. Methods Primers 2021, 1, 13. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Engelhardt, F.A.S.; Praetorius, F.; Wachauf, C.H.; Brüggenthies, G.; Kohler, F.; Kick, B.; Kadletz, K.L.; Pham, P.N.; Behler, K.L.; Gerling, T.; et al. Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS Nano 2019, 13, 5015–5027. [Google Scholar] [CrossRef]
- Tsukanov, R.; Tomov, T.E.; Liber, M.; Berger, Y.; Nir, E. Developing DNA Nanotechnology Using Single-Molecule Fluorescence. Acc. Chem. Res. 2014, 47, 1789–1798. [Google Scholar] [CrossRef]
- Funke, J.J.; Dietz, H. Placing Molecules with Bohr Radius Resolution Using DNA Origami. Nat. Nanotechnol. 2015, 11, 47–52. [Google Scholar] [CrossRef]
- Furubayashi, T.; Ishida, K.; Nakata, E.; Morii, T.; Naruse, K.; Matsushita, M.; Fujiyoshi, S. Cryogenic Far-Field Fluorescence Nanoscopy: Evaluation with DNA Origami. J. Phys. Chem. B 2020, 124, 7525–7536. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.M.; Chen, W.J.; Banal, J.L.; Bricker, W.P.; Dodin, A.; Markova, L.; Vyborna, Y.; Willard, A.P.; Häner, R.; Bathe, M.; et al. Engineering Couplings for Exciton Transport Using Synthetic DNA Scaffolds. Chem 2021, 7, 752–773. [Google Scholar] [CrossRef]
- von Hippel, P.H.; Johnson, N.P.; Marcus, A.H. Fifty Years of DNA “Breathing”: Reflections on Old and New Approaches. Biopolymers 2013, 99, 923–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyerle, E.R.; Dinpajooh, M.; Ji, H.; von Hippel, P.H.; Marcus, A.H.; Guenza, M.G. Dinucleotides as Simple Models of the Base Stacking-Unstacking Component of DNA ‘Breathing’ Mechanisms. Nucleic Acids Res. 2021, 49, 1872–1885. [Google Scholar] [CrossRef]
- Roy, S.K.; Mass, O.A.; Kellis, D.L.; Wilson, C.K.; Hall, J.A.; Yurke, B.; Knowlton, W.B. Exciton Delocalization and Scaffold Stability in Bridged Nucleotide-Substituted, DNA Duplex-Templated Cyanine Aggregates. J. Phys. Chem. B 2021, 125, 13670–13684. [Google Scholar] [CrossRef]
- Mathur, D.; Kim, Y.C.; Díaz, S.A.; Cunningham, P.D.; Rolczynski, B.S.; Ancona, M.G.; Medintz, I.L.; Melinger, J.S. Can a DNA Origami Structure Constrain the Position and Orientation of an Attached Dye Molecule? J. Phys. Chem. C 2021, 125, 1509–1522. [Google Scholar] [CrossRef]
- Hübner, K.; Joshi, H.; Aksimentiev, A.; Stefani, F.D.; Tinnefeld, P.; Acuna, G.P. Determining the In-Plane Orientation and Binding Mode of Single Fluorescent Dyes in DNA Origami Structures. ACS Nano 2021, 15, 5109–5117. [Google Scholar] [CrossRef]
- Jahnke, K.; Grubmüller, H.; Igaev, M.; Göpfrich, K. Choice of Fluorophore Affects Dynamic DNA Nanostructures. Nucleic Acids Res. 2021, 49, 4186–4195. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.; Han, W.; Yin, X.; Liu, X.; Xiao, S.; Liang, H. How Fluorescent Labels Affect the Kinetics of the Toehold-Mediated DNA Strand Displacement Reaction. Chem. Commun. 2022, 58, 5849–5852. [Google Scholar] [CrossRef]
- Sindbert, S.; Kalinin, S.; Nguyen, H.; Kienzler, A.; Clima, L.; Bannwarth, W.; Appel, B.; Müller, S.; Seidel, C.A.M. Accurate Distance Determination of Nucleic Acids via Förster Resonance Energy Transfer: Implications of Dye Linker Length and Rigidity. J. Am. Chem. Soc. 2011, 133, 2463–2480. [Google Scholar] [CrossRef]
- Zacharias, M. Base-Pairing and Base-Stacking Contributions to Double-Stranded DNA Formation. J. Phys. Chem. B 2020, 124, 10345–10352. [Google Scholar] [CrossRef] [PubMed]
- Ariga, K.; Leong, D.T.; Mori, T. Nanoarchitectonics for Hybrid and Related Materials for Bio-Oriented Applications. Adv. Funct. Mater. 2018, 28, 1702905. [Google Scholar] [CrossRef]
- Howorka, S. DNA Nanoarchitectonics: Assembled DNA at Interfaces. Langmuir 2013, 29, 7344–7353. [Google Scholar] [CrossRef]
- Li, B.L.; Zhang, H.; Li, N.B.; Qian, H.; Leong, D.T. Materialistic Interfaces with Nucleic Acids: Principles and Their Impact. Adv. Funct. Mater. 2022, 2201172. [Google Scholar] [CrossRef]
- Iqbal, A.; Arslan, S.; Okumus, B.; Wilson, T.J.; Giraud, G.; Norman, D.G.; Ha, T.; Lilley, D.M.J. Orientation Dependence in Fluorescent Energy Transfer between Cy3 and Cy5 Terminally Attached to Double-Stranded Nucleic Acids. Proc. Natl. Acad. Sci. USA 2008, 105, 11176–11181. [Google Scholar] [CrossRef] [Green Version]
- Deniz, A.A.; Dahan, M.; Grunwell, J.R.; Ha, T.; Faulhaber, A.E.; Chemla, D.S.; Weiss, S.; Schultz, P.G. Single-Pair Fluorescence Resonance Energy Transfer on Freely Diffusing Molecules: Observation of Förster Distance Dependence and Subpopulations. Proc. Natl. Acad. Sci. USA 1999, 96, 3670–3675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, A.; Wang, L.; Thompson, K.C.; Lilley, D.M.J.; Norman, D.G. The Structure of Cyanine 5 Terminally Attached to Double-Stranded DNA: Implications for FRET Studies. Biochemistry 2008, 47, 7857–7862. [Google Scholar] [CrossRef]
- Aguet, F.F.; Geissbühler, S.; Geissbühler, G.; Märki, I.; Lasser, T.; Unser, M.; Patterson, H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; et al. Super-Resolution Orientation Estimation and Localization of Fluorescent Dipoles Using 3-D Steerable Filters. Opt. Express 2009, 17, 6829–6848. [Google Scholar] [CrossRef]
- Mortensen, K.I.; Sung, J.; Flyvbjerg, H.; Spudich, J.A. Optimized Measurements of Separations and Angles between Intra-Molecular Fluorescent Markers. Nat. Commun. 2015, 6, 8621. [Google Scholar] [CrossRef] [Green Version]
- Budde, J.-H.; van der Voort, N.; Felekyan, S.; Folz, J.; Kühnemuth, R.; Lauterjung, P.; Köhler, M.; Schönle, A.; Sindram, J.; Otten, M.; et al. FRET Nanoscopy Enables Seamless Imaging of Molecular Assemblies with Sub-Nanometer Resolution. arXiv 2021, arXiv:2108.00024v2. [Google Scholar] [CrossRef]
- Gidi, Y.; Bayram, S.; Ablenas, C.J.; Blum, A.S.; Cosa, G. Efficient One-Step PEG-Silane Passivation of Glass Surfaces for Single-Molecule Fluorescence Studies. ACS Appl. Mater. Interfaces 2018, 10, 39505–39511. [Google Scholar] [CrossRef] [PubMed]
- Green, C.M.; Hughes, W.L.; Graugnard, E.; Kuang, W. Correlative Super-Resolution and Atomic Force Microscopy of DNA Nanostructures and Characterization of Addressable Site Defects. ACS Nano 2021, 15, 11597–11606. [Google Scholar] [CrossRef] [PubMed]
- Heussman, D.; Kittell, J.; Kringle, L.; Tamimi, A.; von Hippel, P.H.; Marcus, A.H. Measuring Local Conformations and Conformational Disorder of (Cy3)2 Dimer Labeled DNA Fork Junctions Using Absorbance, Circular Dichroism and Two-Dimensional Fluorescence Spectroscopy. Faraday Discuss 2019, 216, 211–235. [Google Scholar] [CrossRef] [PubMed]
- Levitus, M.; Ranjit, S. Cyanine Dyes in Biophysical Research: The Photophysics of Polymethine Fluorescent Dyes in Biomolecular Environments. Q. Rev. Biophys. 2011, 44, 123–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fan, Y.; Li, D.; Sun, C.; Lei, Z.; Lu, L.; Wang, T.; Zhang, F. Anti-Quenching NIR-II Molecular Fluorophores for in Vivo High-Contrast Imaging and PH Sensing. Nat. Commun. 2019, 10, 1058. [Google Scholar] [CrossRef]
- Matikonda, S.S.; Helmerich, D.A.; Meub, M.; Beliu, G.; Kollmannsberger, P.; Greer, A.; Sauer, M.; Schnermann, M.J. Defining the Basis of Cyanine Phototruncation Enables a New Approach to Single-Molecule Localization Microscopy. ACS Cent. Sci. 2021, 7, 1144–1155. [Google Scholar] [CrossRef]
- Kreger, K.; Schmidt, H.-W.; Hildner, R.; Bricks, J.L.; Slominskii, Y.L.; Panas, I.D.; Demchenko, A.P. Fluorescent J-Aggregates of Cyanine Dyes: Basic Research and Applications Review. Methods Appl. Fluoresc. 2017, 6, 012001. [Google Scholar] [CrossRef] [Green Version]
- Jenatsch, S.; Wang, L.; Bulloni, M.; Véron, A.C.; Ruhstaller, B.; Altazin, S.; Nüesch, F.; Hany, R. Doping Evolution and Junction Formation in Stacked Cyanine Dye Light-Emitting Electrochemical Cells. ACS Appl. Mater. Interfaces 2016, 8, 6554–6562. [Google Scholar] [CrossRef]
- Kent, J.T. The Fisher-Bingham Distribution on the Sphere. J. R. Stat.Soc. Ser. B Methodol. 1982, 44, 71–80. [Google Scholar] [CrossRef]
- Fisher, N.I.; Lewis, T.; Embleton, B.J.J. Statistical Analysis of Spherical Data; paperback edition (with errata); Cambridge University Press: Cambridge, UK, 1993; ISBN 9780521242738. [Google Scholar]
- Leong, P.; Carlile, S. Methods for Spherical Data Analysis and Visualization. J. Neurosci. Methods 1998, 80, 191–200. [Google Scholar] [CrossRef]
- Olson, W.K.; Gorin, A.A.; Lu, X.J.; Hock, L.M.; Zhurkin, V.B. DNA Sequence-Dependent Deformability Deduced from Protein-DNA Crystal Complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 11163–11168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Huang, Q.; Qu, Z.; Li, C.; Li, Q.; Shi, J.; Fan, C.; Wang, L.; Zuo, X.; Shen, J.; et al. Probing Transient DNA Conformation Changes with an Intercalative Fluorescent Excimer. Angew. Chem. Int. Ed. 2021, 60, 6624–6630. [Google Scholar] [CrossRef] [PubMed]
- Nicy; Chakraborty, D.; Wales, D.J. Energy Landscapes for Base-Flipping in a Model DNA Duplex. J. Phys. Chem. B 2022, acs.jpcb.2c00340. [Google Scholar] [CrossRef] [PubMed]
- Stennett, E.M.S.; Ma, N.; van der Vaart, A.; Levitus, M. Photophysical and Dynamical Properties of Doubly Linked Cy3–DNA Constructs. J. Phys. Chem. B 2013, 118, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Yarmoluk, S.M.; Lukashov, S.S.; Ogul’Chansky, T.Y.; Losytskyy, M.Y.; Kornyushyna, O.S. Interaction of Cyanine Dyes with Nucleic Acids. XXI. Arguments for Half-Intercalation Model of Interaction. Biopolymers 2001, 62, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Aebi, U.; Pollard, T.D. A Glow Discharge Unit to Render Electron Microscope Grids and Other Surfaces Hydrophilic. J. Electron Microsc. Tech. 1987, 7, 29–33. [Google Scholar] [CrossRef]
- Schueder, F.; Stein, J.; Stehr, F.; Auer, A.; Sperl, B.; Strauss, M.T.; Schwille, P.; Jungmann, R. An Order of Magnitude Faster DNA-PAINT Imaging by Optimized Sequence Design and Buffer Conditions. Nat. Methods 2019, 16, 1101–1104. [Google Scholar] [CrossRef]
- Ovesný, M.; Křížek, P.; Borkovec, J.; Švindrych, Z.; Hagen, G.M. ThunderSTORM: A Comprehensive ImageJ Plug-in for PALM and STORM Data Analysis and Super-Resolution Imaging. Bioinformatics 2014, 30, 2389–2390. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision A.03; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Mignon, P.; Loverix, S.; Steyaert, J.; Geerlings, P. Influence of the π–π Interaction on the Hydrogen Bonding Capacity of Stacked DNA/RNA Bases. Nucleic Acids Res. 2005, 33, 1779–1789. [Google Scholar] [CrossRef] [Green Version]
- van Mourik, T.; Hogan, S.W.L. DNA Base Stacking Involving Adenine and 2-Aminopurine. Struct. Chem. 2016, 27, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Toupkanloo, H.A.; Rahmani, Z. An In-Depth Study on Noncovalent Stacking Interactions between DNA Bases and Aromatic Drug Fragments Using DFT Method and AIM Analysis: Conformers, Binding Energies, and Charge Transfer. Appl. Biol. Chem. 2018, 61, 209–226. [Google Scholar] [CrossRef] [Green Version]
- Hobza, P.; Šponer, J. Toward True DNA Base-Stacking Energies: MP2, CCSD(T), and Complete Basis Set Calculations. J. Am. Chem. Soc. 2002, 124, 11802–11808. [Google Scholar] [CrossRef] [PubMed]
- da Chai, J.; Head-Gordon, M. Long-Range Corrected Hybrid Density Functionals with Damped Atom–Atom Dispersion Corrections. Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, S.; Duran, M.; Dannenberg, J.J. How Does Basis Set Superposition Error Change the Potential Surfaces for Hydrogen-bonded Dimers? J. Chem. Phys. 1998, 105, 11024. [Google Scholar] [CrossRef] [Green Version]
- Boys, S.F.; Bernardi, F. The Calculation of Small Molecular Interactions by the Differences of Separate Total Energies. Some Procedures with Reduced Errors. Mol. Phys. 2006, 19, 553–566. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbová, M.; Šponer, J.; Otyepka, M.; Jurečka, P.; Cheatham, T.E. Assessing the Current State of Amber Force Field Modifications for DNA. J. Chem. Theory Comput. 2016, 12, 4114–4127. [Google Scholar] [CrossRef]
- Yoo, J.; Aksimentiev, A. Improved Parametrization of Li+, Na+, K+, and Mg2+ Ions for All-Atom Molecular Dynamics Simulations of Nucleic Acid Systems. J. Phys. Chem. Lett. 2012, 3, 45–50. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.D.; Kollman, P.A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges: The RESP Model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Yurke, B.; Kuang, W. Passive Linear Nanoscale Optical and Molecular Electronics Device Synthesis from Nanoparticles. Phys. Rev. A 2010, 81, 033814. [Google Scholar] [CrossRef]
- Barclay, M.S.; Roy, S.K.; Huff, J.S.; Mass, O.A.; Turner, D.B.; Wilson, C.K.; Kellis, D.L.; Terpetschnig, E.A.; Lee, J.; Davis, P.H.; et al. Rotaxane Rings Promote Oblique Packing and Extended Lifetimes in DNA-Templated Molecular Dye Aggregates. Commun. Chem. 2021, 4, 19. [Google Scholar] [CrossRef]
- Mass, O.A.; Wilson, C.K.; Roy, S.K.; Barclay, M.S.; Patten, L.K.; Terpetschnig, E.A.; Lee, J.; Pensack, R.D.; Yurke, B.; Knowlton, W.B. Exciton Delocalization in Indolenine Squaraine Aggregates Templated by DNA Holliday Junction Scaffolds. J. Phys. Chem. B 2020, 124, 9636–9647. [Google Scholar] [CrossRef]
- Gorman, J.; Orsborne, S.R.E.; Sridhar, A.; Pandya, R.; Budden, P.; Ohmann, A.; Panjwani, N.A.; Liu, Y.; Greenfield, J.L.; Dowland, S.; et al. Deoxyribonucleic Acid Encoded and Size-Defined π-Stacking of Perylene Diimides. J. Am. Chem. Soc. 2022, 144, 368–376. [Google Scholar] [CrossRef]
- Wang, X.; Sha, R.; Knowlton, W.B.; Seeman, N.C.; Canary, J.W.; Yurke, B. Exciton Delocalization in a DNA-Templated Organic Semiconductor Dimer Assembly. ACS Nano 2022, 16, 1301–1307. [Google Scholar] [CrossRef]
- Filbrun, S.L.; Zhao, F.; Chen, K.; Huang, T.-X.; Yang, M.; Cheng, X.; Dong, B.; Fang, N. Imaging Dynamic Processes in Multiple Dimensions and Length Scales. Annu. Rev. Phys. Chem. 2022, 73, 377–402. [Google Scholar] [CrossRef]
- Cervantes-Salguero, K.; Hamada, S.; Nomura, S.I.M.; Murata, S. Polymorphic Ring-Shaped Molecular Clusters Made of Shape-Variable Building Blocks. Nanomaterials 2015, 5, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Gür, F.N.; Kempter, S.; Schueder, F.; Sikeler, C.; Urban, M.J.; Jungmann, R.; Nickels, P.C.; Liedl, T. Double- to Single-Strand Transition Induces Forces and Motion in DNA Origami Nanostructures. Adv. Mater. 2021, 33, 2101986. [Google Scholar] [CrossRef] [PubMed]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A Self-Assembled Nanoscale Robotic Arm Controlled by Electric Fields. Science 2018, 359, 296–301. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Yang, S.J.; Zheng, Y.C.; Cui, Y.; Zhang, Z.; Zhong, J.Y.; Zhou, J. Electrostatic Effect of Functional Surfaces on the Activity of Adsorbed Enzymes: Simulations and Experiments. ACS Appl. Mater. Interfaces 2020, 12, 35676–35687. [Google Scholar] [CrossRef]
- Kamińska, I.; Bohlen, J.; Yaadav, R.; Schüler, P.; Raab, M.; Schröder, T.; Zähringer, J.; Zielonka, K.; Krause, S.; Tinnefeld, P. Graphene Energy Transfer for Single-Molecule Biophysics, Biosensing, and Super-Resolution Microscopy. Adv. Mater. 2021, 33. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.; Grossi, G.; McRae, E.K.S.; Rothemund, P.W.K.; Andersen, E.S. RNA Origami Design Tools Enable Cotranscriptional Folding of Kilobase-Sized Nanoscaffolds. Nat. Chem. 2021, 13, 549–558. [Google Scholar] [CrossRef]
- Aupič, J.; Strmšek, Ž.; Lapenta, F.; Pahovnik, D.; Pisanski, T.; Drobnak, I.; Ljubetič, A.; Jerala, R. Designed Folding Pathway of Modular Coiled-Coil-Based Proteins. Nat. Commun. 2021, 12, 940. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervantes-Salguero, K.; Biaggne, A.; Youngsman, J.M.; Ward, B.M.; Kim, Y.C.; Li, L.; Hall, J.A.; Knowlton, W.B.; Graugnard, E.; Kuang, W. Strategies for Controlling the Spatial Orientation of Single Molecules Tethered on DNA Origami Templates Physisorbed on Glass Substrates: Intercalation and Stretching. Int. J. Mol. Sci. 2022, 23, 7690. https://doi.org/10.3390/ijms23147690
Cervantes-Salguero K, Biaggne A, Youngsman JM, Ward BM, Kim YC, Li L, Hall JA, Knowlton WB, Graugnard E, Kuang W. Strategies for Controlling the Spatial Orientation of Single Molecules Tethered on DNA Origami Templates Physisorbed on Glass Substrates: Intercalation and Stretching. International Journal of Molecular Sciences. 2022; 23(14):7690. https://doi.org/10.3390/ijms23147690
Chicago/Turabian StyleCervantes-Salguero, Keitel, Austin Biaggne, John M. Youngsman, Brett M. Ward, Young C. Kim, Lan Li, John A. Hall, William B. Knowlton, Elton Graugnard, and Wan Kuang. 2022. "Strategies for Controlling the Spatial Orientation of Single Molecules Tethered on DNA Origami Templates Physisorbed on Glass Substrates: Intercalation and Stretching" International Journal of Molecular Sciences 23, no. 14: 7690. https://doi.org/10.3390/ijms23147690
APA StyleCervantes-Salguero, K., Biaggne, A., Youngsman, J. M., Ward, B. M., Kim, Y. C., Li, L., Hall, J. A., Knowlton, W. B., Graugnard, E., & Kuang, W. (2022). Strategies for Controlling the Spatial Orientation of Single Molecules Tethered on DNA Origami Templates Physisorbed on Glass Substrates: Intercalation and Stretching. International Journal of Molecular Sciences, 23(14), 7690. https://doi.org/10.3390/ijms23147690