Genome-Wide Identification of Glutathione S-Transferase and Expression Analysis in Response to Anthocyanin Transport in the Flesh of the New Teinturier Grape Germplasm ‘Zhongshan-HongYu’
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization of the GST Family
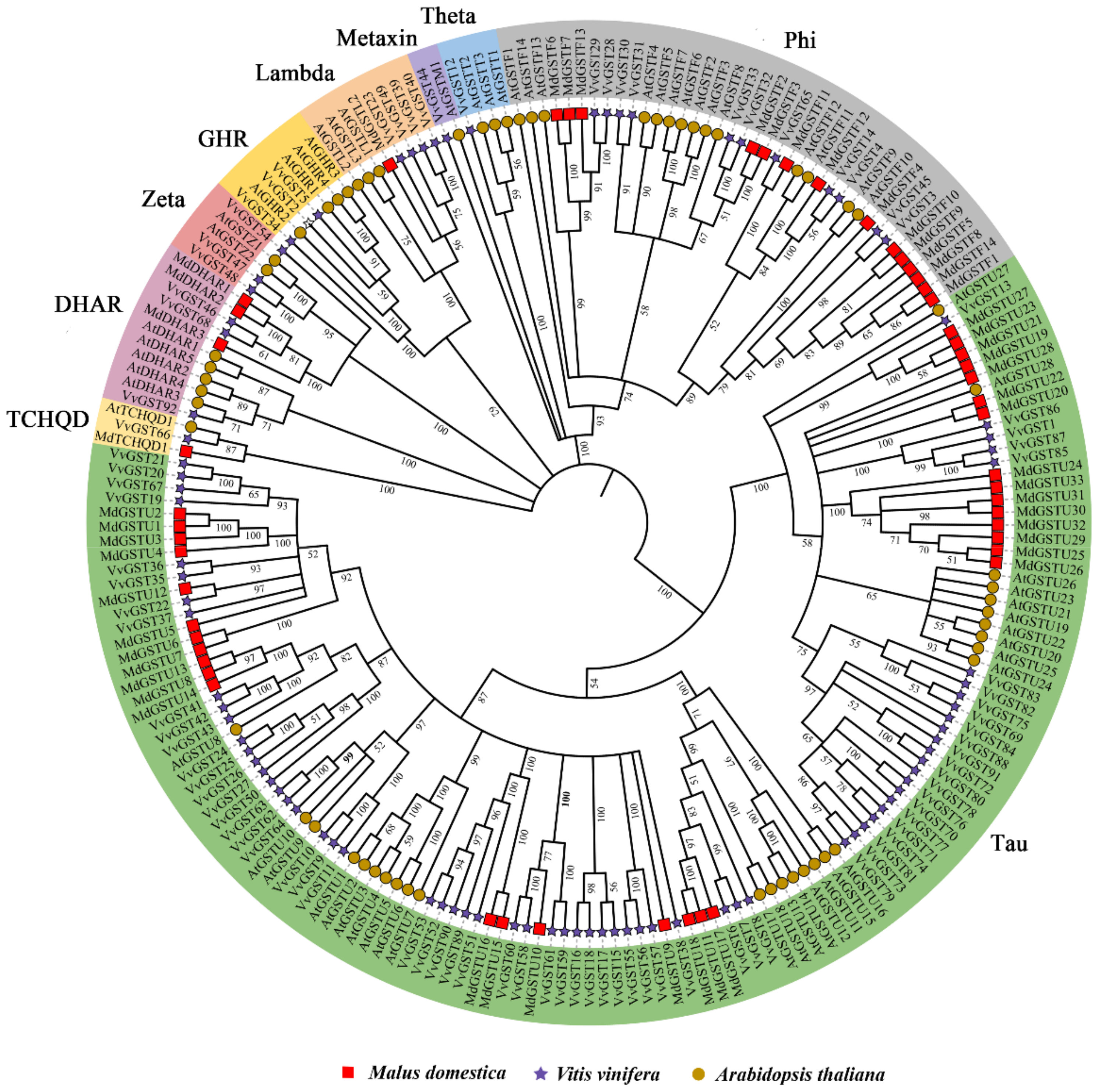
2.2. Phylogenetic Analysis of the GST Gene Family
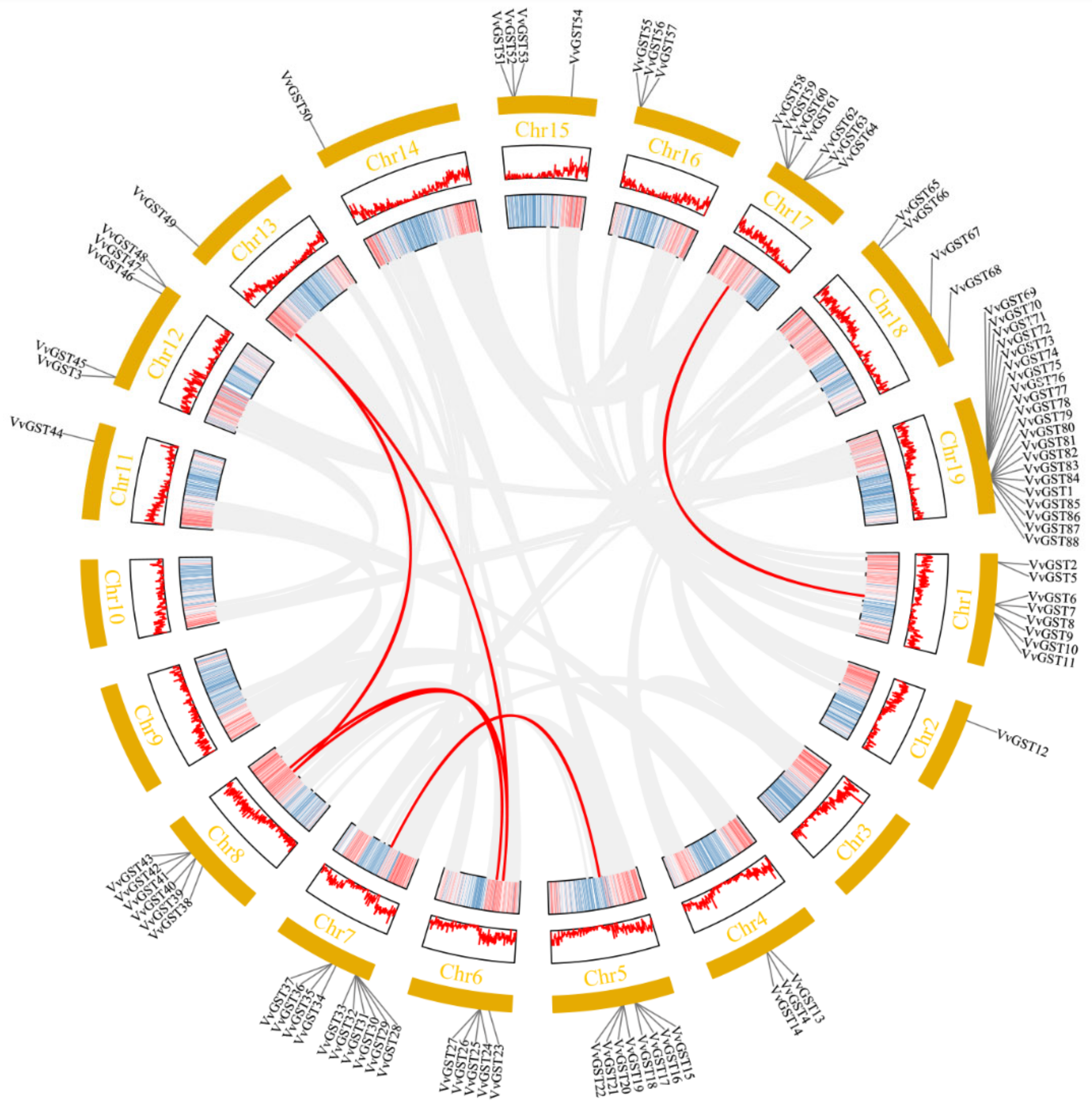
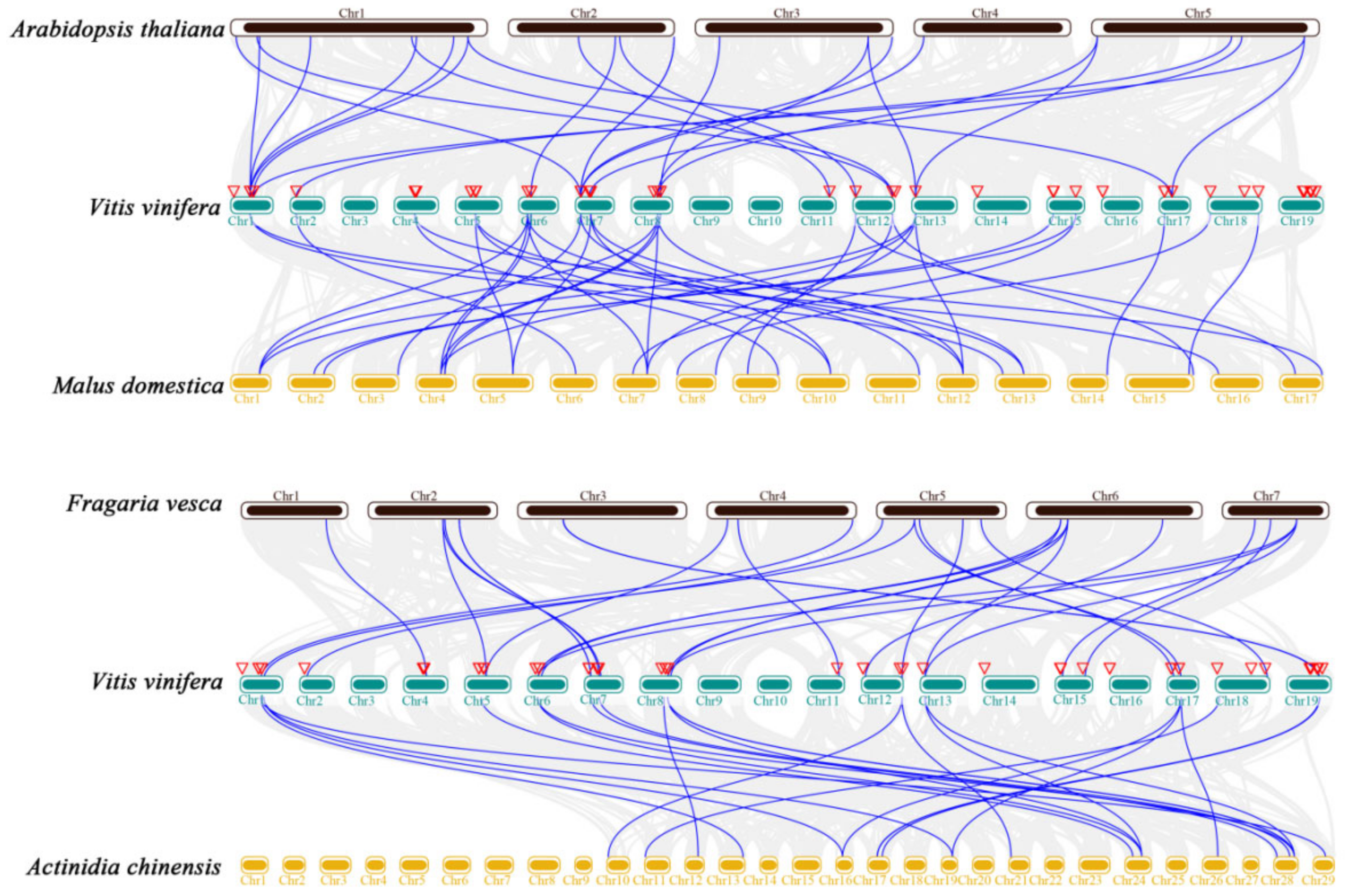
2.3. Chromosomal Location and Collinearity Analysis of GST Genes
2.4. Gene Structure and Motif Analysis of GST Genes
2.5. Identification of cis-Acting Elements in the Promoter Region of GST Genes
2.6. Phenotypic Characterization of the Flesh of ‘ZS-HY’ and ‘ZC’
2.7. Analysis of Transcriptome Data and the Expression Analysis of GST Genes in Different Tissues
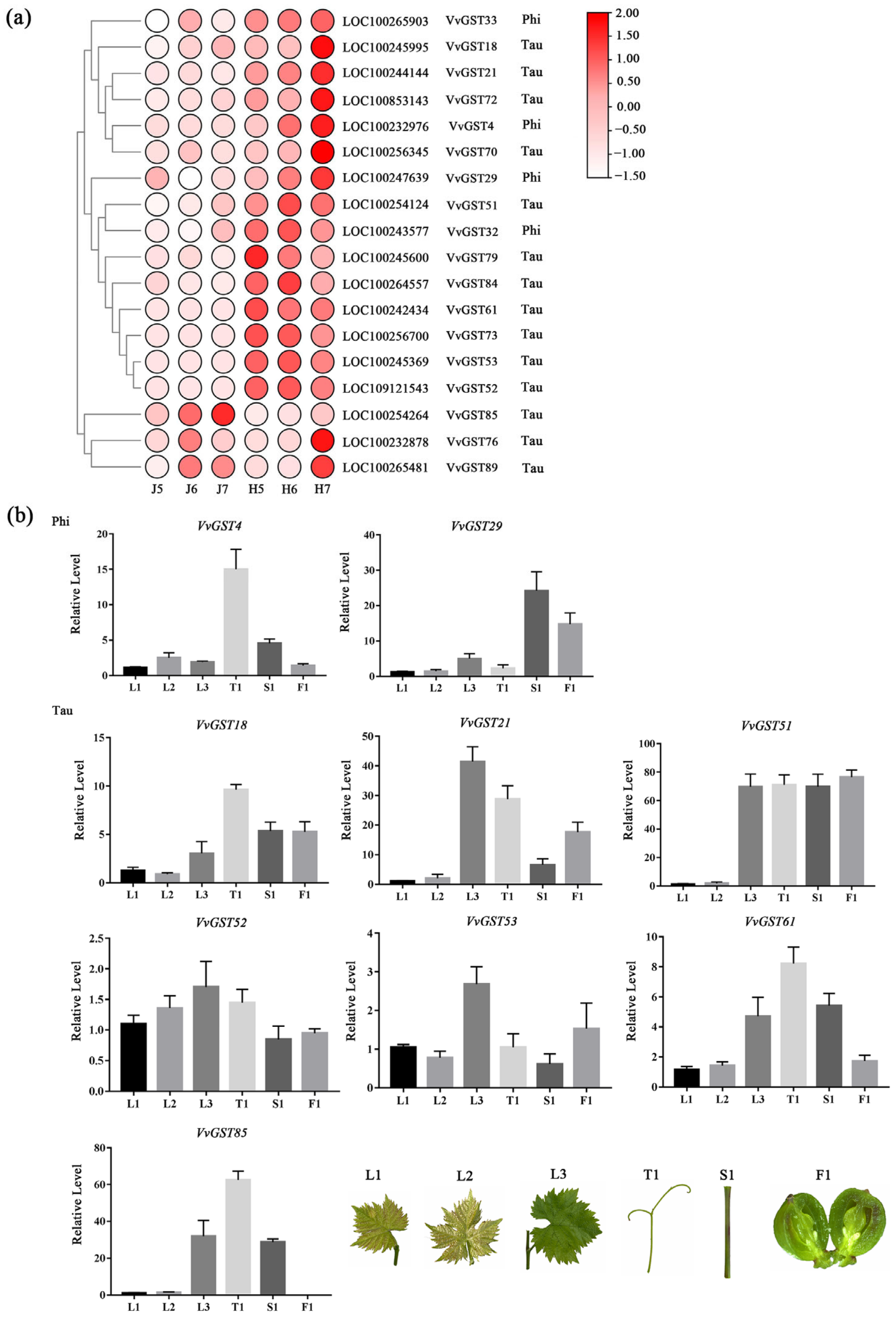
2.8. Analysis of GST Genes Evolutionary Relationships in Transcriptome Data
2.9. qRT-PCR Verification of Transcriptome Data
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of the GST Family
4.3. Phylogenetic Analysis
4.4. Chromosomal Location and Collinearity Analysis of GST
4.5. Gene Structure and Domain, and Motif Analysis
4.6. Analysis of cis-Acting Elements in GST Gene Promoters
4.7. Measurement of Anthocyanin Content
4.8. RNA-Seq Data Analysis of ‘ZS-HY’
4.9. Validation of Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frova, C. The plant glutathione transferase gene family: Genomic structure, functions, expression and evolution. Physiol. Plant. 2003, 119, 469–479. [Google Scholar] [CrossRef]
- Dixon, D.P.; Skipsey, M.; Edwards, R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 2010, 71, 338–350. [Google Scholar] [CrossRef]
- Mannervik, B.; Danielson, U.H. Glutathione Transferases—Structure and Catalytic Activity. Crit. Rev. Biochem. 1988, 23, 283–337. [Google Scholar] [CrossRef] [PubMed]
- Moons, A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTS). Vitam. Horm. 2005, 72, 155–202. [Google Scholar] [CrossRef] [PubMed]
- Cummins, I.; Dixon, D.P.; Freitag-Pohl, S.; Skipsey, M.; Edwards, R. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab. Rev. 2011, 43, 266–280. [Google Scholar] [CrossRef]
- Dixon, D.P.; Hawkins, T.; Hussey, P.J.; Edwards, R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 2009, 60, 1207–1218. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.; Dixon, D.P.; Walbot, V. Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000, 5, 193–198. [Google Scholar] [CrossRef]
- Labrou, N.E.; Papageorgiou, A.C.; Pavli, O.; Flemetakis, E. Plant GSTome: Structure and functional role in xenome network and plant stress response. Curr. Opin. Biotech. 2015, 32, 186–194. [Google Scholar] [CrossRef]
- Dixon, D.P.; Lapthorn, A.; Edwards, R. Plant glutathione transferases. Genome Biol. 2002, 3, reviews3004.1. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Wagner, U.; Edwards, R.; Dixon, D.P.; Mauch, F. Probing the diversity of the arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; An, Y.; Zheng, J.; Shangguan, L.; Wang, L. Genome-wide identification and comparative analysis of GST gene family in apple (Malus domestica) and their expressions under ALA treatment. 3 Biotech 2020, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- McGonigle, B.; Keeler, S.J.; Lau, S.M.; Koeppe, M.K.; O’Keefe, D.P. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000, 124, 1105–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licciardello, C.; D’Agostino, N.; Traini, A.; Recupero, G.R.; Frusciante, L.; Chiusano, M.L. Characterization of the glutathione S-transferase gene family through ESTs and expression analyses within common and pigmented cultivars of Citrus sinensis (L.) Osbeck. BMC Plant Biol. 2014, 14, 39. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Chen, J.; Yue, X.; Wei, X.; Zou, J.; Chen, Y.; Su, N.; Cui, J. The zinc-regulated protein (ZIP) family genes and glutathione s-transferase (GST) family genes play roles in Cd resistance and accumulation of pak choi (Brassica campestris ssp. chinensis). Ecotox. Environ. Saf. 2019, 183, 109571. [Google Scholar] [CrossRef]
- Gao, J.; Chen, B.; Lin, H.; Liu, Y.; Wei, Y.; Chen, F.; Li, W. Identification and characterization of the glutathione S-Transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene 2020, 743, 144484. [Google Scholar] [CrossRef]
- Csiszar, J.; Horvath, E.; Vary, Z.; Galle, A.; Bela, K.; Brunner, S.; Tari, I. Glutathione transferase supergene family in tomato: Salt stress-regulated expression of representative genes from distinct GST classes in plants primed with salicylic acid. Plant Physiol. Bioch. 2014, 78, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Zhou, F.; Shan, C.; Zhang, Q.; Ning, M.; Liu, X.; Zhao, X.; Cai, W.; Yang, X.; Hao, G.; et al. Identification of Glutathione S-Transferase Genes in Hami Melon (Cucumis melo var. saccharinus) and Their Expression Analysis Under Cold Stress. Front. Plant Sci. 2021, 12, 949. [Google Scholar] [CrossRef]
- Islam, S.; Das Sajib, S.; Jui, Z.S.; Arabia, S.; Islam, T.; Ghosh, A. Genome-wide identification of glutathione S-transferase gene family in pepper, its classification, and expression profiling under different anatomical and environmental conditions. Sci. Rep. 2019, 9, 9101. [Google Scholar] [CrossRef]
- Gomez, C.; Conejero, G.; Torregrosa, L.; Cheynier, V.; Terrier, N.; Ageorges, A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 2011, 67, 960–970. [Google Scholar] [CrossRef]
- Perez-Diaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; Gonzalez-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis vinifera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Liao, L.; Zhou, H.; Gu, C.; Wang, L.; Han, Y. A small indel mutation in an anthocyanin transporter causes variegated colouration of peach flowers. J. Exp. Bot. 2015, 66, 7227–7239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Cao, H.; Pan, L.; Niu, L.; Wei, B.; Cui, G.; Wang, L.; Yao, J.; Zeng, W.; Wang, Z. Two loss-of-function alleles of the glutathione S-transferase (GST) gene cause anthocyanin deficiency in flower and fruit skin of peach (Prunus persica). Plant J. 2021, 107, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Qi, Y.; Zhang, A.; Wu, H.; Liu, Z.; Ren, X. Molecular cloning and functional characterization of AcGST1, an anthocyanin-related glutathione S-transferase gene in kiwifruit (Actinidia chinensis). Plant Mol. Biol. 2019, 100, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Dai, Z.; Wu, B.; Wu, J.; Merlin, I.; Hilbert, G.; Renaud, C.; Gomes, E.; Edwards, E.; Li, S.; et al. Anthocyanin biosynthesis is differentially regulated by light in the skin and flesh of white-fleshed and teinturier grape berries. Planta 2016, 243, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Li, J.; Fan, P.; Chen, S.; Fang, J.; Li, S.; Wu, B. Anthocyanin Accumulation in Various Organs of a Teinturier Cultivar (Vitis vinifera L.) during the Growing Season. Am. J. Enol. Viticult. 2012, 63, 177–184. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Pan, Q.; Cui, X.; Duan, C. Different Anthocyanin Profiles of the Skin and the Pulp of Yan73 (Muscat Hamburg × Alicante Bouschet) Grape Berries. Molecules 2010, 15, 1141–1153. [Google Scholar] [CrossRef]
- Wang, Q.; Zheng, H.; Gao, S.; Li, H.; Tao, J. Transcriptomic analysis of berry development and a corresponding analysis of anthocyanin biosynthesis in teinturier grape. J. Plant Interact. 2019, 14, 617–629. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Gambetta, G.A.; Wada, H.; Shackel, K.A.; Matthews, M.A. Fruit ripening in Vitis vinifera: Spatiotemporal relationships among turgor, sugar accumulation, and anthocyanin biosynthesis. J. Exp. Bot. 2011, 62, 4345–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, C.; Huang, X.; Hu, D. Genome-Wide Analysis of the Glutathione S-Transferase (GST) Genes and Functional Identification of MdGSTU12 Reveals the Involvement in the Regulation of Anthocyanin Accumulation in Apple. Genes 2021, 12, 1733. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Phys. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.A.; Goodman, C.D.; Silady, R.A.; Walbot, V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000, 123, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Marrs, K.A.; Alfenito, M.R.; Lloyd, A.M.; Walbot, V. A Glutathione-S-Transferase Involved in Vacuolar Transfer Encoded by the Maize Gene Bronze-2. Nature 1995, 375, 397–400. [Google Scholar] [CrossRef]
- Larsen, E.S.; Alfenito, M.R.; Briggs, W.R.; Walbot, V. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell. Rep. 2003, 21, 900–904. [Google Scholar] [CrossRef]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Millar, A.H.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef]
- Soranzo, N.; Gorla, M.S.; Mizzi, L.; De Toma, G.; Frova, C. Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol. Genet. Genom. 2004, 271, 511–521. [Google Scholar] [CrossRef]
- Hu, B.; Zhao, J.; Lai, B.; Qin, Y.; Wang, H.; Hu, G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell. Rep. 2016, 35, 831–843. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z.; Wu, J.; Han, X.; Wang-Pruski, G.; Zhang, Z. Genome-wide identification, characterization, and expression analysis related to autotoxicity of the GST gene family in Cucumis melo L. Plant Physiol. Bioch. 2020, 155, 59–69. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, L.; Zhang, J.; Zhang, Y.; Wang, Y.; Chen, Q.; Luo, Y.; Zhang, Y.; Li, M.; Wang, X.; et al. Identification of Anthocyanins-Related Glutathione S-Transferase (GST) Genes in the Genome of Cultivated Strawberry (Fragaria × ananassa). Int. J. Mol. Sci. 2020, 21, 8708. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Li, C.; Zhang, Y.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Glutathione S-Transferase Gene Family in Gossypium raimondii and G. arboreum: Comparative Genomic Study and their Expression under Salt Stress. Front. Plant Sci. 2016, 7, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yin, X.; Li, X.; Wang, L.; Zheng, Y.; Xu, X.; Zhang, Y.; Wang, X. Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 2014, 15, 5730–5748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, M.; Deutsch, M. Association of intron phases with conservation at splice site sequences and evolution of spliceosomal introns. Mol. Biol. Evol. 1999, 16, 1528–1534. [Google Scholar] [CrossRef] [Green Version]
- Conn, S.; Curtin, C.; Bezier, A.; Franco, C.; Zhang, W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [Green Version]
- Varanasi, A.; Worthington, M.; Nelson, L.; Brown, A.; Chizk, T.M.; Threlfall, R.; Howard, L.; Conner, P.; Figueroa-Balderas, R.; Massonnet, M.; et al. Glutathione S-transferase: A candidate gene for berry color in muscadine grapes (Vitis rotundifolia). G3 2022, 12, jkac060. [Google Scholar] [CrossRef]
- Wangwattana, B.; Koyama, Y.; Nishiyama, Y.; Kitayama, M.; Yamazaki, M.; Saito, K. Characterization of PAP1-upregulated Glutathione S-transferase genes in Arabidopsis thaliana. Plant Biotechnol. Nar. 2008, 25, 191–196. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Dai, C.; Li, Y.; Feng, J.; Liu, Z.; Kang, C. Reduced Anthocyanins in Petioles codes for a GST anthocyanin transporter that is essential for the foliage and fruit coloration in strawberry. J. Exp. Bot. 2018, 69, 2595–2608. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Chen, M.; He, N.; Chen, X.; Wang, N.; Sun, Q.; Zhang, T.; Xu, H.; Fang, H.; Wang, Y.; et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Hortic. Res. 2019, 6, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Dong, W.; Wang, K.; Zhang, B.; Allan, A.C.; Lin-Wang, K.; Chen, K.; Xu, C. Differential Sensitivity of Fruit Pigmentation to Ultraviolet Light between Two Peach Cultivars. Front. Plant Sci. 2017, 8, 1552. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, Y.S.; Jia, Y.; Wang, J.Y.; Yuan, Y.; Yu, Y.; Tao, J.M. Effect of floral cluster pruning on anthocyanin levels and anthocyanain-related gene expression in ‘Houman’ grape. Hortic. Res. 2016, 3, 16037. [Google Scholar] [CrossRef]






| Component Name | Content (μg/g−1) FW | |||||
|---|---|---|---|---|---|---|
| J5 | J6 | J7 | H5 | H6 | H7 | |
| Malvidin 3-O-glucoside | ---- | ---- | ---- | 0.65 | 0.67 | 1.21 |
| Malvidin 3-O-(6″-p-coumaroyl-glucoside) | ---- | ---- | ---- | 0.65 | 0.70 | ---- |
| Peonidin 3-O-glucoside | ---- | ---- | ---- | 0.62 | 0.73 | 1.33 |
| Malvidin 3-O-(6″-caffeoyl-glucoside) | ---- | ---- | ---- | 0.60 | 0.63 | 0.90 |
| Peonidin 3-O-(6″-caffeoyl-glucoside) | ---- | ---- | ---- | ---- | 0.61 | 0.72 |
| Peonidin 3-O-feruloyl-glucoside | ---- | ---- | ---- | ---- | ---- | 1.49 |
| Cyanidin 3-O-glucoside | ---- | ---- | ---- | ---- | ---- | 0.65 |
| Petunidin 3-O-(6″-p-coumaroyl-glucoside) | ---- | ---- | ---- | ---- | ---- | 0.61 |
| Cyanidin 3-O-(6″-p-coumaroyl-glucoside) | ---- | ---- | ---- | ---- | ---- | 0.60 |
| Total anthocyanins | 0 | 0 | 0 | 2.52 | 3.34 | 7.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Yang, Y.; Li, H.; Wang, W.; Zheng, H.; Tao, J. Genome-Wide Identification of Glutathione S-Transferase and Expression Analysis in Response to Anthocyanin Transport in the Flesh of the New Teinturier Grape Germplasm ‘Zhongshan-HongYu’. Int. J. Mol. Sci. 2022, 23, 7717. https://doi.org/10.3390/ijms23147717
Li H, Yang Y, Li H, Wang W, Zheng H, Tao J. Genome-Wide Identification of Glutathione S-Transferase and Expression Analysis in Response to Anthocyanin Transport in the Flesh of the New Teinturier Grape Germplasm ‘Zhongshan-HongYu’. International Journal of Molecular Sciences. 2022; 23(14):7717. https://doi.org/10.3390/ijms23147717
Chicago/Turabian StyleLi, Hui, Yaxin Yang, Haoran Li, Wu Wang, Huan Zheng, and Jianmin Tao. 2022. "Genome-Wide Identification of Glutathione S-Transferase and Expression Analysis in Response to Anthocyanin Transport in the Flesh of the New Teinturier Grape Germplasm ‘Zhongshan-HongYu’" International Journal of Molecular Sciences 23, no. 14: 7717. https://doi.org/10.3390/ijms23147717
APA StyleLi, H., Yang, Y., Li, H., Wang, W., Zheng, H., & Tao, J. (2022). Genome-Wide Identification of Glutathione S-Transferase and Expression Analysis in Response to Anthocyanin Transport in the Flesh of the New Teinturier Grape Germplasm ‘Zhongshan-HongYu’. International Journal of Molecular Sciences, 23(14), 7717. https://doi.org/10.3390/ijms23147717





