Complex Diagnostics of Non-Specific Intellectual Developmental Disorder
Abstract
:1. Introduction
2. Results
2.1. Patients
2.2. Range of Variants Identified by ES
2.3. Candidate Variants
2.4. Spectrum of Variants Identified by CMA
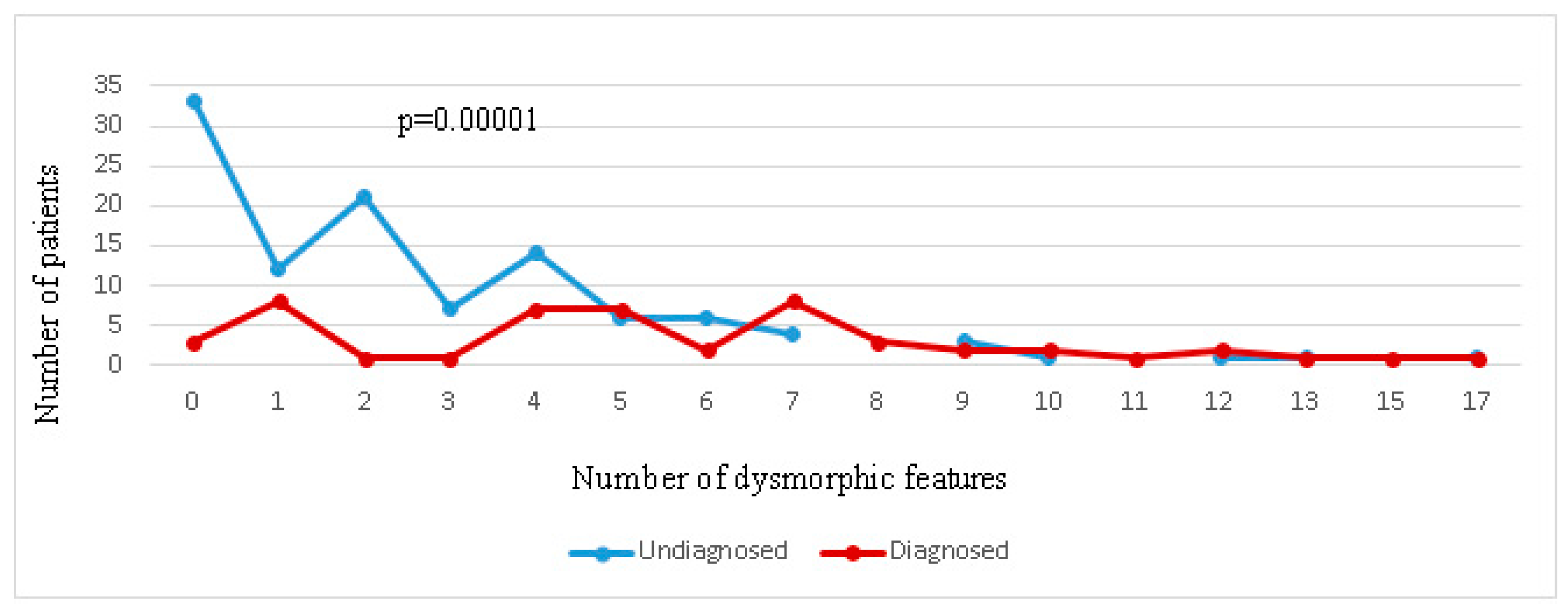
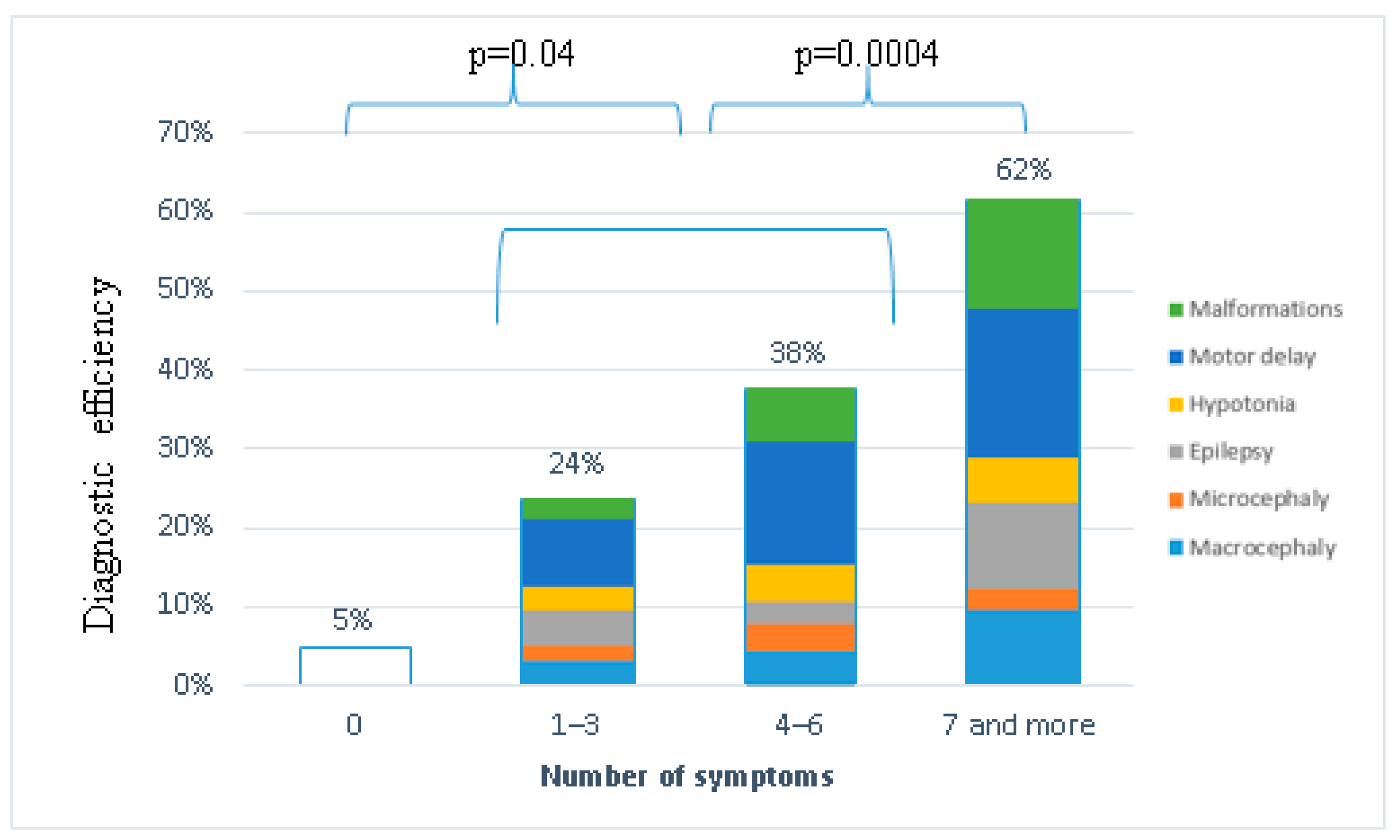
2.5. Genotype–Phenotype Correlation
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Isolation of Genomic DNA
4.3. NGS
4.4. Bioinformatic Analysis
4.5. Chromosomal Microarray Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- King, B.H.; Toth, K.E.; Hodapp, R.M.D.E. Intellectual Disability. In Comprehensive Textbook of Psychiatry, 9th ed.; Sadock, B.J., Sadock, V.A., Ruiz, P., Eds.; Lippincott: Philadelphia, PA, USA, 2009; pp. 3444–3474. [Google Scholar]
- Leonard, H.; Wen, X. The Epidemiology of Mental Retardation: Challenges and Opportunities in the New Millennium. Ment. Retard. Dev. Disabil. Res. Rev. 2002, 8, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Carulla, L.S.; Reed, G.M.; Vaez-Azizi, L.M.; Cooper, S.-A.; Leal, R.M.; Bertelli, M.; Adnams, C.; Cooray, S.; Deb, S.; Dirani, L.A.; et al. Intellectual Developmental Disorders: Towards a New Name, Definition and Framework for “ Mental Retardation /Intellectual Disability ” in ICD-11. World Psychiatry 2011, 10, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Bass, N.; Skuse, D. Genetic Testing in Children and Adolescents with Intellectual Disability. Curr. Opin. Psychiatry 2018, 31, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redin, C.; Gérard, B.; Lauer, J.; Herenger, Y.; Muller, J.; Quartier, A.; Masurel-Paulet, A.; Willems, M.; Lesca, G.; El-Chehadeh, S.; et al. Efficient Strategy for the Molecular Diagnosis of Intellectual Disability Using Targeted High-Throughput Sequencing. J. Med. Genet. 2014, 51, 724–736. [Google Scholar] [CrossRef]
- van der Sluijs, P.J.; Jansen, S.; Vergano, S.A.; Adachi-Fukuda, M.; Alanay, Y.; AlKindy, A.; Baban, A.; Bayat, A.; Beck-Wödl, S.; Berry, K.; et al. The ARID1B Spectrum in 143 Patients: From Nonsyndromic Intellectual Disability to Coffin–Siris Syndrome. Genet. Med. 2019, 21, 1295–1307. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.F.; Fitzgerald, T.W.; Jones, W.D.; Clayton, S.; McRae, J.F.; Van Kogelenberg, M.; King, D.A.; Ambridge, K.; Barrett, D.M.; Bayzetinova, T.; et al. Genetic Diagnosis of Developmental Disorders in the DDD Study: A Scalable Analysis of Genome-Wide Research Data. Lancet 2015, 385, 1305–1314. [Google Scholar] [CrossRef] [Green Version]
- Bélanger, S.A.; Caron, J. Evaluation of the Child with Global Developmental Delay and Intellectual Disability. Paediatr. Child Health 2018, 23, 403–410. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [Green Version]
- Vissers, L.E.L.M.; De Ligt, J.; Gilissen, C.; Janssen, I.; Steehouwer, M.; De Vries, P.; Van Lier, B.; Arts, P.; Wieskamp, N.; Rosario, M.; et al. A de Novo Paradigm for Mental Retardation. Nat. Genet. 2010, 42, 1109–1112. [Google Scholar] [CrossRef]
- Van Dijck, A.; Silfhout, A.T.V.; Cappuyns, E.; Van Der Werf, I.M.; Mancini, M.; Tzschach, A.; Bernier, R.; Gozes, I.; Eichler, E.E.; Lindstrand, A.; et al. Clinical Presentation of a Complex Neurodevelopmental Disorder Caused by Mutations in ADNP. Biol. Psychiatry 2019, 85, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Voisin, N.; Schnur, R.E.; Douzgou, S.; Hiatt, S.M.; Rustad, C.F.; Brown, N.J.; Earl, D.L.; Keren, B.; Levchenko, O.; Geuer, S.; et al. Variants in the Degron of AFF3 Are Associated with Intellectual Disability, Mesomelic Dysplasia, Horseshoe Kidney, and Epileptic Encephalopathy. Am. J. Hum. Genet. 2021, 108, 857–873. [Google Scholar] [CrossRef] [PubMed]
- De Ligt, J.; Willemsen, M.H.; van Bon, B.W.M.; Kleefstra, T.; Yntema, H.G.; Kroes, T.; Vulto-van Silfhout, A.T.; Koolen, D.A.; de Vries, P.; Gilissen, C.; et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N. Engl. J. Med. 2012, 367, 1921–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldhammer, M.; Durand, S.; Pshezhetsky, A.V. Protein Misfolding as an Underlying Molecular Defect in Mucopolysaccharidosis III Type C. PLoS ONE 2009, 4, e743. [Google Scholar] [CrossRef] [PubMed]
- Kosmicki, J.A.; Samocha, K.E.; Howrigan, D.P.; Stephan, J.; Slowikowski, K.; Lek, M.; Karczewski, K.J.; David, J.; Devlin, B.; Roeder, K.; et al. Refining the Role of de Novo Protein Truncating Variants in Neurodevelopmental Disorders Using Population Reference Samples. Nat. Genet. 2017, 49, 504–510. [Google Scholar] [CrossRef] [Green Version]
- Snijders Blok, L.; Hiatt, S.M.; Bowling, K.M.; Prokop, J.W.; Engel, K.L.; Cochran, J.N.; Bebin, E.M.; Bijlsma, E.K.; Ruivenkamp, C.A.L.; Terhal, P.; et al. De Novo Mutations in MED13, a Component of the Mediator Complex, Are Associated with a Novel Neurodevelopmental Disorder. Hum. Genet. 2018, 137, 375–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reuter, M.S.; Musante, L.; Hu, H.; Diederich, S.; Sticht, H.; Ekici, A.B.; Uebe, S.; Wienker, T.F.; Bartsch, O.; Zechner, U.; et al. NDST1 Missense Mutations in Autosomal Recessive Intellectual Disability. Am. J. Med. Genet. Part A 2014, 164, 2753–2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uusimaa, J.; Kaarteenaho, R.; Paakkola, T.; Tuominen, H.; Karjalainen, M.K.; Nadaf, J.; Varilo, T.; Uusi-Mäkelä, M.; Suo-Palosaari, M.; Pietilä, I.; et al. NHLRC2 Variants Identified in Patients with Fibrosis, Neurodegeneration, and Cerebral Angiomatosis (FINCA): Characterisation of a Novel Cerebropulmonary Disease. Acta Neuropathol. 2018, 135, 727–742. [Google Scholar] [CrossRef] [Green Version]
- Kondakova, O.B.; Krasnenko, A.Y.; Tsukanov, K.Y.; Klimchuk, O.I.; Korostin, D.O.; Davidova, A.I.; Batysheva, T.T.; Zhurkova, N.V.; Ekaterina Ivanovna Surkova, P.A.S.; Ilinsk, V.V. Compound Heterozygous POMGNT1 Mutations Leading to Muscular Dystrophy- Dystroglycanopathy Type A3: A Case Report. BMC Pediatr. 2019, 19, 98. [Google Scholar]
- Lenaerts, L.; Reynhout, S.; Verbinnen, I.; Laumonnier, F.; Toutain, A.; Bonnet-Brilhault, F.; Hoorne, Y.; Joss, S.; Chassevent, A.K.; Smith-Hicks, C.; et al. The Broad Phenotypic Spectrum of PPP2R1A-Related Neurodevelopmental Disorders Correlates with the Degree of Biochemical Dysfunction. Genet. Med. 2021, 23, 352–362. [Google Scholar] [CrossRef]
- Houge, G.; Haesen, D.; Vissers, L.E.L.M.; Mehta, S.; Parker, M.J.; Wright, M.; Vogt, J.; McKee, S.; Tolmie, J.L.; Cordeiro, N.; et al. B56δ-Related Protein Phosphatase 2A Dysfunction Identified in Patients with Intellectual Disability. J. Clin. Investig. 2015, 125, 3051–3062. [Google Scholar] [CrossRef] [Green Version]
- Marsh, D.J.; Kum, J.B.; Lunetta, K.L.; Bennett, M.J.; Gorlin, R.J.; Ahmed, S.F.; Bodurtha, J.; Crowe, C.; Curtis, M.A.; Dasouki, M.; et al. PTEN Mutation Spectrum and Genotype-Phenotype Correlations in Bannayan-Riley-Ruvalcaba Syndrome Suggest a Single Entity with Cowden Syndrome. Hum. Mol. Genet. 1999, 8, 1461–1472. [Google Scholar] [CrossRef] [PubMed]
- Nelen, M.R.; Van Staveren, W.C.G.; Peeters, E.A.J.; Ben Hassel, M.; Gorlin, R.J.; Hamm, H.; Lindboe, C.F.; Fryns, J.P.; Sijmons, R.H.; Woods, D.G.; et al. Germline Mutations in the PTEN/MMAC1 Gene in Patients with Cowden Disease. Hum. Mol. Genet. 1997, 6, 1383–1387. [Google Scholar] [CrossRef] [PubMed]
- Monies, D.; Abouelhoda, M.; AlSayed, M.; Alhassnan, Z.; Alotaibi, M.; Kayyali, H.; Al-Owain, M.; Shah, A.; Rahbeeni, Z.; Al-Muhaizea, M.A.; et al. The Landscape of Genetic Diseases in Saudi Arabia Based on the First 1000 Diagnostic Panels and Exomes. Hum. Genet. 2017, 136, 921–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamberger, H.; Nikanorova, M.; Willemsen, M.H.; Accorsi, P.; Angriman, M.; Baier, H.; Benkel-Herrenbrueck, I.; Benoit, V.; Budetta, M.; Caliebe, A.; et al. STXBP1 Encephalopathy. Neurology 2016, 86, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Polla, D.L.; Farazi Fard, M.A.; Tabatabaei, Z.; Habibzadeh, P.; Levchenko, O.A.; Nikuei, P.; Makrythanasis, P.; Hussain, M.; von Hardenberg, S.; Zeinali, S.; et al. Biallelic Variants in TMEM222 Cause a New Autosomal Recessive Neurodevelopmental Disorder. Genet. Med. 2021, 23, 1246–1254. [Google Scholar] [CrossRef]
- Hennies, H.C.; Rauch, A.; Seifert, W.; Schumi, C.; Moser, E.; Al-taji, E.; Tariverdian, G.; Chrzanowska, K.H.; Krajewska-, M.; Rajab, A.; et al. Allelic Heterogeneity in the COH1 Gene Explains Clinical Variability in Cohen Syndrome. Am. J. Hum. Genet. 2004, 75, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Wang, T.; Wu, H.; Long, M.; Coe, B.P.; Li, H.; Xun, G.; Ou, J.; Chen, B.; Duan, G.; et al. Inherited and Multiple de Novo Mutations in Autism/Developmental Delay Risk Genes Suggest a Multifactorial Model. Mol. Autism 2018, 9, 64. [Google Scholar] [CrossRef]
- Dadali, E.L.; Markova, T.V.; Levchenko, O.A.; Chukhrova, A.L.; Shchagina, O.A. Clinical and Genetic Characteristics of X-Linked Mental Retardation 102 Type Caused by Novel Mutations in the DDX3X Gene (OMIM:300958). Neuromuscul. Dis. 2020, 10, 75–80. [Google Scholar] [CrossRef]
- Méjécase, C.; Way, C.M.; Owen, N.; Moosajee, M. Ocular Phenotype Associated with Dyrk1a Variants. Genes 2021, 12, 324. [Google Scholar] [CrossRef]
- Levchenko, O.A.; Rudenskaya, G.E.; Markova, T.V.; Bessonova, L.A.; Marakhonov, A.V.; Nagieva, S.E.; Shchagina, O.A.; Lavrov, A. Autosomal Dominant Intellectual Disability Associated with the MED13L Gene. Ross. Vestn. Perinatol. Pediatr. Russ. Bull. Perinatol. Pediatr. 2022, 67, 101–107. [Google Scholar] [CrossRef]
- Lopes, F.; Barbosa, M.; Ameur, A.; Soares, G.; De Sá, J.; Dias, A.I.; Oliveira, G.; Cabral, P.; Temudo, T.; Calado, E.; et al. Identification of Novel Genetic Causes of Rett Syndrome-like Phenotypes. J. Med. Genet. 2016, 53, 190–199. [Google Scholar] [CrossRef] [Green Version]
- Kashevarova, A.A.; Nazarenko, L.P.; Skryabin, N.A.; Salyukova, O.A.; Chechetkina, N.N.; Tolmacheva, E.N.; Sazhenova, E.A.; Magini, P.; Graziano, C.; Romeo, G.; et al. Array CGH Analysis of a Cohort of Russian Patients with Intellectual Disability. Gene 2014, 536, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Soden, S.E.; Saunders, C.J.; Willig, L.K.; Farrow, E.G.; Smith, L.D.; Petrikin, J.E.; LePichon, J.-B.B.; Miller, N.A.; Thiffault, I.; Dinwiddie, D.L.; et al. Effectiveness of Exome and Genome Sequencing Guided by Acuity of Illness for Diagnosis of Neurodevelopmental Disorders. Sci. Transl. Med. 2014, 6, 265ra168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenderski, W.; Wang, L.; Krokhotin, A.; Walsh, J.J.; Li, H.; Shoji, H.; Ghosh, S.; George, R.D.; Miller, E.L.; Elias, L.; et al. Loss of the Neural-Specific BAF Subunit ACTL6B Relieves Repression of Early Response Genes and Causes Recessive Autism. Proc. Natl. Acad. Sci. USA 2020, 117, 10055–10066. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.; Rousseau, J.; Peng, H.; Aouabed, Z.; Priam, P.; Theroux, J.F.; Jefri, M.; Tanti, A.; Wu, H.; Kolobova, I.; et al. Mutations in ACTL6B Cause Neurodevelopmental Deficits and Epilepsy and Lead to Loss of Dendrites in Human Neurons. Am. J. Hum. Genet. 2019, 104, 815–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.A.; Lattier, J.; Zhu, W.; Rosenfeld, J.; Wang, L.; Scott, T.M.; Du, H.; Patel, V.; Dang, A.; Magoulas, P.; et al. Retrospective Analysis of a Clinical Exome Sequencing Cohort Reveals the Mutational Spectrum and Identifies Candidate Disease–Associated Loci for BAFopathies. Genet. Med. 2022, 24, 364–373. [Google Scholar] [CrossRef]
- Garavelli, L.; Zollino, M.; Cerruti Mainardi, P.; Gurrieri, F.; Rivieri, F.; Soli, F.; Verri, R.; Albertini, E.; Favaron, E.; Zignani, M.; et al. Mowat-Wilson Syndrome: Facial Phenotype Changing with Age: Study of 19 Italian Patients and Review of the Literature. Am. J. Med. Genet. Part A 2009, 149, 417–426. [Google Scholar] [CrossRef]
- Kuil, L.E.; MacKenzie, K.C.; Tang, C.S.; Windster, J.D.; Le, T.L.; Karim, A.; de Graaf, B.M.; van der Helm, R.; van Bever, Y.; Sloots, C.E.J.; et al. Size Matters: Large Copy Number Losses in Hirschsprung Disease Patients Reveal Genes Involved in Enteric Nervous System Development. PLoS Genet. 2021, 17, e1009698. [Google Scholar] [CrossRef]
- Borlot, F.; Regan, B.M.; Bassett, A.S.; Stavropoulos, D.J.; Andrade, D.M. Prevalence of Pathogenic Copy Number Variation in Adults with Pediatric-Onset Epilepsy and Intellectual Disability. JAMA Neurol. 2017, 74, 1301–1311. [Google Scholar] [CrossRef]
- Árvai, K.; Horváth, P.; Balla, B.; Tobiás, B.; Kató, K.; Kirschner, G.; Klujber, V.; Lakatos, P.; Kósa, J.P. Next-Generation Sequencing of Common Osteogenesis Imperfecta-Related Genes in Clinical Practice. Sci. Rep. 2016, 6, 28417. [Google Scholar] [CrossRef] [Green Version]
- Sule, G.; Campeau, P.M.; Zhang, V.W.; Nagamani, S.C.S.; Dawson, B.C.; Grover, M.; Bacino, C.A.; Sutton, V.R.; Brunetti-Pierri, N.; Lu, J.T.; et al. Next-Generation Sequencing for Disorders of Low and High Bone Mineral Density. Osteoporos. Int. 2013, 24, 2253–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Deignan, J.L.; Dorrani, N.; Strom, S.P.; Kantarci, S.; Quintero-Rivera, F.; Das, K.; Toy, T.; Harry, B.; Yourshaw, M.; et al. Clinical Exome Sequencing for Genetic Identification of Rare Mendelian Disorders. JAMA 2014, 312, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Chérot, E.; Keren, B.; Dubourg, C.; Carré, W.; Fradin, M.; Lavillaureix, A.; Afenjar, A.; Burglen, L.; Whalen, S.; Charles, P.; et al. Using Medical Exome Sequencing to Identify the Causes of Neurodevelopmental Disorders: Experience of 2 Clinical Units and 216 Patients. Clin. Genet. 2018, 93, 567–576. [Google Scholar] [CrossRef]
- Fan, Y.; Wu, Y.; Wang, L.; Wang, Y.; Gong, Z.; Qiu, W.; Wang, J.; Zhang, H.; Ji, X.; Ye, J.; et al. Chromosomal Microarray Analysis in Developmental Delay and Intellectual Disability with Comorbid Conditions. BMC Med. Genom. 2018, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Ta, E.Z.; Karaosmano, B.; Ko, C.; Akgün-do, Ö.; Şimşek-Kiper, P.Ö.; Alika, M.; Boduro, K.; Utine, G.E. Diagnostic Yield of Whole-Exome Sequencing in Non-Syndromic Intellectual Disability. J. Intellect. Disabil. Res. 2021, 65, 577–588. [Google Scholar] [CrossRef]
- Martínez, F.; Caro-Llopis, A.; Roselló, M.; Oltra, S.; Mayo, S.; Sandra Monfort, C.O. High Diagnostic Yield of Syndromic Intellectual Disability by Targeted Next-Generation Sequencing. J. Med. Genet. 2017, 54, 87–92. [Google Scholar] [CrossRef]
- Oğuz, S.; Arslan, U.E.; Kiper, P.Ö.Ş.; Alikaşifoğlu, M.; Boduroğlu, K.; Utine, G.E. Diagnostic Yield of Microarrays in Individuals with Non-Syndromic Developmental Delay and Intellectual Disability. J. Intellect. Disabil. Res. 2021, 65, 1033–1048. [Google Scholar] [CrossRef]
- Kingsmore, S.F.; Cakici, J.A.; Clark, M.M.; Gaughran, M.; Feddock, M.; Batalov, S.; Bainbridge, M.N.; Carroll, J.; Caylor, S.A.; Clarke, C.; et al. A Randomized, Controlled Trial of the Analytic and Diagnostic Performance of Singleton and Trio, Rapid Genome and Exome Sequencing in Ill Infants. Am. J. Hum. Genet. 2019, 105, 719–733. [Google Scholar] [CrossRef]
- Lavrov, A.V.; Bannikov, A.V.; Chausheva, A.I.; Dadali, E.L. Genetics of Mental Retardation. Ross. Vestn. Perinatol. Pediatr. 2016, 61, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Mironovich, O.; Dadali, E.; Malmberg, S.; Markova, T.; Ryzhkova, O.; Poliakov, A. Identification of a Novel de Novo Variant in the Syt2 Gene Causing a Rare Type of Distal Hereditary Motor Neuropathy. Genes 2020, 11, 1238. [Google Scholar] [CrossRef]
- Cappuccio, G.; Sayou, C.; Tanno, P.L.; Tisserant, E.; Bruel, A.L.; Kennani, S.E.; Sá, J.; Low, K.J.; Dias, C.; Havlovicová, M.; et al. De Novo SMARCA2 Variants Clustered Outside the Helicase Domain Cause a New Recognizable Syndrome with Intellectual Disability and Blepharophimosis Distinct from Nicolaides–Baraitser Syndrome. Genet. Med. 2020, 22, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.B.; Abdul-Rahman, O.A.; Buttanu, A.; Cormier-Daire, V.; Frayer, A.; Gillessen-Kaesbach, G.; Horn, D.; Josifoa, D.; Kuechler, A.; Lees, M.; et al. Nicolaides-Baraitser Syndrome: Delineation of the Phenotype. Am. J. Med. Genet. Part A 2009, 149, 1628–1640. [Google Scholar] [CrossRef] [PubMed]
- Santen, G.W.E.; Aten, E.; Vulto-van Silfhout, A.T.; Pottinger, C.; van Bon, B.W.M.; van Minderhout, I.J.H.M.; Snowdowne, R.; van der Lans, C.A.C.; Boogaard, M.; Linssen, M.M.L.; et al. Coffin-Siris Syndrome and the BAF Complex: Genotype-Phenotype Study in 63 Patients. Hum. Mutat. 2013, 34, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Fichera, M.; Failla, P.; Saccuzzo, L.; Miceli, M.; Salvo, E.; Castiglia, L.; Galesi, O.; Grillo, L.; Calì, F.; Greco, D.; et al. Mutations in ACTL6B, Coding for a Subunit of the Neuron-Specific Chromatin Remodeling Complex NBAF, Cause Early Onset Severe Developmental and Epileptic Encephalopathy with Brain Hypomyelination and Cerebellar Atrophy. Hum. Genet. 2019, 138, 187–198. [Google Scholar] [CrossRef]
- Bögershausen, N.; Wollnik, B. Mutational Landscapes and Phenotypic Spectrum of SWI/SNF-Related Intellectual Disability Disorders. Front. Mol. Neurosci. 2018, 11, 252. [Google Scholar] [CrossRef]
- Seranski, P.; Heiss, N.S.; Dhorne-Pollet, S.; Radelof, U.; Korn, B.; Hennig, S.; Backes, E.; Schmidt, S.; Wiemann, S.; Schwarz, C.E.; et al. Transcription Mapping in a Medulloblastoma Breakpoint Interval and Smith-Magenis Syndrome Candidate Region: Identification of 53 Transcriptional Units and New Candidate Genes. Genomics 1999, 56, 1–11. [Google Scholar] [CrossRef]
- Bi, W.; Saifi, G.M.; Shaw, C.J.; Walz, K.; Fonseca, P.; Wilson, M.; Potocki, L.; Lupski, J.R. Mutations of RAI1, a PHD-Containing Protein, in Nondeletion Patients with Smith-Magenis Syndrome. Hum. Genet. 2004, 115, 515–524. [Google Scholar] [CrossRef]
- Gilissen, C.; Hehir-Kwa, J.Y.; Thung, D.T.; Van De Vorst, M.; Van Bon, B.W.M.; Willemsen, M.H.; Kwint, M.; Janssen, I.M.; Hoischen, A.; Schenck, A.; et al. Genome Sequencing Identifies Major Causes of Severe Intellectual Disability. Nature 2014, 511, 344–347. [Google Scholar] [CrossRef]
- Brusco, A.; Bybjerg-grauholm, J.; Carracedo, A.; Chan, M.C.Y.; Chioc-chetti, A.G.; Chung, B.H.Y.; Cuccaro, M.L.; Curró, A.; Bernardina, B.D.; Doan, R.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2021, 180, 568–584. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, W.; Amin, J.B.; Perozo, E.; Kannan, V.; Keller, S.R.; Wilcox, W.R.; Lemke, J.R.; Myers, J.; Swanger, S.A.; et al. De Novo GRIN Variants in NMDA Receptor M2 Channel Pore-Forming Loop Are Associated with Neurological Diseases. Hum. Mutat. 2020, 40, 2393–2413. [Google Scholar] [CrossRef]
- Lemke, J.R.; Geider, K.; Helbig, K.L.; Heyne, H.O.; Schütz, H.; Hentschel, J.; Courage, C.; Depienne, C.; Nava, C.; Heron, D.; et al. Delineating the GRIN1 Phenotypic Spectrum: A Distinct Genetic NMDA Receptor Encephalopathy. Neurology 2016, 86, 2171–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamberger, H.; Hammer, T.B.; Gardella, E.; Vlaskamp, D.R.M.; Bertelsen, B.; Mandelstam, S.; de Lange, I.; Zhang, J.; Myers, C.T.; Fenger, C.; et al. NEXMIF Encephalopathy: An X-Linked Disorder with Male and Female Phenotypic Patterns. Genet. Med. 2021, 23, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Magome, T.; Hattori, T.; Taniguchi, M.; Ishikawa, T.; Miyata, S.; Yamada, K.; Takamura, H.; Matsuzaki, S.; Ito, A.; Tohyama, M.; et al. XLMR Protein Related to Neurite Extension (Xpn/KIAA2022) Regulates Cell-Cell and Cell-Matrix Adhesion and Migration. Neurochem. Int. 2013, 63, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Bramswig, N.C.; Lüdecke, H.J.; Pettersson, M.; Albrecht, B.; Bernier, R.A.; Cremer, K.; Eichler, E.E.; Falkenstein, D.; Gerdts, J.; Jansen, S.; et al. Identification of New TRIP12 Variants and Detailed Clinical Evaluation of Individuals with Non-Syndromic Intellectual Disability with or without Autism. Hum. Genet. 2017, 136, 179–192. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Gambin, T.; Yuan, B.; Szafranski, P.; Rosenfeld, J.A.; Balwi, M.A.; Alswaid, A.; Al-Gazali, L.; Al Shamsi, A.; Komara, M.; et al. Haploinsufficiency of the E3 Ubiquitin-Protein Ligase Gene TRIP12 Causes Intellectual Disability with or without Autism Spectrum Disorders, Speech Delay, and Dysmorphic Features. Hum. Genet. 2017, 136, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Knaus, A.; Awaya, T.; Helbig, I.; Afawi, Z.; Pendziwiat, M.; Abu-Rachma, J.; Thompson, M.D.; Cole, D.E.; Skinner, S.; Annese, F.; et al. Rare Noncoding Mutations Extend the Mutational Spectrum in the PGAP3 Subtype of Hyperphosphatasia with Mental Retardation Syndrome. Hum. Mutat. 2016, 37, 737–744. [Google Scholar] [CrossRef] [Green Version]
- Howard, M.F.; Murakami, Y.; Pagnamenta, A.T.; Daumer-Haas, C.; Fischer, B.; Hecht, J.; Keays, D.A.; Knight, S.J.L.; Kölsch, U.; Krüger, U.; et al. Mutations in PGAP3 Impair GPI-Anchor Maturation, Causing a Subtype of Hyperphosphatasia with Mental Retardation. Am. J. Hum. Genet. 2014, 94, 278–287. [Google Scholar] [CrossRef] [Green Version]
- Courcet, J.B.; Faivre, L.; Malzac, P.; Masurel-Paulet, A.; Lopez, E.; Callier, P.; Lambert, L.; Lemesle, M.; Thevenon, J.; Gigot, N.; et al. The DYRK1A Gene Is a Cause of Syndromic Intellectual Disability with Severe Microcephaly and Epilepsy. J. Med. Genet. 2012, 49, 731–736. [Google Scholar] [CrossRef]
- Gozes, I.; Van Dijck, A.; Hacohen-Kleiman, G.; Grigg, I.; Karmon, G.; Giladi, E.; Eger, M.; Gabet, Y.; Pasmanik-Chor, M.; Cappuyns, E.; et al. Premature Primary Tooth Eruption in Cognitive/Motor-Delayed ADNP-Mutated Children. Transl. Psychiatry 2017, 7, e1043. [Google Scholar] [CrossRef]
- Rentas, S.; Rathi, K.S.; Kaur, M.; Raman, P.; Krantz, I.D.; Sarmady, M.; Tayoun, A.A. Diagnosing Cornelia de Lange Syndrome and Related Neurodevelopmental Disorders Using RNA Sequencing. Genet. Med. 2020, 22, 927–936. [Google Scholar] [CrossRef]
- Olley, G.; Ansari, M.; Bengani, H.; Grimes, G.R.; Rhodes, J.; von Kriegsheim, A.; Blatnik, A.; Stewart, F.J.; Wakeling, E.; Carroll, N.; et al. BRD4 Interacts with NIPBL and BRD4 Is Mutated in a Cornelia de Lange–like Syndrome. Nat. Genet. 2019, 51, 1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.H.; Yuen, R.K.C.C.; Jin, X.; Wang, M.; Chen, N.; Wu, X.; Ju, J.; Mei, J.; Shi, Y.; He, M.; et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am. J. Hum. Genet. 2013, 93, 249–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, K.; Kato, M.; Osaka, H.; Yamashita, S.; Nakagawa, E.; Haginoya, K.; Tohyama, J.; Okuda, M.; Wada, T.; Shimakawa, S.; et al. Clinical Spectrum of SCN2A Mutations Expanding to Ohtahara Syndrome. Neurology 2013, 81, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Rauch, A.; Wieczorek, D.; Graf, E.; Wieland, T.; Endele, S.; Schwarzmayr, T.; Albrecht, B.; Bartholdi, D.; Beygo, J.; Di Donato, N.; et al. Range of Genetic Mutations Associated with Severe Non-Syndromic Sporadic Intellectual Disability: An Exome Sequencing Study. Lancet 2012, 380, 1674–1682. [Google Scholar] [CrossRef]
- Reynolds, C.; King, M.D.; Gorman, K.M. The Phenotypic Spectrum of SCN2A-Related Epilepsy. Eur. J. Paediatr. Neurol. 2020, 24, 117–122. [Google Scholar] [CrossRef]
- Xia, F.; Bainbridge, M.N.; Tan, T.Y.; Wangler, M.F.; Scheuerle, A.E.; Zackai, E.H.; Harr, M.H.; Sutton, V.R.; Nalam, R.L.; Zhu, W.; et al. De Novo Truncating Mutations in AHDC1 in Individuals with Syndromic Expressive Language Delay, Hypotonia, and Sleep Apnea. Am. J. Hum. Genet. 2014, 94, 784–789. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Douglas, G.; Monaghan, K.G.; Retterer, K.; Cho, M.T.; Escobar, L.F.; Tucker, M.E.; Stoler, J.; Rodan, L.H.; Stein, D.; et al. De Novo Truncating Variants in the AHDC1 Gene Encoding the AT-Hook DNA-Binding Motif-Containing Protein 1 Are Associated with Intellectual Disability and Developmental Delay. Mol. Case Stud. 2015, 1, a000562. [Google Scholar] [CrossRef] [Green Version]
- Tatton-Brown, K.; Zachariou, A.; Loveday, C.; Renwick, A.; Mahamdallie, S.; Aksglaede, L.; Baralle, D.; Barge-Schaapveld, D.; Blyth, M.; Bouma, M.; et al. The Tatton-Brown-Rahman Syndrome: A Clinical Study of 55 Individuals with de Novo Constitutive DNMT3A Variants. Wellcome Open Res. 2018, 3, 1–17. [Google Scholar] [CrossRef]
- White, J.; Beck, C.R.; Harel, T.; Posey, J.E.; Jhangiani, S.N.; Tang, S.; Farwell, K.D.; Powis, Z.; Mendelsohn, N.J.; Baker, J.A.; et al. POGZ Truncating Alleles Cause Syndromic Intellectual Disability. Genome Med. 2016, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Stessman, H.A.F.; Willemsen, M.H.; Fenckova, M.; Penn, O.; Hoischen, A.; Xiong, B.; Wang, T.; Hoekzema, K.; Vives, L.; Vogel, I.; et al. Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. Am. J. Hum. Genet. 2016, 98, 541–552. [Google Scholar] [CrossRef] [Green Version]
- Zarate, Y.A.; Fish, J.L. SATB2-Associated Syndrome: Mechanisms, Phenotype, and Practical Recommendations. Am. J. Med. Genet. Part A 2017, 173, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Farwell, K.D.; Shahmirzadi, L.; El-Khechen, D.; Powis, Z.; Chao, E.C.; Tippin Davis, B.; Baxter, R.M.; Zeng, W.; Mroske, C.; Parra, M.C.; et al. Enhanced Utility of Family-Centered Diagnostic Exome Sequencing with Inheritance Model-Based Analysis: Results from 500 Unselected Families with Undiagnosed Genetic Conditions. Genet. Med. 2015, 17, 578–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.S.; Yoo, Y.; Lim, B.C.; Kim, K.J.; Choi, M.; Chae, J.H. SATB2-Associated Syndrome Presenting with Rett-like Phenotypes. Clin. Genet. 2016, 89, 728–732. [Google Scholar] [CrossRef]
- Breuss, M.; Heng, J.I.T.; Poirier, K.; Tian, G.; Jaglin, X.H.; Qu, Z.; Braun, A.; Gstrein, T.; Ngo, L.; Haas, M.; et al. Mutations in the β-Tubulin Gene TUBB5 Cause Microcephaly with Structural Brain Abnormalities. Cell Rep. 2012, 2, 1554–1562. [Google Scholar] [CrossRef] [Green Version]
- Dentici, M.L.; Terracciano, A.; Bellacchio, E.; Capolino, R.; Novelli, A.; Digilio, M.C.; Dallapiccola, B. Intrafamiliar Clinical Variability of Circumferential Skin Creases Kunze Type Caused by a Novel Heterozygous Mutation of N-Terminal TUBB Gene. Clin. Genet. 2018, 93, 1223–1228. [Google Scholar] [CrossRef]
- Cohen, J.S.; Srivastava, S.; Farwell Hagman, K.D.; Shinde, D.N.; Huether, R.; Darcy, D.; Wallerstein, R.; Houge, G.; Berland, S.; Monaghan, K.G.; et al. Further Evidence That de Novo Missense and Truncating Variants in ZBTB18 Cause Intellectual Disability with Variable Features. Clin. Genet. 2017, 91, 697–707. [Google Scholar] [CrossRef]
- Piard, J.; Hu, J.H.; Campeau, P.M.; Rzońca, S.; Van Esch, H.; Vincent, E.; Han, M.; Rossignol, E.; Castaneda, J.; Chelly, J.; et al. FRMPD4 Mutations Cause X-Linked Intellectual Disability and Disrupt Dendritic Spine Morphogenesis. Hum. Mol. Genet. 2018, 27, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Kamien, B.; Lionel, A.C.; Bain, N.; Scherer, S.W.; Hunter, M. Outfoxed by RBFOX1-A Caution about Ascertainment Bias. Am. J. Med. Genet. Part A 2014, 164, 1411–1418. [Google Scholar] [CrossRef]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [Green Version]
- Zawerton, A.; Yao, B.; Yeager, J.P.; Pippucci, T.; Haseeb, A.; Smith, J.D.; Wischmann, L.; Kühl, S.J.; Dean, J.C.S.; Pilz, D.T.; et al. De Novo SOX4 Variants Cause a Neurodevelopmental Disease Associated with Mild Dysmorphism. Am. J. Hum. Genet. 2019, 104, 246–259. [Google Scholar] [CrossRef] [Green Version]
- Moortgat, S.; Berland, S.; Aukrust, I.; Maystadt, I.; Baker, L.; Benoit, V.; Caro-Llopis, A.; Cooper, N.S.; Debray, F.G.; Faivre, L.; et al. HUWE1 Variants Cause Dominant X-Linked Intellectual Disability: A Clinical Study of 21 Patients. Eur. J. Hum. Genet. 2018, 26, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Aref-Eshghi, E.; Bend, E.G.; Hood, R.L.; Schenkel, L.C.; Carere, D.A.; Chakrabarti, R.; Nagamani, S.C.S.; Cheung, S.W.; Campeau, P.M.; Prasad, C.; et al. BAFopathies’ DNA Methylation Epi-Signatures Demonstrate Diagnostic Utility and Functional Continuum of Coffin–Siris and Nicolaides–Baraitser Syndromes. Nat. Commun. 2018, 9, 4885. [Google Scholar] [CrossRef]
- Latypova, X.; Matsumoto, N.; Vinceslas-Muller, C.; Bézieau, S.; Isidor, B.; Miyake, N. Novel KCNB1 Mutation Associated with Non-Syndromic Intellectual Disability. J. Hum. Genet. 2017, 62, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Torkamani, A.; Bersell, K.; Jorge, B.S.; Bjork, R.L., Jr.; Friedman, J.R.; Bloss, C.S.; Cohen, J.; Gupta, S.; Naidu, S.; Vanoye, C.G.; et al. De Novo KCNB1 Mutations in Epileptic Encephalopathy. Bone 2008, 23, 1–7. [Google Scholar] [CrossRef]
- Gong, X.; Jiang, Y.W.; Zhang, X.; An, Y.; Zhang, J.; Wu, Y.; Wang, J.; Sun, Y.; Liu, Y.; Gao, X.; et al. High Proportion of 22q13 Deletions and SHANK3 Mutations in Chinese Patients with Intellectual Disability. PLoS ONE 2012, 7, e34739. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Chen, H.I.; Liao, H.M.; Chen, Y.J.; Fang, J.S.; Lee, K.F.; Gau, S.S.F. Clinical and Molecular Characterization of Three Genomic Rearrangements at Chromosome 22q13.3 Associated with Autism Spectrum Disorder. Psychiatr. Genet. 2017, 27, 23–33. [Google Scholar] [CrossRef]
- Marin-Valencia, I.; Novarino, G.; Johansen, A.; Rosti, B.; Issa, M.Y.; Musaev, D.; Bhat, G.; Scott, E.; Silhavy, J.L.; Stanley, V.; et al. A Homozygous Founder Mutation in TRAPPC6B Associates with a Neurodevelopmental Disorder Characterised by Microcephaly, Epilepsy and Autistic Features. J. Med. Genet. 2018, 55, 48–54. [Google Scholar] [CrossRef]
- Anazi, S.; Maddirevula, S.; Salpietro, V.; Asi, Y.T.; Alsahli, S.; Alhashem, A. Expanding the Genetic Heterogeneity of Intellectual Disability. Hum. Genet. 2017, 136, 1419–1429. [Google Scholar] [CrossRef]
- Whalen, S.; Shaw, M.; Mignot, C.; Héron, D.; Bastaraud, S.C.; Walti, C.C.; Liebelt, J.; Elmslie, F.; Yap, P.; Hurst, J.; et al. Further Delineation of BCAP31-Linked Intellectual Disability: Description of 17 New Families with LoF and Missense Variants. Eur. J. Hum. Genet. 2021, 29, 1405–1417. [Google Scholar] [CrossRef]

| DNA id | Gene | GRCh37 | Zygosity | References | |
|---|---|---|---|---|---|
| 1 | D553 | ADNP | NC_000020.10:g.49510461G > A NM_015339.5:c.790C > T NP_056154.1:p.(Arg264Ter) | het | [11] |
| 2 | D467 | AFF3 | NC_000002.11:g.100623270C > T NM_001025108.2:c.772G > A NP_001020279.1:p.(Ala258Thr) | Het | * [12] |
| 3 | D656 | ALG13 | NC_000023.10:g.110928268A > G NM_001099922.3:c.320A > G NP_001093392.1:p.(Asn107Ser) | het | [13] |
| 4 | D232 | GRIA1 | NC_000005.9:g.153144076G > A NM_001114183.2:c.1906G > A NP_001107655.1:p.(Ala636Thr) | het | [13] |
| 5 | D352 | HGSNAT | NC_000008.10:g.43037306G > A NM_152419.3:c.1031G > A NP_689632.2:p.(Arg344His) | hom | [14] |
| 6 | D475 | KCNQ2 | NC_000020.10:g.62073782C > T NM_172107.4:c.793G > A NP_742105.1:p.(Ala265Thr) | het | [15] |
| 7 | D645 | MED13 | NC_000017.10:g.60108837G > A NM_005121.3:c.977C > T NP_005112.2:p.(Thr326Ile) | het | [16] |
| 8 | D835 | NDST1 | NC_000005.9:g.149921213G > A NM_001543.5:c.1831G > A NP_001534.1:p.(Gly611Ser) | hom | [17] |
| 9 | D1060 | NHLRC2 | NC_000010.10:g.115636390G > T NM_198514.4:c.442G > T NP_940916.2:p.(Asp148Tyr) | hom | [18] |
| 10 | D923 | POMGNT1 | NC_000001.10:g.46661719G > A NM_017739.4:c.385C > T NP_060209.4:p.(Arg129Trp) | hom | [19] |
| 11 | D542 | PPP2R1A | NC_000019.9:g.52715968C > A NM_014225.6:c.533C > A NP_055040.2:p.(Thr178Asn) | het | [20] |
| 12 | D428 | PPP2R5D | NC_000006.11:g.42975009G > A NM_006245.4:c.598G > A NP_006236.1:p.(Glu200Lys) | het | [21] |
| 13 | D189 | PTEN | NC_000010.10:g.89717712C > T NM_000314.8:c.737C > T NP_000305.3:p.(Pro246Leu) | het | [22] |
| 14 | D341 | PTEN | NC_000010.10:g.89692904C > T NM_000314.8:c.388C > T NP_000305.3:p.(Arg130Ter) | het | [23] |
| 15 | D957 | SMARCA4 | NC_000019.9:g.11132465C > T NM_001128849.3:c.2681C > T NP_001122321.1:p.(Thr894Met) | het | [24] |
| 16 | D293 | STXBP1 | NC_000009.11:g.130430439G > A NM_003165.6:c.875G > A NP_003156.1:p.(Arg292His) | het | [25] |
| 17 | D808 | TMEM222 | NC_000001.10:g.27660774T > C NM_032125.3:c.539 + 2T > C NP_115501.2:p.? | hom | * [26] |
| 18 | S3 | VPS13B | NC_000008.10:g.100791008C > T NM_017890.5:c.7603C > T NP_060360.3:p.(Arg2535Ter) | het | * [27] |
| DNA id | Gene | GRCh37 | Zygosity | ACMG Criteria | Gnomad Frequency | Structural Functional Impact # | ACMG Classification | |
|---|---|---|---|---|---|---|---|---|
| 1 | D177 | ACTL6B | NC_000007.13:g.100246360A > G NM_016188.5:c.554T > C NP_057272.1:p.(Leu185Pro) | hom | PM2, PP1(M), PP2, PP3. | n/d | LoF? | LP |
| 2 | D410 | ADNP | NC_000020.10:g.49509097del NM_015339.5:c.2155del NP_056154.1:p.(Tyr719ThrfsTer9) | het | PVS1, PS2, PM2, PP5 [28] | n/d | LoF | P |
| 3 | D594 | AHDC1 | NC_000001.10:g.27877448_27877449del NM_001029882.3:c.1181_1182del NP_001025053.1:p. (Cys394SerfsTer122) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 4 | D473 | BRD4 | NC_000019.9:g.15349980_15349986dup NM_058243.3:c.3666_3672dup NP_490597.1:p.(Glu1225GlnfsTer16) | het | PVS1, PM2 | n/d | LoF | LP |
| 5 | D424 | DDX3X | NC_000023.10:g.41198298A > G NM_001193416.3:c.113A > G NP_001180345.1:p.(Tyr38Cys) | het | PS2, PM2, PP2, PP3 * [29] | n/d | interacting with EIF4E region | LP |
| 6 | D659 | DNMT3A | NC_000002.11:g.25468920G > T NM_175629.2:c.1443C > A NP_783328.1:p.(Tyr481Ter) | het | PVS1, PS2, PM2, PP5 (ClinVar:872726) | n/d | LoF | P |
| 7 | D375 | DYRK1A | NC_000021.8:g.38858824_38858827del NM_001396.5:c.572_575del NP_001387.2:p.(Lys191ThrfsTer6) | het | PVS1, PS2, PM2, PP5 [30] | n/d | LoF | P |
| 8 | D843 | FRMPD4 | NC_000023.10:g.12725711G > T NM_014728.3:c.1411G > T NP_055543.2:p.(Glu471Ter) | hemi | PVS1, PS2, PM2 | n/d | LoF | P |
| 9 | D289 | GRIN1 | NC_000009.11:g.140057096G > C NM_007327.4:c.1918G > C NP_015566.1:p.(Ala640Pro) | het | PS2, PM2, PP2, PP3 | n/d | transmembrane helix | LP |
| 10 | D971 | HUWE1 | NC_000023.10:g.53561589G > A NM_031407.7:c.12719C > T NP_113584.3:p.(Ser4240Phe) | hemi | PS2, PM2, PP2, PP3 | n/d | LoF | LP |
| 11 | D682 | MED13L | NC_000012.11:g.116403946del NM_015335.5:c.6331del NP_056150.1:p.(Gln2111SerfsTer18) | het | PVS1, PS2, PM2 * [31] | n/L | LoF | P |
| 12 | D332 | NEXMIF | NC_000023.10:g.73961725C > T NM_001008537.3:c.2667G > A NP_001008537.1:p.(Trp889Ter) | hemi | PVS1, PS2, PM2 | n/d | LoF | P |
| 13 | D1020 | NFIX | NC_000019.9:g.13183833_13264696del | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 14 | D364 | PGAP3 | NC_000017.10:g.37829376G > A NM_033419.5:c.827C > T NP_219487.3:p.(Pro276Leu) | hom | PM2, PP1, PP3, PP4, PP5 (ClinVar:426134) | 6.35 × 10−5 | transmembrane helix | LP |
| 15 | D680 | POGZ | NC_000001.10:g.151400859dup NM_015100.4:c.600dup NP_055915.2:p.(Gly201TrpfsTer114) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 16 | D198 | RAI1 | NC_000017.10:g.17696891C > G NM_030665.4:c.629C > G NP_109590.3:p.(Pro210Arg) | het | PS2, PM2, PP3. | n/d | “region” (Uniprot) | LP |
| 17 | D837 | RBFOX1 | NC_000016.9:g.7760742T > G NM_145891.3:c.1252T > G NP_665898.1:p.(Tyr418Asp) | het | PS2, PM2, PP3. | n/d | gaining of acceptor splice site | LP |
| 18 | D685 | SATB2 | NC_000002.11:g.200246475_200246476del NM_015265.4:c.414_415del NP_056080.1:p.(Val139GlyfsTer69) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 19 | D886 | SATB2 | NC_000002.11:g.200233333dup NM_015265.4:c.696dup NP_056080.1:p.(Lys233Ter) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 20 | D543 | SCN2A | NC_000002.11:g.166172096_166172097del NM_021007.3:c.1499_1500del NP_066287.2:p.(Glu500AlafsTer21) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 21 | D601 | SCN2A | NC_000002.11:g.166188070G > A NM_021007.3:c.2380G > A NP_066287.2:p.(Gly794Arg) | het | PS2, PM2, PP2, PP3 | n/d | repeated domain II | LP |
| 22 | D171 | SMARCA2 | NC_000009.11:g.2056756C > T NM_003070.5:c.1258C > T NP_003061.3:p.(Arg420Cys) | het | PS2, PM2, PP2, PP3 | 3.98 × 10−6 | HSA domain | LP |
| 23 | D1059 | SMARCA4 | NC_000019.9:g.11134267G > A NM_001128849.3:c.2933G > A NP_001122321.1:p.(Arg978Gln) | het | PS2, PM2, PM5, PP2, PP3 | n/d | no | P |
| 24 | D495 | SON | NC_000021.8:g.34926178_34926179del NM_032195.3:c.4641_4642del NP_115571.3:p.(His1547GlnfsTer9) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 25 | D965 | SOX4 | NC_000006.11:g.21595046G > A NM_003107.3:c.281G > A NP_003098.1:p.(Gly94Asp) | het | PS2, PM2, PP3 | n/d | DNA-binding region | LP |
| 26 | D336 | TRIP12 | NC_000002.11:g.230657847_230657848del NM_004238.3:c.3759_3760del NP_004229.1:p.(Gly1254IlefsTer36) | het | PVS1, PS2, PM2.PP5 (ClinVar:521198) | n/d | LoF | P |
| 27 | D737 | TUBB | NC_000006.11:g.30691462_30691463del NM_178014.4:c.623_624del NP_821133.1:p.(Tyr208Ter) | het | PVS1, PS2, PM2 | n/d | LoF | P |
| 28 | D755 | ZBTB18 | NC_000001.10:g.244217659C > T NM_205768.3:c.583C > T NP_991331.1:p.(Arg195Ter) | het | PVS1, PS2, PM2, PP5 [32] | n/d | LoF | P |
| DNA id | Gene | Position (hg19) | Zygosity | ACMG Criteria | Gnomad Frequency | Structural Functional Impact # | ACMG Classification |
|---|---|---|---|---|---|---|---|
| S4 | DYNC1H1 | NC_000014.8:g.102446128_102446130del NM_001376.5:c.591_593del NP_001367.2:p.(Gln198del) | Het | PM2, PM4, PP1 | n/d | coiled coil structural motif | VUS |
| D381 | TRAPPC6B | NC_000014.8:g.39628717C > A NM_177452.4:c.119G > T NP_803235.1:p.(Gly40Val) | Hom | PM2, PP1 (moderate, PP3 | n/d | cryptic donor splice site | VUS |
| D954 | BCAP31 | NC_000023.10:g.152966417C > T NM_001139441.1:c.716G > A NP_001132913.1:p.(Gly239Asp) | Hem | PM2, PP1 (moderate, PP3 | 5.74 × 10−6 | no | VUS |
| DNA id | Molecular Karyotype | Size Mb | IDD Genes | |
|---|---|---|---|---|
| 1 | S3 | 46,XX,arr(hg19) 8q22.2(100286202_100287976)x3 | 0.002 | VPS13B |
| 2 | D212 | 46,XX,arr(hg19) 1p21.2p13.3(101493397_111245231)x1 | 9.7 | KCNA2 |
| 3 | D254 | 46,XX,arr(hg19) 5q31.2q31.3(138736957_139616749)x1 | 0.88 | PURA |
| 4 | D296 | 46,XX,arr(hg19) 17p11.2(16745570-20449778)x3 | 3.7 | RAI1 |
| 5 | D362 | 46,XX,arr(hg19) 16q23.1q24.3(78036006_90155062)x3, 18q23(77840421_78013728)x1 | 12.1 0.17 | ANKRD11 |
| 6 | D494 | 46,XY,arr(hg19) 5q14.3(88018426_88641953)x1 | 0.62 | MEF2C |
| 7 | D600 | arr(hg19)16q22.3-q21.3 (73919969_75197862)x1 | 1.2 | GLG1 |
| 8 | D801 | 46,XY,arr(hg19) 18p11.21 (12360754_12425280)x1 | 0.06 | AFG3L2 |
| 9 | D856 | 46,XX,arr(hg19) 10q26.3(133728056-135427143)x1 16p13.3(85881_5019217)x3 | 1.6; 4.9 | TSC2, CREBBP |
| 10 | D872 | 46,XX,arr(hg19) 22q13.31q13.33(45676621_51177928)x1, 21q22.3(44879067_48097372)x3 | 5.5; 3.2 | SHANK3 |
| 11 | D898 | 46,XX,arr(hg19) 5p15.33p15.32(113577_5170554)x3, 18q22.3q23(70734990_78014123)x1 | 5 7.3 | TERT, NDUFS6 ZNF407, CTDP1 |
| 12 | D904 | 46,XY,arr(hg19) 17p12(14087934_15484858)x3, 22q13.32q13.33(48571448_51197838)x1 | 1.4 2.6 | SHANK3 |
| 13 | D928 | 46,XY,arr(hg19) 2q33.1q34(202909263_211154254)x1 | 8.2 | MAP2 |
| 14 | D948 | 46,XY,arr(hg19) Xq21.1q21.31(80848988_90921090)x1 | 10 | ZNF711 |
| 15 | D951 | 46,XY,arr(hg19) 7q36.1(151934936_151936775)x3 | 0.002 | KMT2C |
| 16 | D1011 | 46,XY,arr(hg19) 8p23.3p23.1(158049_6999220)x1 8p22p23.1(11895232_39651909)x3 | 6.8 27.8 | DLGAP2 KAT6A, NEFL |
| Features | Undiagnosed | Diagnosed by ES | Diagnosed by CMA | |||
|---|---|---|---|---|---|---|
| Amount | Fraction | Amount | Fraction | Amount | Fraction | |
| Motor developmental delay | 34 | 31% | 23 | 50% | 9 | 60% |
| Malformations | 31 | 28% | 13 | 28% | 8 | 53% |
| Dysplastic ears | 20 | 18% | 12 | 26% | 2 | 13% |
| Microcephaly (p = 0.0016) | 11 | 10% | 16 | 35% | 1 | 7% |
| Seizurs | 26 | 24% | 13 | 28% | 2 | 13% |
| Low-set ears | 12 | 11% | 8 | 17% | 2 | 13% |
| Hypotonia | 9 | 8% | 9 | 20% | 2 | 13% |
| Macrotia | 15 | 14% | 4 | 9% | 0 | 0% |
| Valgus feet | 6 | 5% | 9 | 20% | 3 | 20% |
| Number of patients | 110 | 46 | 15 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levchenko, O.; Dadali, E.; Bessonova, L.; Demina, N.; Rudenskaya, G.; Matyushchenko, G.; Markova, T.; Anisimova, I.; Semenova, N.; Shchagina, O.; et al. Complex Diagnostics of Non-Specific Intellectual Developmental Disorder. Int. J. Mol. Sci. 2022, 23, 7764. https://doi.org/10.3390/ijms23147764
Levchenko O, Dadali E, Bessonova L, Demina N, Rudenskaya G, Matyushchenko G, Markova T, Anisimova I, Semenova N, Shchagina O, et al. Complex Diagnostics of Non-Specific Intellectual Developmental Disorder. International Journal of Molecular Sciences. 2022; 23(14):7764. https://doi.org/10.3390/ijms23147764
Chicago/Turabian StyleLevchenko, Olga, Elena Dadali, Ludmila Bessonova, Nina Demina, Galina Rudenskaya, Galina Matyushchenko, Tatiana Markova, Inga Anisimova, Natalia Semenova, Olga Shchagina, and et al. 2022. "Complex Diagnostics of Non-Specific Intellectual Developmental Disorder" International Journal of Molecular Sciences 23, no. 14: 7764. https://doi.org/10.3390/ijms23147764








