Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein
Abstract
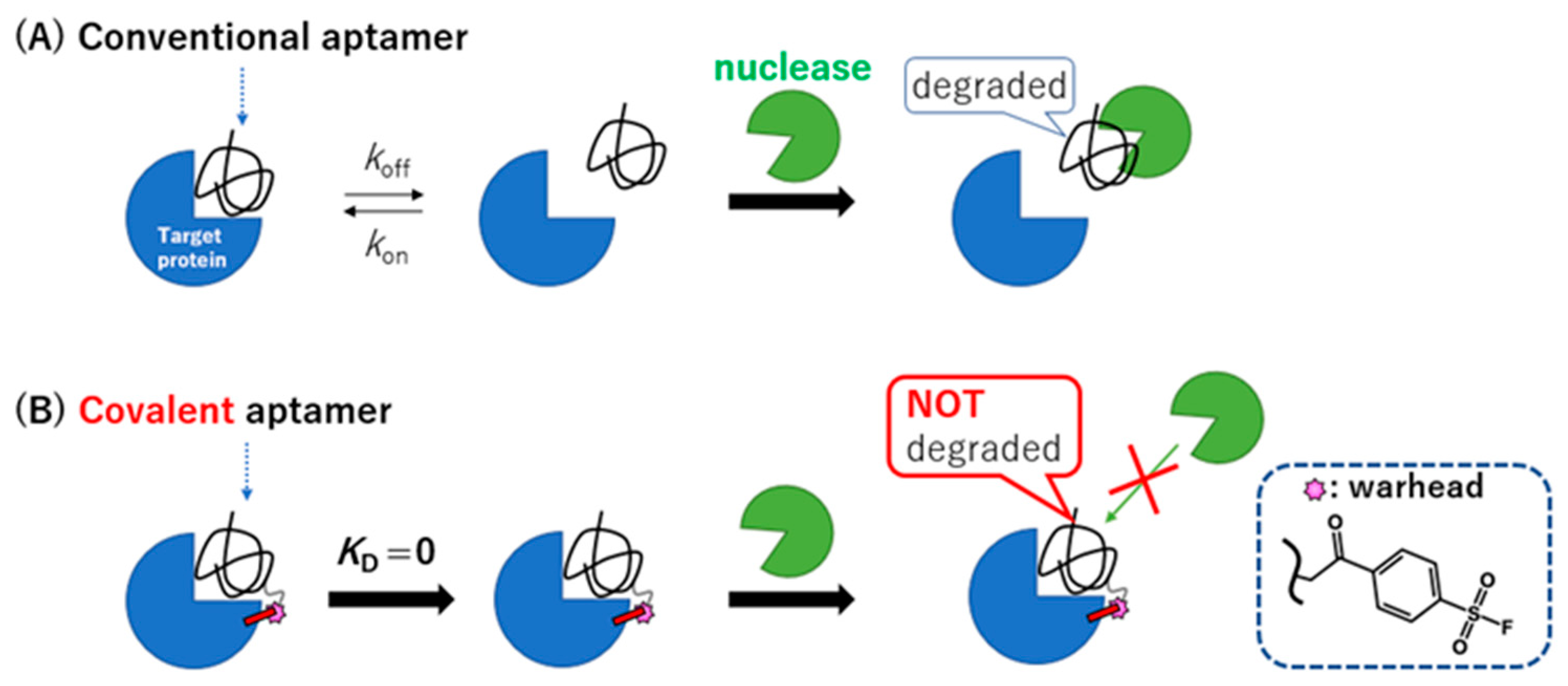
:1. Introduction
2. Results and Discussion
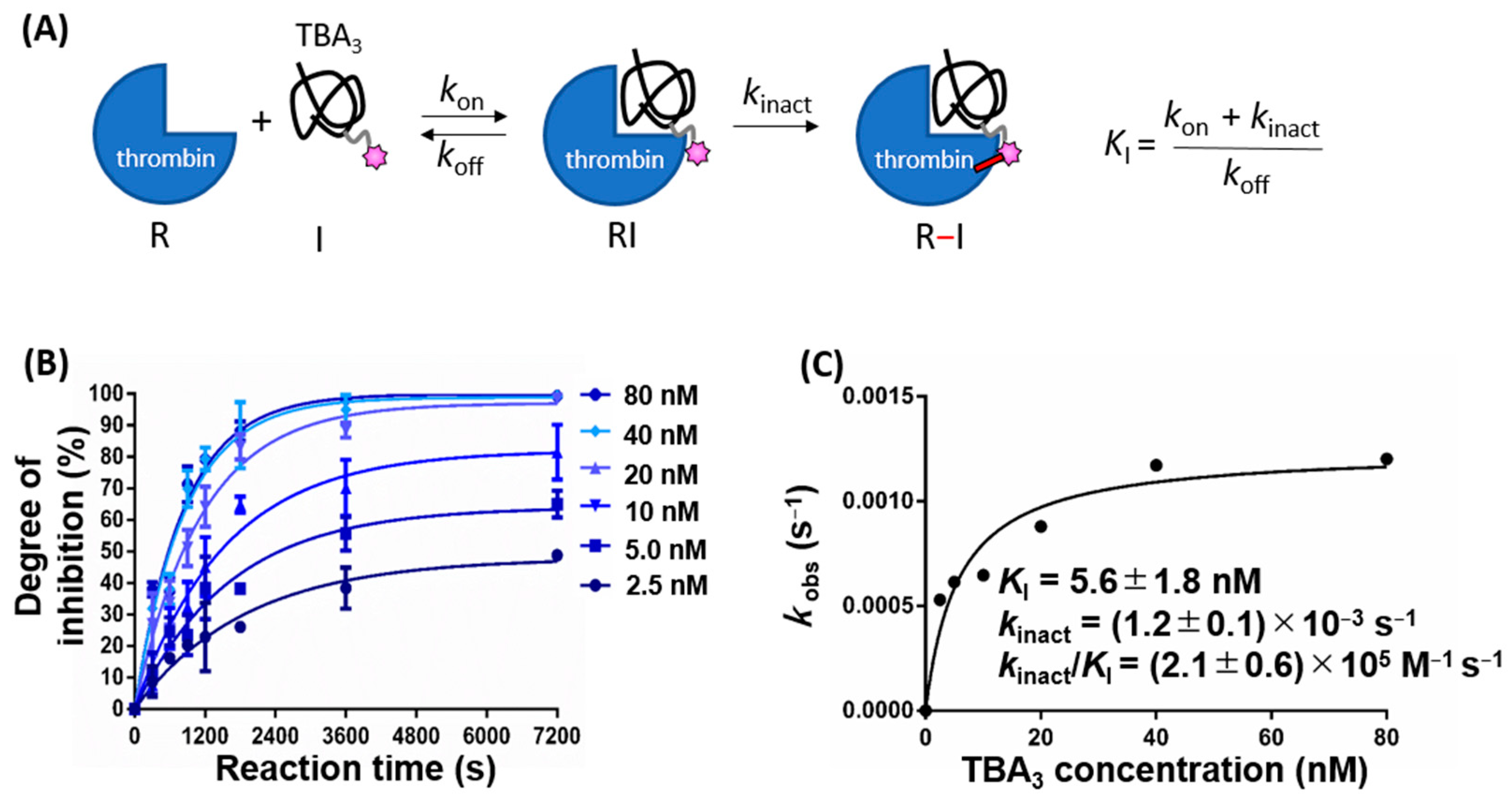
2.1. Kinetics of Thrombin Inhibition by TBA3 Are Consistent with a Two-Step Process
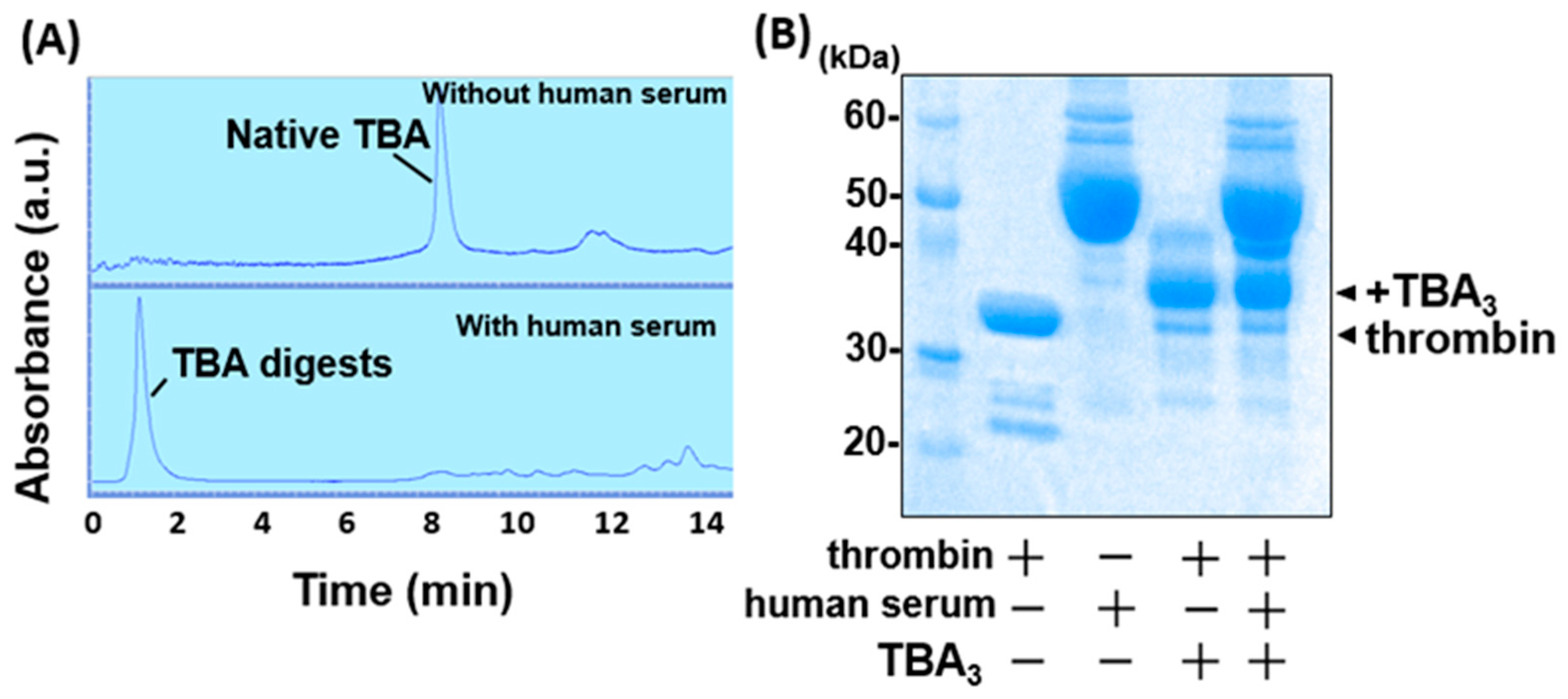
2.2. TBA3 Covalently Bound to Thrombin Resists Degradation by Human Serum
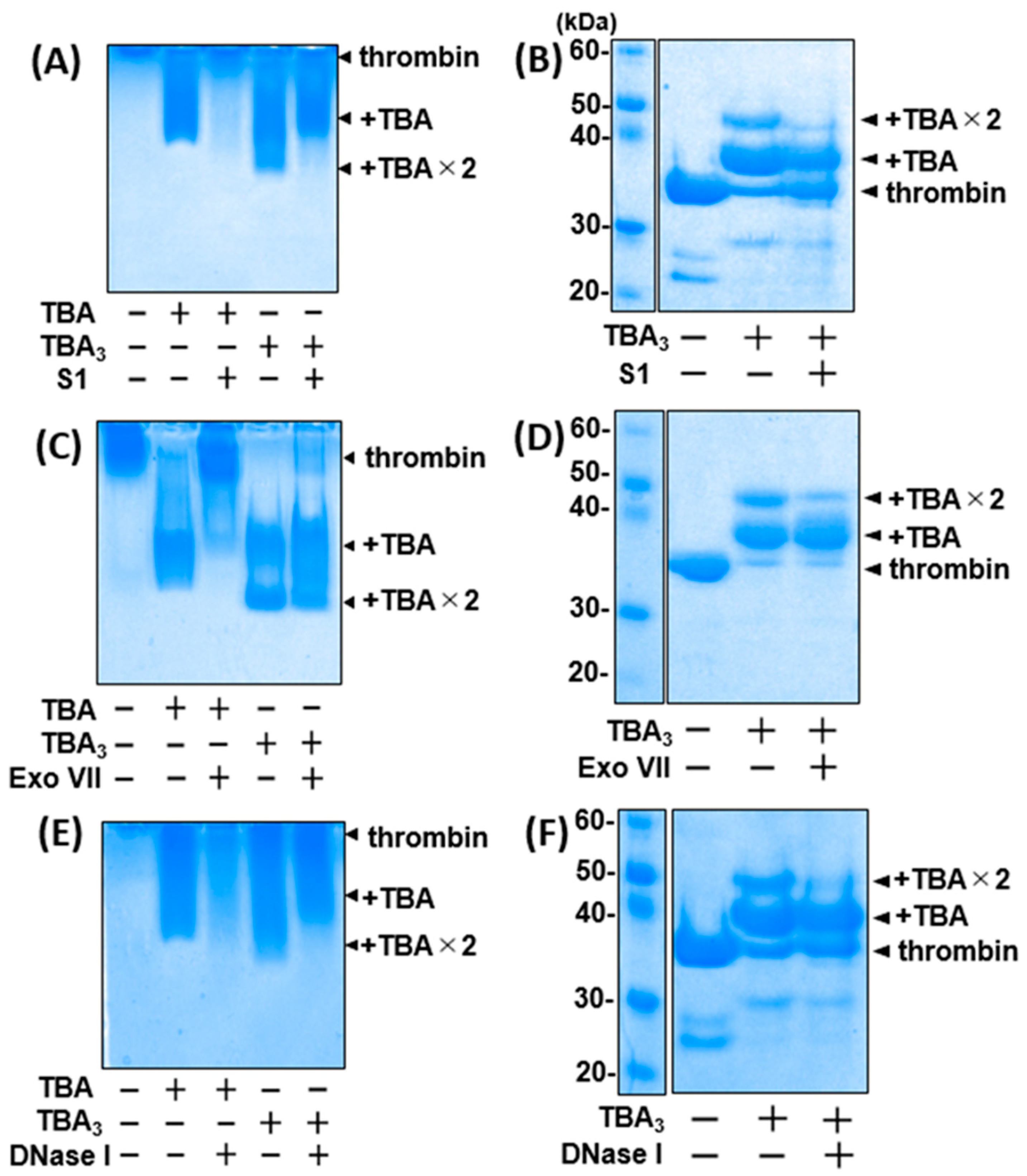
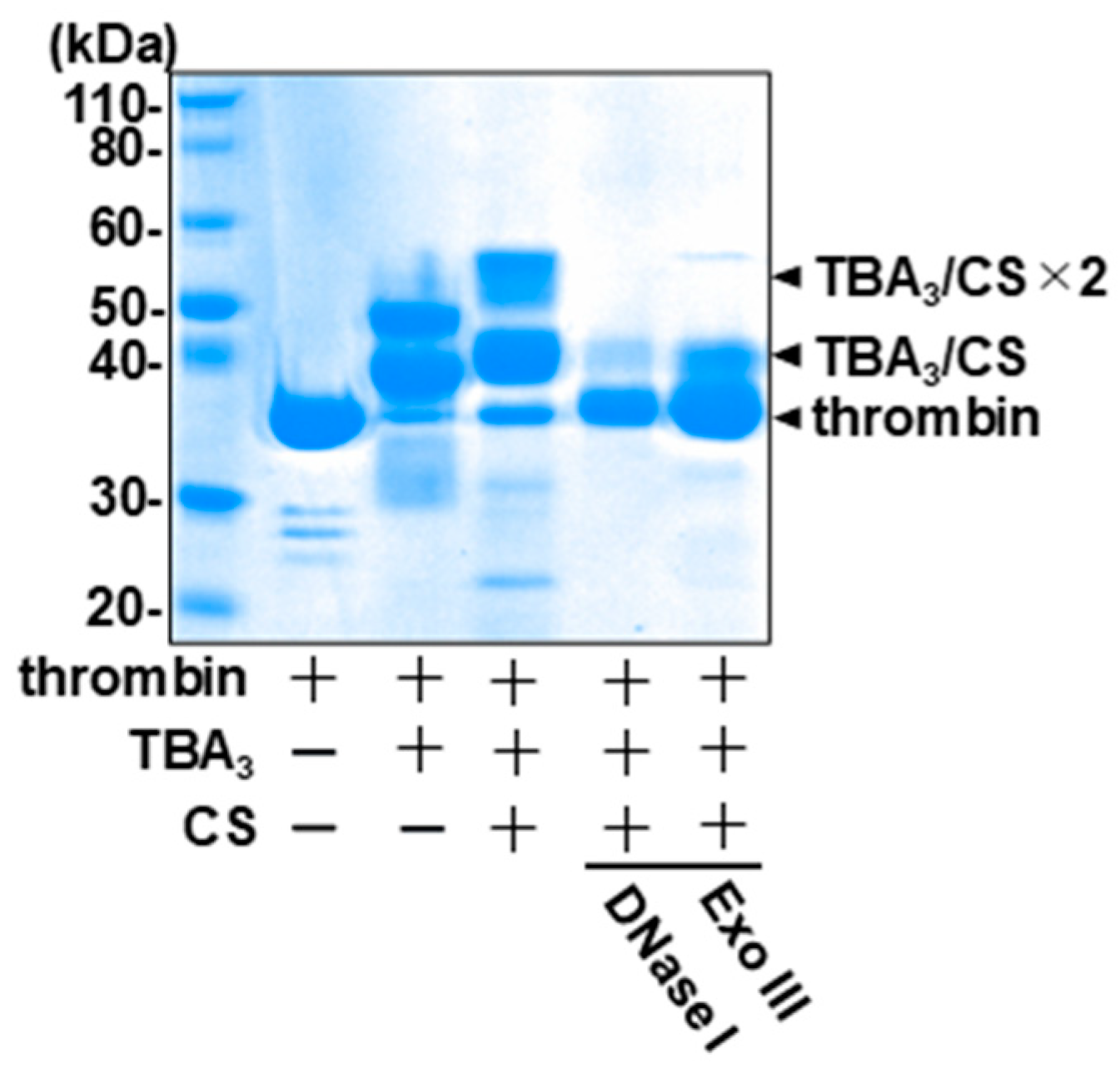
2.3. TBA3 Covalently Bound to Thrombin Resists Degradation by Both Exonucleases and Endonucleases
2.4. The Complementary-Strand Antidote Restores the Nuclease Sensitivity
2.5. Sustained Inhibition of Thrombin Activity by TBA3 in the Presence of Nucleases
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. High-Pressure Liquid Chromatography (HPLC) Analysis
3.3. Image Capturing
3.4. Synthesis of a Covalent Aptamer: TBA3
3.5. Kinetics Evaluation of TBA3
3.6. Evaluation of Nuclease Resistance by Liquid Chromatography or Gel Electrophoresis
3.7. Evaluation of Nuclease Resistance of TBA3 after the Addition of the Complementary Strand (CS) by Gel Electrophoresis
3.8. Evaluation of Thrombin Inhibition Activity in the Presence of Human Serum or Nucleases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kasahara, Y.; Kuwahara, M. Artificial specific binders directly recovered from chemically modified nucleic acid libraries. J. Nucleic Acids 2012, 2012, 156482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusconi, C.P.; Scardino, E.; Layzer, J.; Pitoc, G.A.; Ortel, T.L.; Monroe, D.; Sullenger, B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 2002, 419, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [Green Version]
- Bompiani, K.M.; Woodruff, R.S.; Becker, R.C.; Nimjee, S.M.; Sullenger, B.A. Antidote control of aptamer therapeutics: The road to a safer class of drug agents. Curr. Pharm. Biotechnol. 2012, 13, 1924–1934. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Qiu, L.; Cai, R.; Cui, C.; Li, L.; Jiang, J.H.; Tan, W. Aptamer-directed protein-specific multiple modifications of membrane glycoproteins on living cells. ACS Appl. Mater. Interfaces 2020, 12, 37845–37850. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [Green Version]
- Byun, J. Recent progress and opportunities for nucleic acid aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef]
- Tivon, Y.; Falcone, G.; Deiters, A. Protein labeling and crosslinking by covalent aptamers. Angew. Chem. 2021, 60, 15899–15904. [Google Scholar] [CrossRef]
- Smith, S.E.; Papavassiliou, A.G. A coupled Southwestern-DNase I footprinting assay. Nucleic Acids Res. 1992, 20, 5239–5240. [Google Scholar] [CrossRef] [Green Version]
- Galas, D.J.; Schmitz, A. DNAse footprinting: A simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978, 5, 3157–3170. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Li, F.; Su, Y.H.; Sun, X.; Li, X.B.; Schluesener, H.J.; Tang, F.; Xu, S.Q. Ultrasensitive detection of protein using an aptamer-based exonuclease protection assay. Anal. Chem. 2004, 76, 5605–5610. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Li, F.; Shen, S.; Tyrrell, D.L.; Le, X.C.; Li, X.F. DNase-mediated single-cycle selection of aptamers for proteins blotted on a membrane. Anal. Chem. 2012, 84, 7603–7606. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.W.; Ding, Y.J.; Gao, Z.H.; Li, H. S1 nuclease digestion-based rational truncation of PD-L1 aptamer and establishment of a signal dual amplification aptasensor. Sens. Actuators B Chem. 2021, 331, 129442. [Google Scholar] [CrossRef]
- Tabuchi, Y.; Yang, J.; Taki, M. Inhibition of thrombin activity by a covalent-binding aptamer and reversal by the complementary strand antidote. Chem. Commun. 2021, 57, 2483–2486. [Google Scholar] [CrossRef]
- Zheng, Q.; Woehl, J.L.; Kitamura, S.; Santos-Martins, D.; Smedley, C.J.; Li, G.; Forli, S.; Moses, J.E.; Wolan, D.W.; Sharpless, K.B. SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase. Proc. Natl. Acad. Sci. USA 2019, 116, 18808–18814. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Krasnova, L.; Finn, M.G.; Sharpless, K.B. Sulfur(VI) fluoride exchange (SuFEx): Another good reaction for click chemistry. Angew. Chem. 2014, 53, 9430–9448. [Google Scholar] [CrossRef]
- Barrow, A.S.; Smedley, C.J.; Zheng, Q.; Li, S.; Dong, J.; Moses, J.E. The growing applications of SuFEx click chemistry. Chem. Soc. Rev. 2019, 48, 4731–4758. [Google Scholar] [CrossRef]
- Narayanan, A.; Jones, L.H. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci. 2015, 6, 2650–2659. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, H.; Debreczeni, J.; Breed, J.; Tentarelli, S.; Aquila, B.; Dowling, J.E.; Whitty, A.; Grimster, N.P. A study of the reactivity of S-(VI)-F containing warheads with nucleophilic amino-acid side chains under physiological conditions. Org. Biomol. Chem. 2017, 15, 9685–9695. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, N.; Schnier, P.D.; Zheng, F.; Zhu, H.; Polizzi, N.F.; Ittuveetil, A.; Saikam, V.; DeGrado, W.F.; Wang, Q.; et al. Genetically introducing biochemically reactive amino acids dehydroalanine and dehydrobutyrine in proteins. J. Am. Chem. Soc. 2019, 141, 7698–7703. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Xu, T.; Yang, M.; Cao, T.; Liao, S. A rapid access to aliphatic sulfonyl fluorides. Nat. Commun. 2019, 10, 3752. [Google Scholar] [CrossRef]
- Shishido, Y.; Tomoike, F.; Kimura, Y.; Kuwata, K.; Yano, T.; Fukui, K.; Fujikawa, H.; Sekido, Y.; Murakami-Tonami, Y.; Kameda, T.; et al. A covalent G-site inhibitor for glutathione S-transferase Pi (GSTP1-1). Chem. Commun. 2017, 53, 11138–11141. [Google Scholar] [CrossRef]
- Baillie, T.A. Targeted covalent inhibitors for drug design. Angew. Chem. 2016, 55, 13408–13421. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Zhang, H.; Humphreys, W.G. Drug-protein adducts: Chemistry, mechanisms of toxicity, and methods of characterization. Chem. Res. Toxicol. 2016, 29, 2040–2057. [Google Scholar] [CrossRef]
- Smith, A.J.; Zhang, X.; Leach, A.G.; Houk, K.N. Beyond picomolar affinities: Quantitative aspects of non-covalent and covalent binding of drugs to proteins. J. Med. Chem. 2009, 52, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.S.; Weerapana, E.; Cravatt, B.F. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med. Chem. 2010, 2, 949–964. [Google Scholar] [CrossRef] [Green Version]
- Lagoutte, R.; Patouret, R.; Winssinger, N. Covalent inhibitors: An opportunity for rational target selectivity. Curr. Opin. Chem. Biol. 2017, 39, 54–63. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.; Petter, R.C.; Baillie, T.A.; Whitty, A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011, 10, 307–317. [Google Scholar] [CrossRef]
- Bauer, R.A. Covalent inhibitors in drug discovery: From accidental discoveries to avoided liabilities and designed therapies. Drug Discov. Today 2015, 20, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Copeland, R.E. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Schwartz, P.A.; Kuzmic, P.; Solowiej, J.; Bergqvist, S.; Bolanos, B.; Almaden, C.; Nagata, A.; Ryan, K.; Feng, J.; Dalvie, D.; et al. Covalent EGFR inhibitor analysis reveals the importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crisafuli, F.A.P.; Ramos, E.B.; Rocha, M.S. Characterizing the interaction between DNA and GelRed fluorescent stain. Eur. Biophys. J. Biophys. Lett. 2015, 44, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, S.; Kitahara, Y.; Okamatsu, Y.; Fujii, T.; Nakayama, A.; Ueno, S.; Ijichi, C.; Futaki, F.; Nakata, K.; Taki, M. Facile and Efficient Chemoenzymatic Semisynthesis of Fc-Fusion Compounds for Half-Life Extension of Pharmaceutical Components. Bioconjug. Chem. 2019, 30, 2323–2331. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabuchi, Y.; Yang, J.; Taki, M. Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein. Int. J. Mol. Sci. 2022, 23, 7778. https://doi.org/10.3390/ijms23147778
Tabuchi Y, Yang J, Taki M. Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein. International Journal of Molecular Sciences. 2022; 23(14):7778. https://doi.org/10.3390/ijms23147778
Chicago/Turabian StyleTabuchi, Yudai, Jay Yang, and Masumi Taki. 2022. "Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein" International Journal of Molecular Sciences 23, no. 14: 7778. https://doi.org/10.3390/ijms23147778
APA StyleTabuchi, Y., Yang, J., & Taki, M. (2022). Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein. International Journal of Molecular Sciences, 23(14), 7778. https://doi.org/10.3390/ijms23147778






