Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action
Abstract
:1. Introduction
2. Results and Discussion
2.1. Physicochemical Properties of NP-6
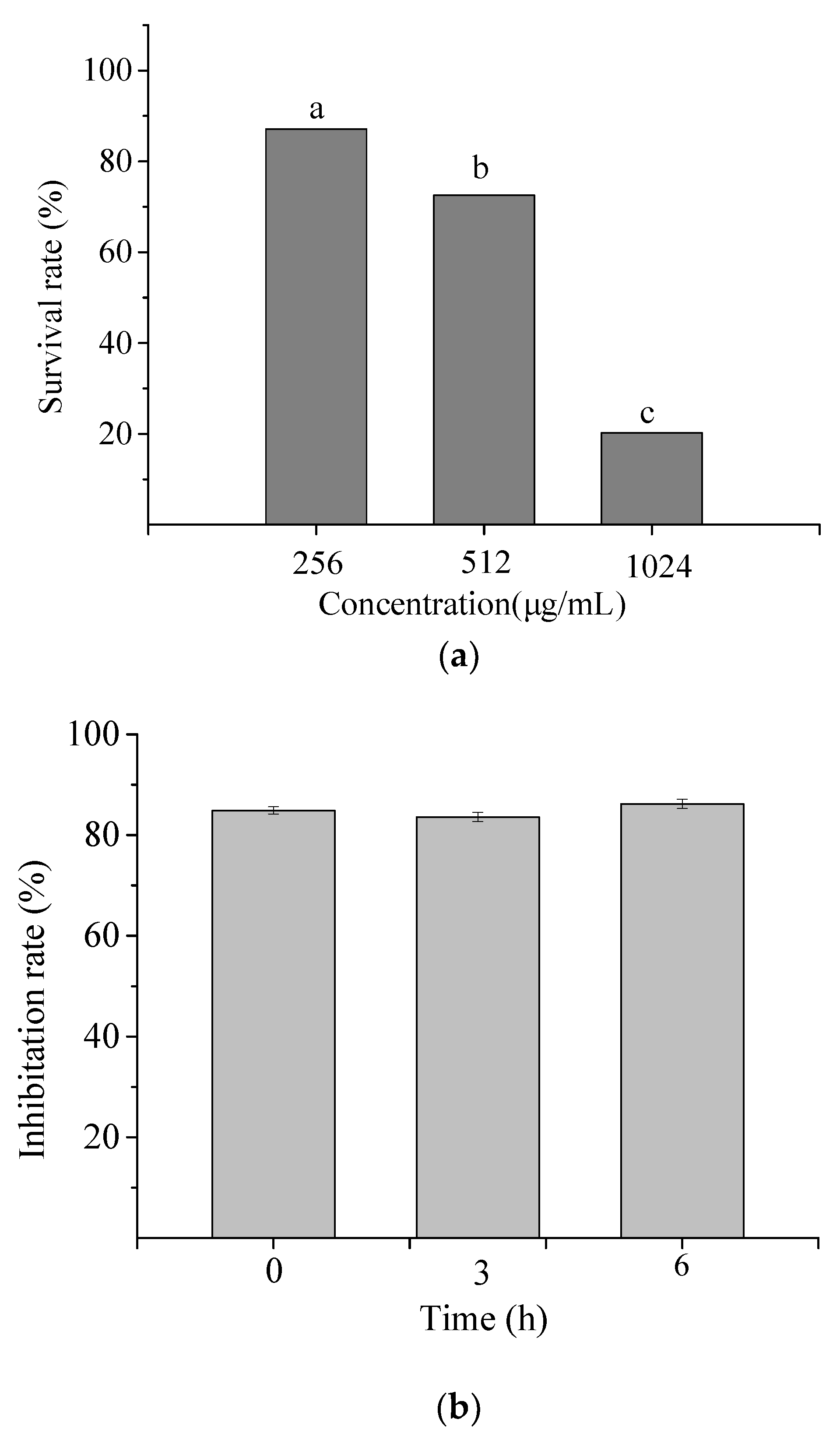
2.1.1. Hemolysis Activity of NP-6
2.1.2. Cytotoxicity of NP-6
2.1.3. Serum Stability of NP-6
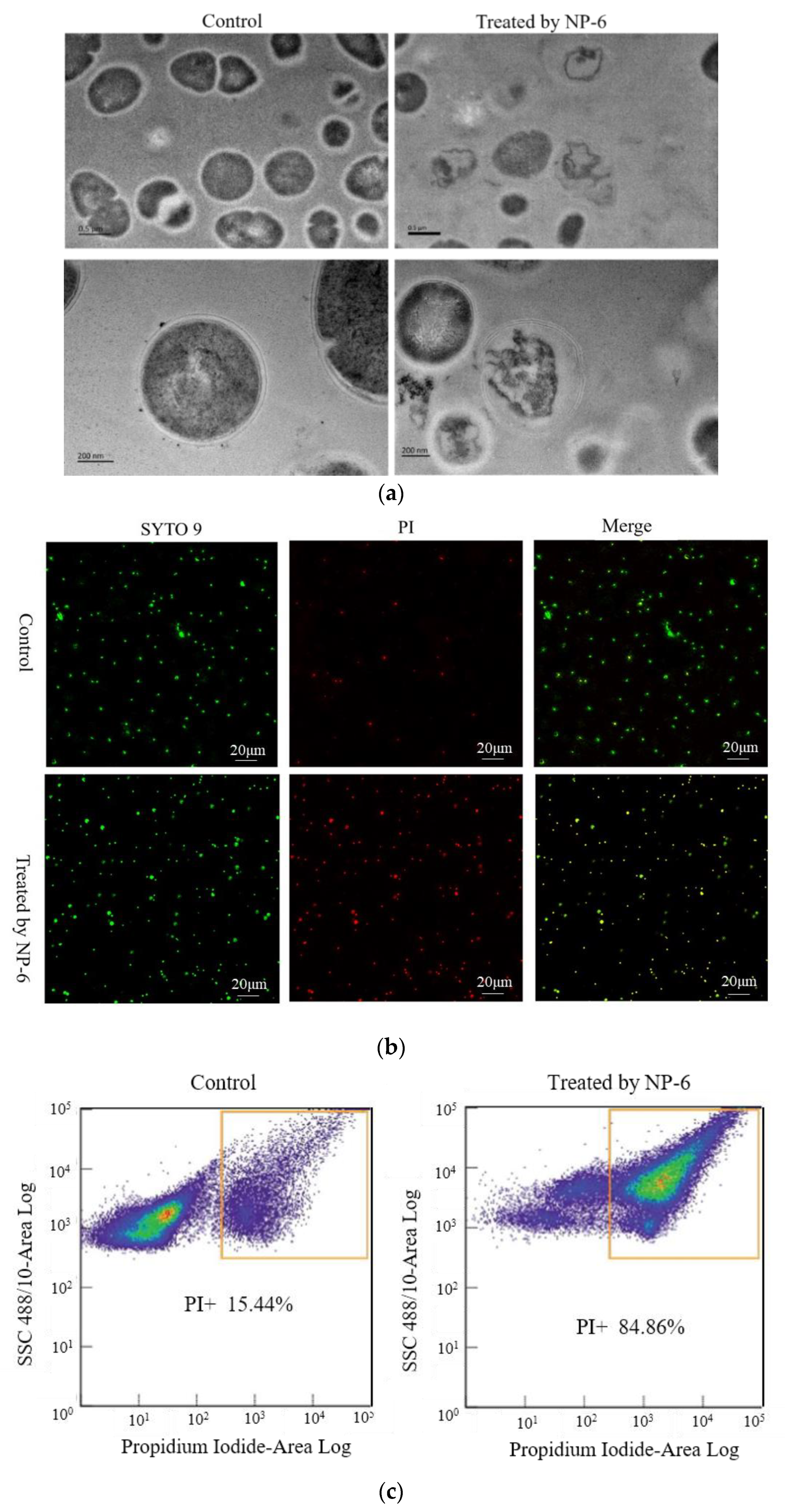
2.2. TEM Observations
2.3. PI Uptake Analysis
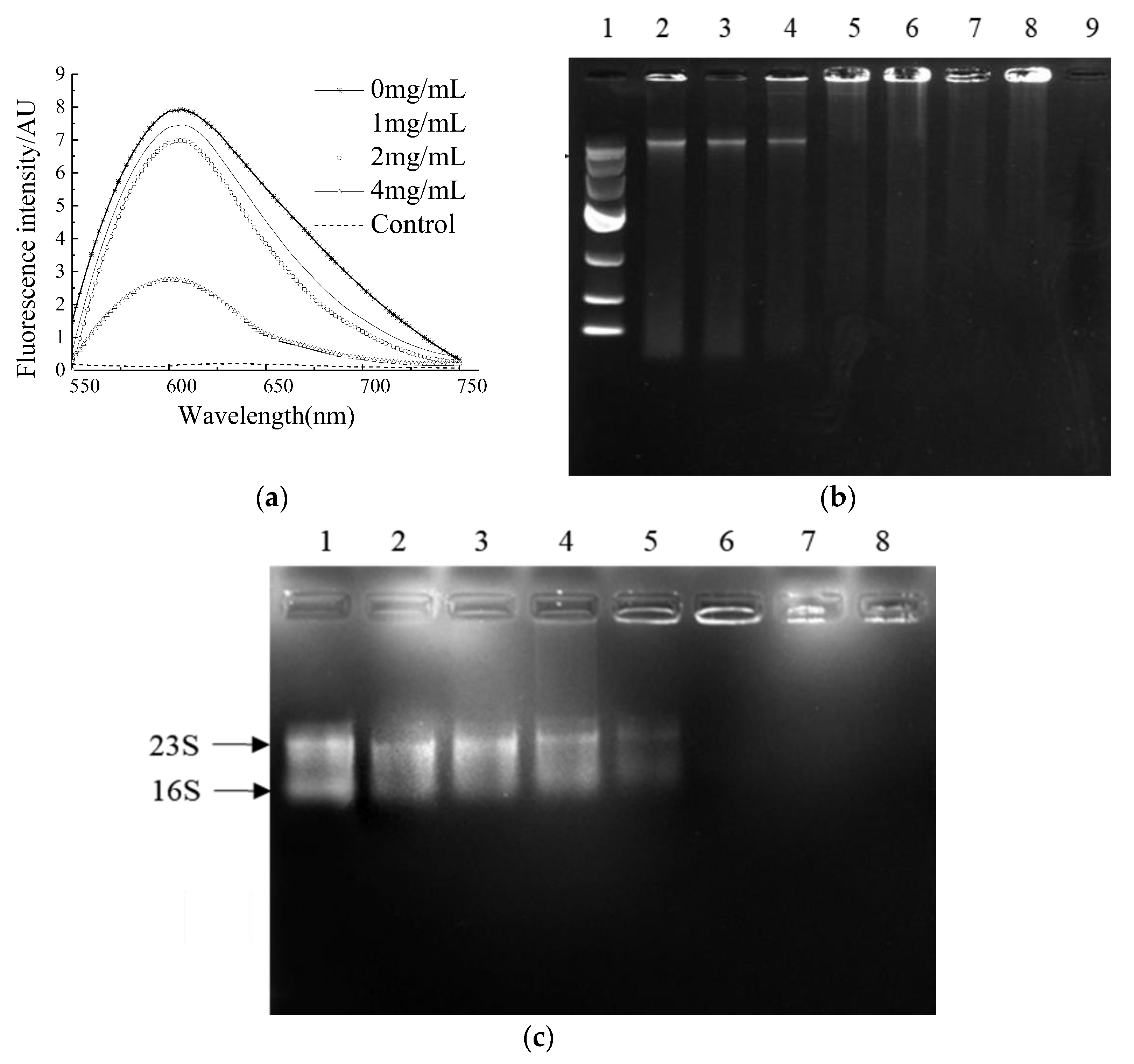
2.4. Binding Ability of NP-6 with Bacterial DNA and RNA
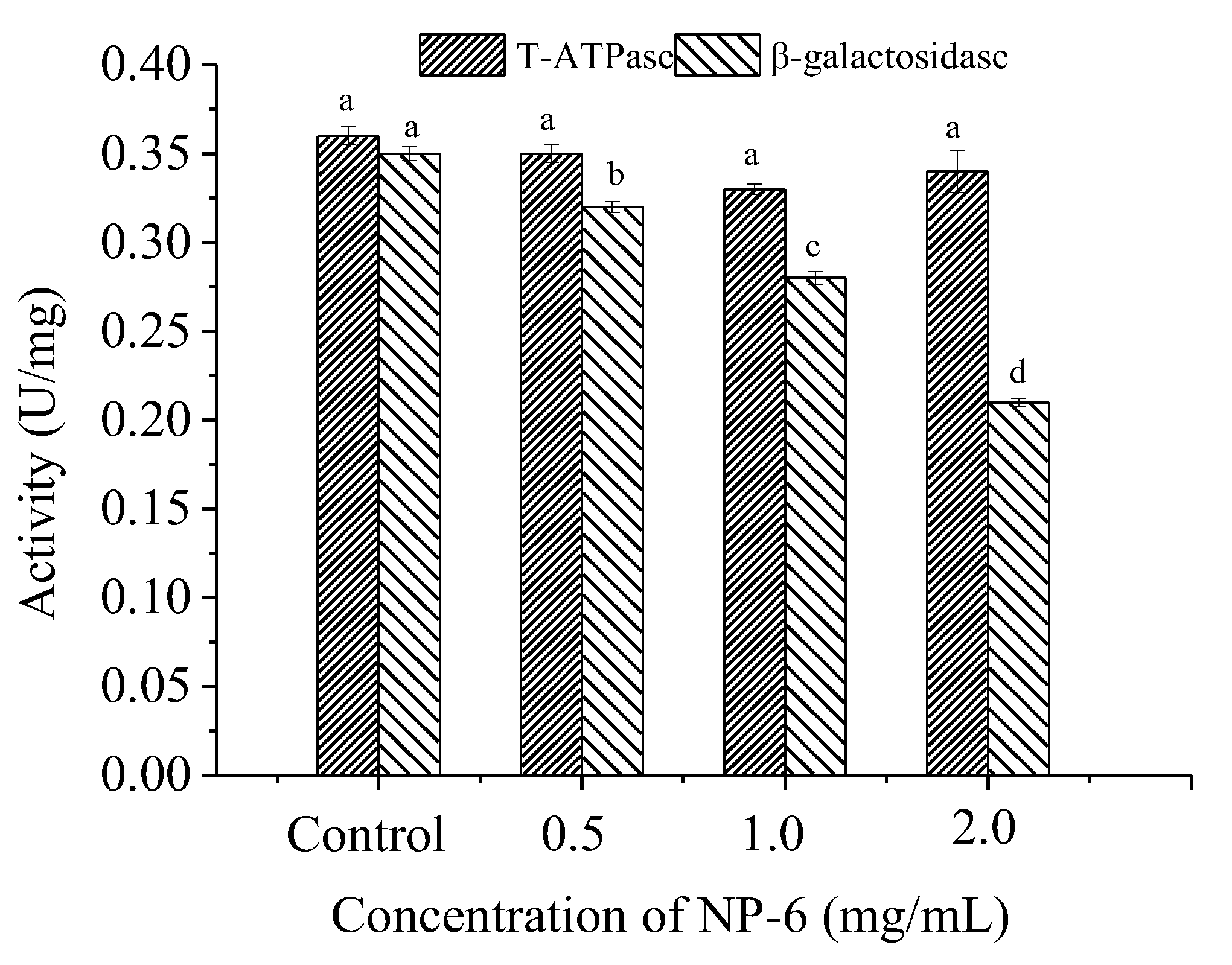
2.5. Effect of NP-6 on the Activities of Intracellular T-ATPase and β-Galactosidase
3. Material and Methods
3.1. Materials
3.2. Physicochemical Properties of NP-6
3.2.1. Hemolysis Assay
3.2.2. Cytotoxicity Assay
3.2.3. Serum Stability
3.3. Observation of Transmission Electron Microscopy (TEM)
3.4. Confocal Laser Scanning Microscopy (CLSM) Test
3.5. Flow Cytometric Analysis
3.6. Competitive Binding of NP-6 and EB with Bacterial DNA
3.7. DNA Gel Retardation
3.8. RNA Gel Retardation
3.9. Effect of NP-6 on the Activity of T-ATPase
3.10. Effect of NP-6 on the Activity of β-Galactosidase
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hultmark, D.; Steiner, H.; Rasmuson, T.; Boman, H. Insect Immunity. Purification and Properties of Three Inducible Bactericidal Proteins from Hemolymph of Immunized Pupae of Hyalophora cecropia. Eur. J. Biochem. 1980, 106, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Hall, K.N.; Aguilar, M.I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2016, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Y.; Xue, Z.; Jia, Y.; Li, R.; He, C.; Chen, H. The structure-mechanism relationship and mode of actions of antimicrobial peptides: A review. Trends Food Sci. Technol. 2021, 109, 103–115. [Google Scholar] [CrossRef]
- Cardoso, M.H.; Meneguetti, B.T.; Costa, B.O.; Buccini, D.F.; Oshiro, K.G.N.; Preza, S.L.E.; Carvalho, C.M.E.; Migliolo, L.; Franco, O.L. Non-Lytic Antibacterial Peptides That Translocate Through Bacterial Membranes to Act on Intracellular Targets. Int. J. Mol. Sci. 2019, 20, 4877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madrazo, A.L.; Campos, M.R.S. Review of antimicrobial peptides as promoters of food safety: Limitations and possibilities within the food industry. J. Food Saf. 2020, 40, e12854. [Google Scholar] [CrossRef]
- Lee, T.H.; Hofferek, V.; Separovic, F.; Reid, G.E.; Aguilar, M.I. The role of bacterial lipid diversity and membrane properties in modulating antimicrobial peptide activity and drug resistance. Curr. Opin. Chem. Biol. 2019, 52, 85–92. [Google Scholar] [CrossRef]
- da Silva, A.; Teschke, O. Effects of the antimicrobial peptide PGLa on live Escherichia coli. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2003, 1643, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Zamora-Carreras, H.; Strandberg, E.; Mühlhäuser, P.; Bürck, J.; Wadhwani, P.; Jiménez, M.Á.; Bruix, M.; Ulrich, A.S. Alanine scan and 2 H NMR analysis of the membrane-active peptide BP100 point to a distinct carpet mechanism of action. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 1328–1338. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Mechanisms, M.P. Antimicrobial Peptides: Basics for Clinical Application; Matsuzaki, K., Ed.; Springer: Singapore, 2019; pp. 9–16. [Google Scholar]
- Matsuzaki, K.; Yoneyama, S.; Murase, O.; Miyajima, K. Transbilayer Transport of Ions and Lipids Coupled with Mastoparan X Translocation. Biochemistry 1996, 35, 8450–8456. [Google Scholar] [CrossRef]
- Kobayashi, S.; Chikushi, A.; Tougu, S.; Imura, Y.; Nishida, M.; Yano, Y.; Matsuzaki, K. Membrane Translocation Mechanism of the Antimicrobial Peptide Buforin 2. Biochemistry 2004, 43, 15610–15616. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Laver, D.R. The barrel-stave model as applied to alamethicin and its analogs reevaluated. Biophys. J. 1994, 66, 355–359. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, A.; Lee, D.; Narasimhaswamy, T.; Nanga, R.P.R. Cholesterol reduces pardaxin’s dynamics—A barrel-stave mechanism of membrane disruption investigated by solid-state NMR. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 223–227. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Le, C.; Fang, C.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef] [Green Version]
- Graf, M.; Mardirossian, M.; Nguyen, F.; Seefeldt, A.C.; Guichard, G.; Scocchi, M.; Innis, C.A.; Wilson, D.N. Proline-rich antimicrobial peptides targeting protein synthesis. Nat. Prod. Rep. 2017, 34, 702–711. [Google Scholar] [CrossRef]
- Roy, R.N.; Lomakin, I.B.; Gagnon, M.G.; Steitz, T.A. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 2015, 22, 466–469. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Sun, J.; Xia, S.; Tian, X.; Cheserek, M.J.; Le, G. Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: Intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 2016, 100, 3245–3253. [Google Scholar] [CrossRef]
- Miao, J.; Zhou, J.; Liu, G.; Chen, F.; Chen, Y.; Gao, X.; Dixon, W.; Song, M.; Xiao, H.; Cao, Y. Membrane disruption and DNA binding of Staphylococcus aureus cell induced by a novel antimicrobial peptide produced by Lactobacillus paracasei subsp. tolerans FX-6. Food Control. 2016, 59, 609–613. [Google Scholar] [CrossRef]
- Hou, X.; Li, S.; Luo, Q.; Shen, G.; Wu, H.; Li, M.; Liu, X.; Chen, A.; Ye, M.; Zhang, Z. Discovery and identification of antimicrobial peptides in Sichuan pepper (Zanthoxylum bungeanum Maxim) seeds by peptidomics and bioinformatics. Appl. Microbiol. Biotechnol. 2019, 103, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Aronica, P.G.A.; Reid, L.M.; Desai, N.; Li, J.; Fox, S.J.; Yadahalli, S.; Essex, J.W.; Verma, C.S. Computational methods and tools in antimicrobial peptide research. J. Chem. Inf. Model. 2021, 61, 3172–3196. [Google Scholar] [CrossRef] [PubMed]
- Pushpanathan, M.; Pooja, S.; Gunasekaran, P.; Rajendhran, J. Critical evaluation and compilation of physicochemical determinants and membrane interactions of MMGP1 antifungal peptide. Mol. Pharm. 2016, 13, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, F.; Asadi, A.; Afsharpour, M.; Robab, H.J. In vitro characterization and evaluation of the cytotoxicity effects of nisin and nisin-loaded PLA-PEG-PLA nanoparticles on gastrointestinal (AGS and KYSE-30), hepatic (HepG2) and blood (K562) cancer cell lines. AAPS PharmSciTech 2018, 19, 1554–1566. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, C.; Liang, G.; Zhang, M.; Zheng, J. Engineering antimicrobial peptides with improved antimicrobial and hemolytic activities. J. Chem. Inf. Model. 2013, 53, 3280–3296. [Google Scholar] [CrossRef]
- Sahariah, P.; Sørensen, K.K.; Hjálmarsdóttir, M.Á.; Sigurjonsson, O.E.; Jensen, K.J.; Masson, M.; Thygesen, M.B. Antimicrobial peptide shows enhanced activity and reduced toxicity upon grafting to chitosan polymers. Chem. Commun. 2015, 51, 11611–11614. [Google Scholar] [CrossRef]
- Hou, X.; Feng, C.; Li, S.; Luo, Q.; Shen, G.; Wu, H.; Li, M.; Liu, X.; Chen, A.; Ye, M.; et al. Mechanismof antimicrobial peptide NP-6 from Sichuan pepper seeds against E. coli and effects of different environmental factors on its activity. Appl. Microbiol. Biotechnol. 2019, 103, 6593–6604. [Google Scholar] [CrossRef]
- Xu, L.; Chou, S.; Wang, J.; Shao, C.; Li, W.; Zhu, X.; Shan, A. Antimicrobial activity and membrane-active mechanism of tryptophan zipper-like β-hairpin antimicrobial peptides. Amino Acids 2015, 47, 2385–2397. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, Y.; Li, S.; Li, H.; Tian, L.; Wang, H.; Shang, D. Anticancer mechanisms of temporin-1CEa, an amphipathic α-helical antimicrobial peptide, in Bcap-37 human breast cancer cells. Life Sci. 2013, 92, 1004–1014. [Google Scholar] [CrossRef]
- Song, W.; Kong, X.; Hua, Y.; Chen, Y.; Zhang, C.; Chen, Y. Identification of antibacterial peptides generated from enzymatic hydrolysis of cottonseed proteins. LWT 2020, 125, 109199. [Google Scholar] [CrossRef]
- Anunthawan, T.; de la Fuente-Núñez, C.; Hancock, R.E.W.; Klaynongsruang, S. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 1352–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Liu, J.; Li, J.; Xia, L.; Yang, J.; Sun, S.; Ma, J.; Zhang, F. A potential food biopreservative, CecXJ-37N, non-covalently intercalates into the nucleotides of bacterial genomic DNA beyond membrane attack. Food Chem. 2017, 217, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Séverine, D.; Chiara, G.; Frédérique, L.; Heinz, S.; Vassilios, I.; Christine, D. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [Green Version]
- Kuriata, A.; Gierut, A.M.; Oleniecki, T.; Ciemny, M.P.; Kolinski, A.; Kurcinski, M.; Kmiecik, S. CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018, 46, W338–W343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds. Curr. Opin. Struct. Biol. 2006, 1, 118–126. [Google Scholar] [CrossRef]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Tang, W.; Zhang, H.; Wang, L.; Qian, H. New cationic antimicrobial peptide screened from boiled-dried anchovies by immobilized bacterial membrane liposome chromatography. J. Agric. Food Chem. 2014, 62, 1564–1571. [Google Scholar] [CrossRef]
- Kim, M.K.; Kang, N.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H.; et al. Antibacterial and antibiofilm activity and mode of action of magainin 2 against drug-resistant acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 3041. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Liu, K.; Cao, S.; Sun, J.; Zhong, B.; Jiong, C. Chemical composition, antimicrobial, antioxidant, and antiproliferative properties of grapefruit essential oil prepared by molecular distillation. Molecules 2020, 25, 217. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Shao, C.; Fang, Y.; Wang, J.; Dong, N.; Shan, A. Binding loop of sunflower trypsin inhibitor 1 serves as a design motif for proteolysis-resistant antimicrobial peptides. Acta Biomater. 2021, 124, 254–269. [Google Scholar] [CrossRef]
- Shi, W.; Li, C.; Li, M.; Zong, X.; Han, D.; Chen, Y. Antimicrobial peptide melittin against Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen in rice. Appl. Microbiol. Biotechnol. 2016, 100, 5059–5067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.; Bisht, G.S.; Rawat, D.S.; Kumar, A.; Kumar, R.; Maiti, S.; Pasha, S. Interaction studies of novel cell selective antimicrobial peptides with model membranes and E. coli ATCC 11775. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 1864–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef]
- Nam, J.; Yun, H.; Rajasekaran, G.; Kumar, S.D.; Kim, J.I.; Min, H.J.; Shin, S.Y.; Lee, C.W. Structural and Functional Assessment of mBjAMP1, an Antimicrobial Peptide from Branchiostoma japonicum, Revealed a Novel α-Hairpinin-like Scaffold with Membrane Permeable and DNA Binding Activity. J. Med. Chem. 2018, 61, 11101–11113. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, X.; Li, J.; Tang, H.; Li, Q.; Shen, G.; Li, S.; Chen, A.; Peng, Z.; Zhang, Y.; Li, C.; et al. Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action. Int. J. Mol. Sci. 2022, 23, 7812. https://doi.org/10.3390/ijms23147812
Hou X, Li J, Tang H, Li Q, Shen G, Li S, Chen A, Peng Z, Zhang Y, Li C, et al. Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action. International Journal of Molecular Sciences. 2022; 23(14):7812. https://doi.org/10.3390/ijms23147812
Chicago/Turabian StyleHou, Xiaoyan, Jianlong Li, Huaqiao Tang, Qingye Li, Guanghui Shen, Shanshan Li, Anjun Chen, Zixin Peng, Yu Zhang, Chaowei Li, and et al. 2022. "Antibacterial Peptide NP-6 Affects Staphylococcus aureus by Multiple Modes of Action" International Journal of Molecular Sciences 23, no. 14: 7812. https://doi.org/10.3390/ijms23147812





