O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations
Abstract
:1. Introduction
2. Results and Discussion
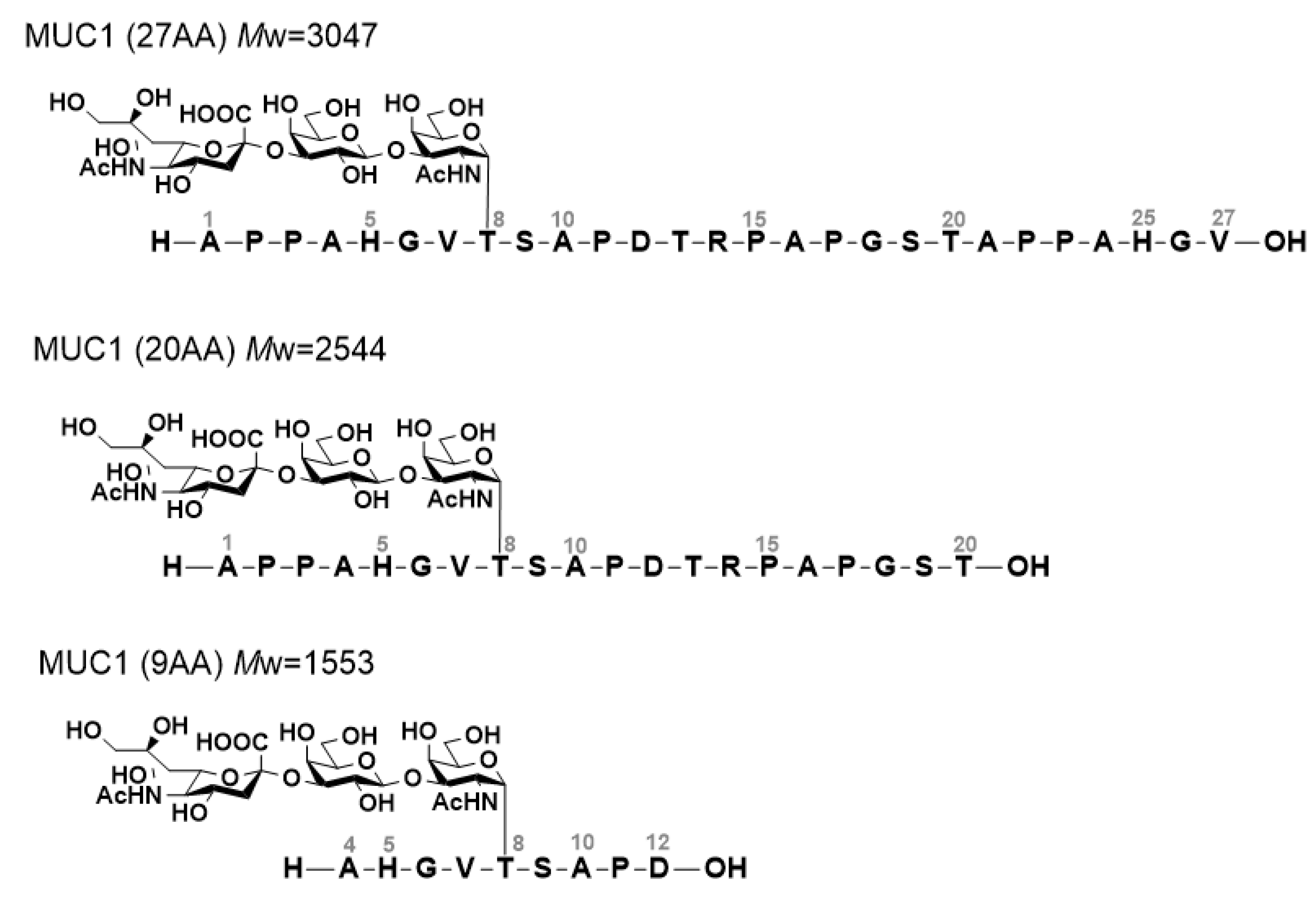
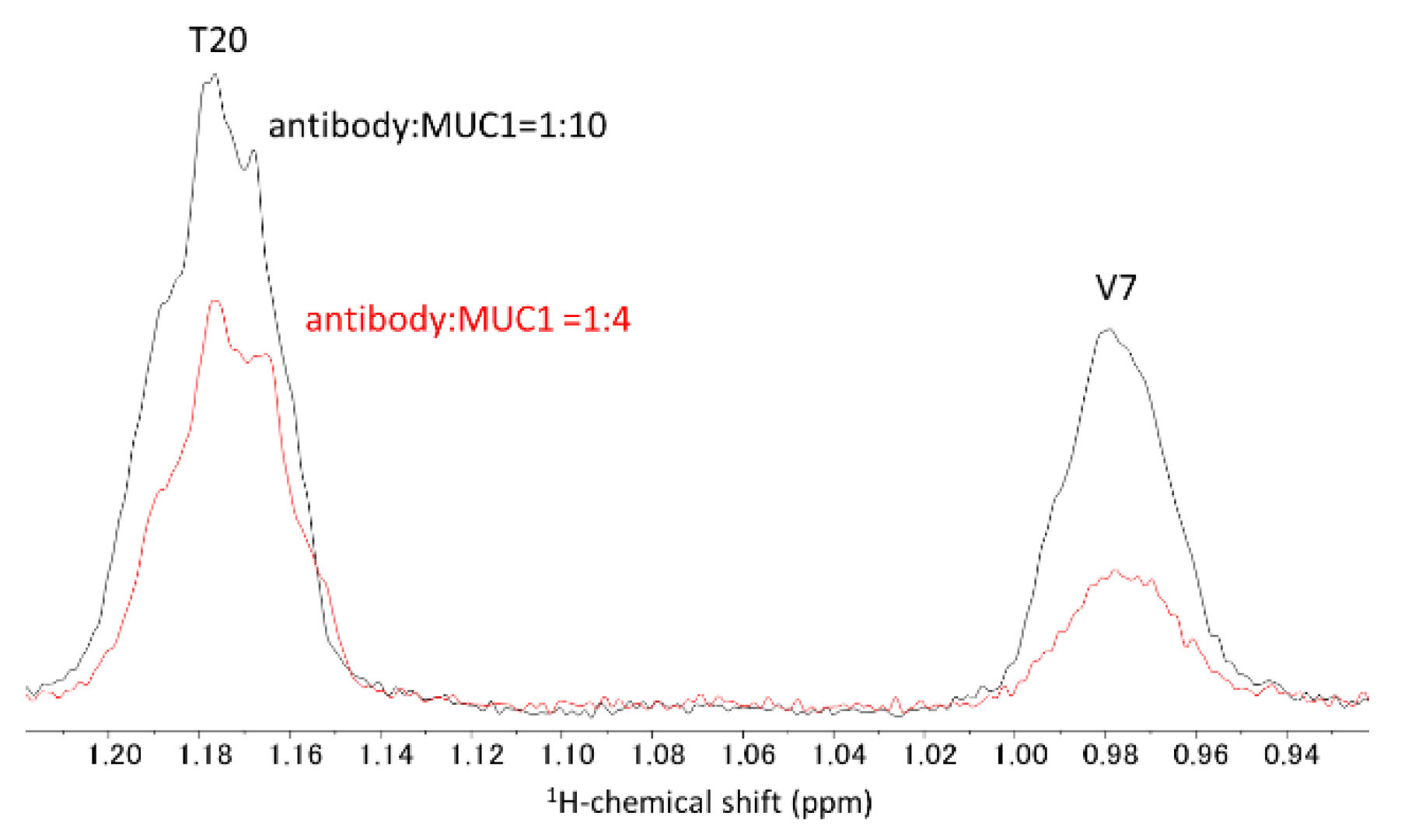
2.1. NMR Titration Study
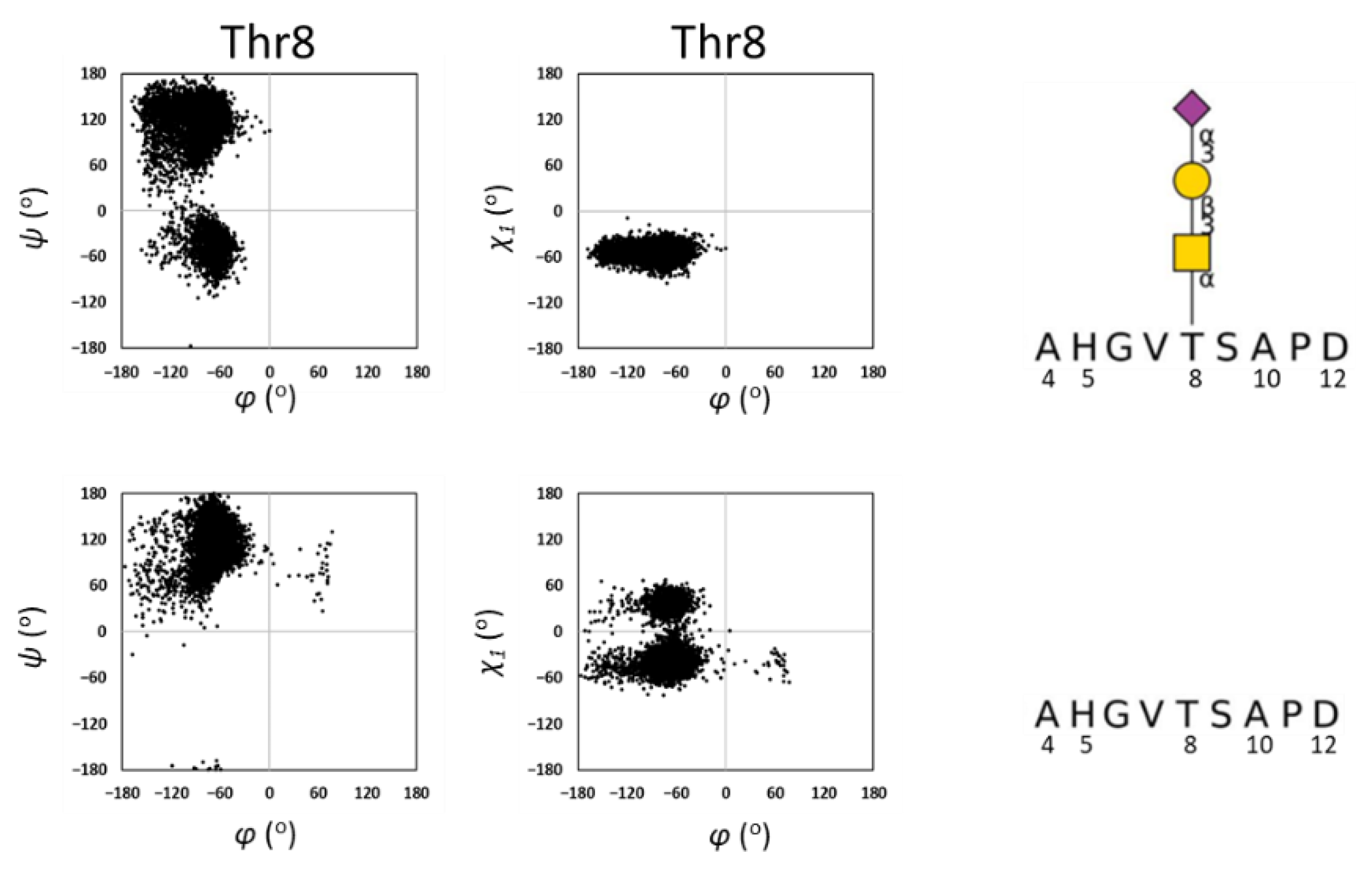
2.2. MD Simulations of MUC1
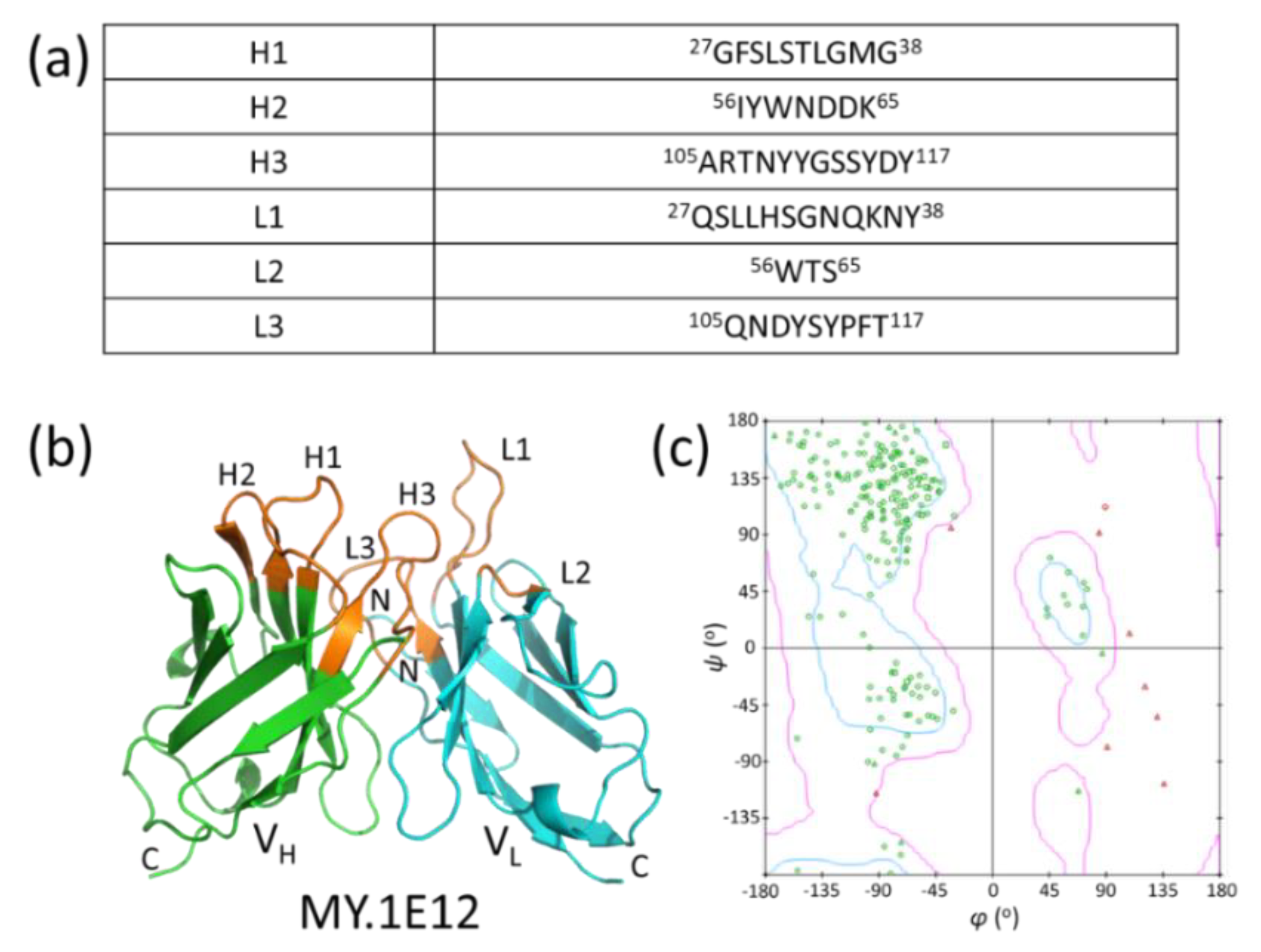
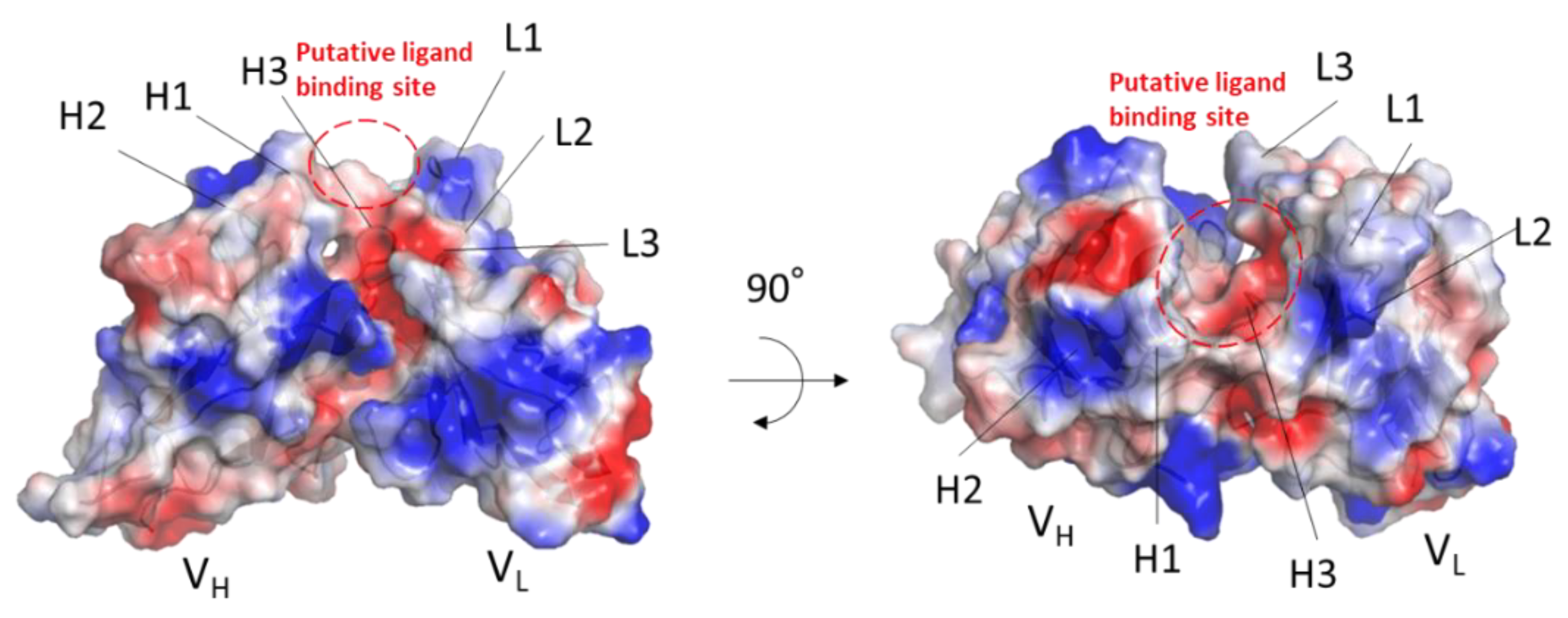
2.3. Modeling of MY.1E12
2.4. Docking of MUC1 Glycopeptide to MY.1E12
3. Materials and Methods
3.1. NMR Analysis
3.2. MD Simulation of MUC1
3.3. Modeling of MY.1E12 Fv Domain
3.4. Docking Simulation of Glycopeptide and Antibody
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Gendler, S.J.; Spicer, A.P. Epithelial Mucin Genes. Annu. Rev. Physiol. 1995, 57, 607–634. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, F.G. O-glycosylation of the mucin type. Biol. Chem. 2001, 382, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.; Goh, G.; Bard, F. Short O-GalNAc glycans: Regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta 2016, 1860, 1623–1639. [Google Scholar] [CrossRef] [Green Version]
- Ju, T.; Wang, Y.; Aryal, R.P.; Lehoux, S.D.; Ding, X.; Kudelka, M.R.; Cutler, C.; Zeng, J.; Wang, J.; Sun, X.; et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteom.-Clin. Appl. 2013, 7, 618–631. [Google Scholar] [CrossRef] [Green Version]
- Fujita-Yamaguchi, Y. Renewed interest in basic and applied research involving monoclonal antibodies against an oncofetal Tn-antigen. J. Biochem. 2013, 154, 103–105. [Google Scholar] [CrossRef] [Green Version]
- Springer, G.F. Tn epitope (N-acetyl-d-galactosamineα-O-serine/threonine) density in primary breast carcinoma: A functional predictor of aggressiveness. Mol. Immunol. 1989, 26, 1–5. [Google Scholar] [CrossRef]
- Hanisch, F.G.; Ninkovic, T. Immunology of O-glycosylated proteins: Approaches to the design of a MUC1 glycopeptide-based tumor vaccine. Curr. Protein Pept. Sci. 2006, 7, 307–315. [Google Scholar] [CrossRef]
- Zhou, D.; Xu, L.; Huang, W.; Tonn, T. Epitopes of MUC1 Tandem Repeats in Cancer as Revealed by Antibody Crystallography: Toward Glycopeptide Signature-Guided Therapy. Molecules 2018, 23, 1326. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Dai, D.; Chen, B.; Tang, H.; Xie, X.; Wei, W. Clinicopathological and Prognostic Significance of Cancer Antigen 15-3 and Carcinoembryonic Antigen in Breast Cancer: A Meta-Analysis including 12,993 Patients. Dis. Markers 2018, 2018, 9863092. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Sáez, N.; Castro-López, J.; Valero-González, J.; Madariaga, D.; Compañón, I.; Somovilla, V.J.; Salvadó, M.; Asensio, J.L.; Jiménez-Barbero, J.; Avenoza, A.; et al. Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an anti-MUC1 Antibody. Angew. Chem. Int. Ed. Engl. 2015, 54, 9830–9834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Movahedin, M.; Brooks, T.M.; Supekar, N.T.; Gokanapudi, N.; Boons, G.J.; Brooks, C.L. Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen. Glycobiology 2017, 27, 677–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Bhavanandan, V.P.; Nakamori, S.; Irimura, T. A Novel Monoclonal Antibody Specific for Sialylated MUC1 Mucin. Jpn. J. Cancer Res. 1996, 87, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Kato, K.; Denda-Nagai, K.; Hanisch, F.G.; Clausen, H.; Irimura, T. The epitope recognized by the unique anti-MUC1 monoclonal antibody MY.1E12 involves sialyl alpha 2-3galactosyl beta 1-3N-acetylgalactosaminide linked to a distinct threonine residue in the MUC1 tandem repeat. J. Immunol. Methods 2002, 270, 199–209. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Denda-Nagai, K.; Takahashi, Y.; Nagashima, I.; Shimizu, H.; Kishimoto, T.; Noji, M.; Shichino, S.; Chiba, Y.; Irimura, T. Products of Chemoenzymatic Synthesis Representing MUC1 Tandem Repeat Unit with T-, ST- or STn-antigen Revealed Distinct Specificities of Anti-MUC1 Antibodies. Sci. Rep. 2019, 9, 16641. [Google Scholar] [CrossRef]
- Coltart, D.M.; Royyuru, A.K.; Williams, L.J.; Glunz, P.W.; Sames, D.; Kuduk, S.D.; Schwarz, J.B.; Chen, X.T.; Danishefsky, S.J.; Live, D.H. Principles of mucin architecture: Structural studies on synthetic glycopeptides bearing clustered mono-, di-, tri-, and hexasaccharide glycodomains. J. Am. Chem. Soc. 2002, 124, 9833–9844. [Google Scholar] [CrossRef]
- Mallajosyula, S.S.; MacKerell, A.D., Jr. Influence of solvent and intramolecular hydrogen bonding on the conformational properties of o-linked glycopeptides. J. Phys. Chem. B 2011, 115, 11215–11229. [Google Scholar] [CrossRef] [Green Version]
- Borgert, A.; Foley, B.L.; Live, D. Contrasting the conformational effects of α-O-GalNAc and α-O-Man glycan protein modifications and their impact on the mucin-like region of alpha-dystroglycan. Glycobiology 2020, 31, 649–661. [Google Scholar] [CrossRef]
- Neelamegham, S.; Aoki-Kinoshita, K.; Bolton, E.; Frank, M.; Lisacek, F.; Lütteke, T.; O'Boyle, N.; Packer, N.H.; Stanley, P.; Toukach, P.; et al. Updates to the Symbol Nomenclature for Glycans guidelines. Glycobiology 2019, 29, 620–624. [Google Scholar] [CrossRef]
- Wormald, M.R.; Petrescu, A.J.; Pao, Y.L.; Glithero, A.; Elliott, T.; Dwek, R.A. Conformational studies of oligosaccharides and glycopeptides: Complementarity of NMR, X-ray crystallography, and molecular modelling. Chem. Rev. 2002, 102, 371–386. [Google Scholar] [CrossRef]
- Jo, S.; Im, W. Glycan fragment database: A database of PDB-based glycan 3D structures. Nucleic Acids Res. 2012, 41, D470–D474. [Google Scholar] [CrossRef] [PubMed]
- Lefranc, M.P.; Pommié, C.; Kaas, Q.; Duprat, E.; Bosc, N.; Guiraudou, D.; Jean, C.; Ruiz, M.; Da Piédade, I.; Rouard, M.; et al. IMGT unique numbering for immunoglobulin and T cell receptor constant domains and Ig superfamily C-like domains. Dev. Comp. Immunol. 2005, 29, 185–203. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Carton, J.M.; Gao, W.; Zhao, Y.; Gilliland, G.L. On the domain pairing in chimeric antibodies. Mol. Immunol. 2010, 47, 2422–2426. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Dassault Systèmes. Discovery Studio 2021; Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Momany, F.A.; Rone, R. Validation of the general purpose QUANTA ®3.2/CHARMm® force field. J. Comput. Chem. 1992, 13, 888–900. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokubu, R.; Ohno, S.; Kuratani, H.; Takahashi, Y.; Manabe, N.; Shimizu, H.; Chiba, Y.; Denda-Nagai, K.; Tsuiji, M.; Irimura, T.; et al. O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations. Int. J. Mol. Sci. 2022, 23, 7855. https://doi.org/10.3390/ijms23147855
Kokubu R, Ohno S, Kuratani H, Takahashi Y, Manabe N, Shimizu H, Chiba Y, Denda-Nagai K, Tsuiji M, Irimura T, et al. O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations. International Journal of Molecular Sciences. 2022; 23(14):7855. https://doi.org/10.3390/ijms23147855
Chicago/Turabian StyleKokubu, Ryoka, Shiho Ohno, Hirohide Kuratani, Yuka Takahashi, Noriyoshi Manabe, Hiroki Shimizu, Yasunori Chiba, Kaori Denda-Nagai, Makoto Tsuiji, Tatsuro Irimura, and et al. 2022. "O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations" International Journal of Molecular Sciences 23, no. 14: 7855. https://doi.org/10.3390/ijms23147855
APA StyleKokubu, R., Ohno, S., Kuratani, H., Takahashi, Y., Manabe, N., Shimizu, H., Chiba, Y., Denda-Nagai, K., Tsuiji, M., Irimura, T., & Yamaguchi, Y. (2022). O-Glycan-Dependent Interaction between MUC1 Glycopeptide and MY.1E12 Antibody by NMR, Molecular Dynamics and Docking Simulations. International Journal of Molecular Sciences, 23(14), 7855. https://doi.org/10.3390/ijms23147855





