Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates
Abstract
:1. Introduction
2. Results
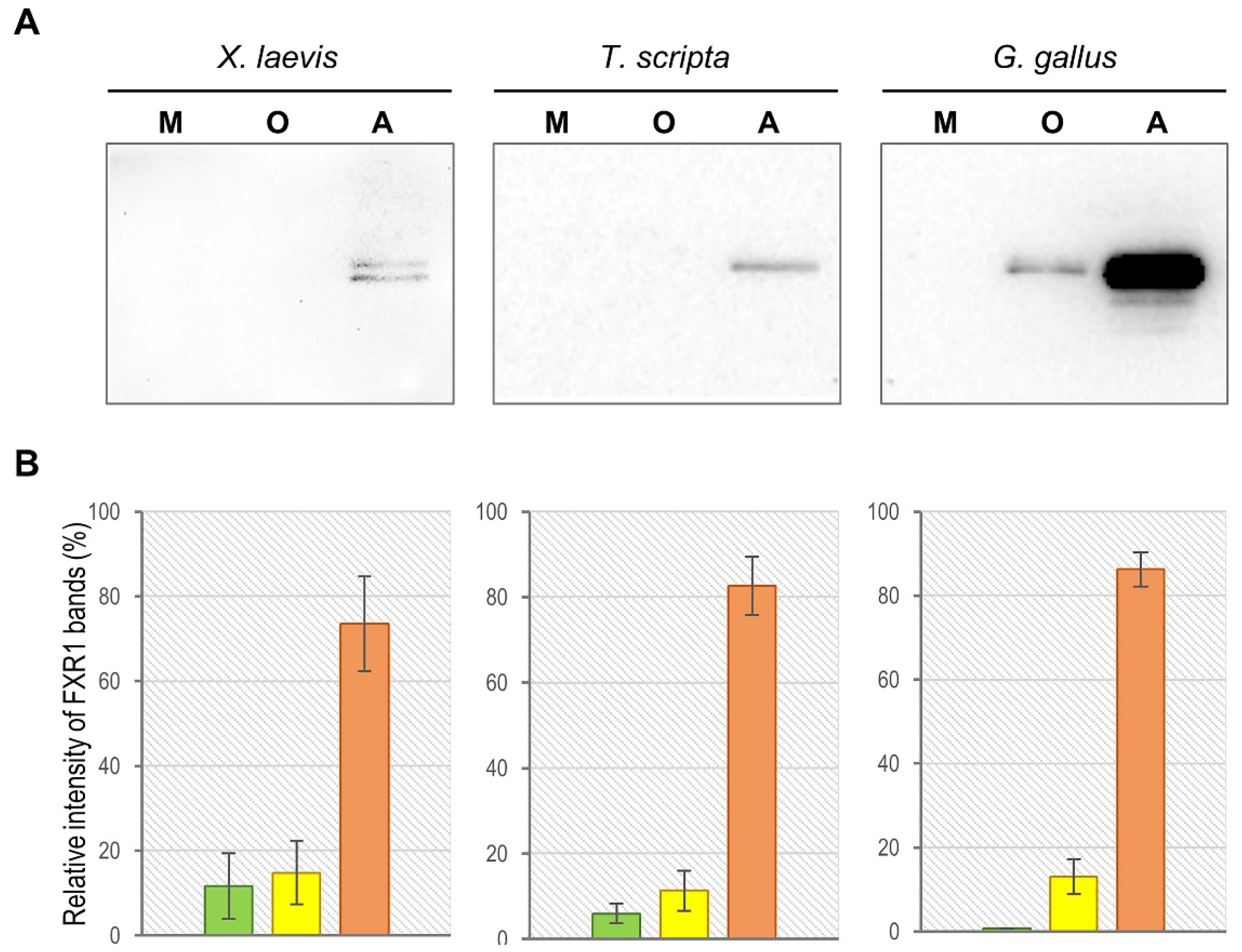
2.1. FXR1 Forms SDS-Resistant Amyloid-like Aggregates in the Brains of Amphibians, Reptiles, and Birds
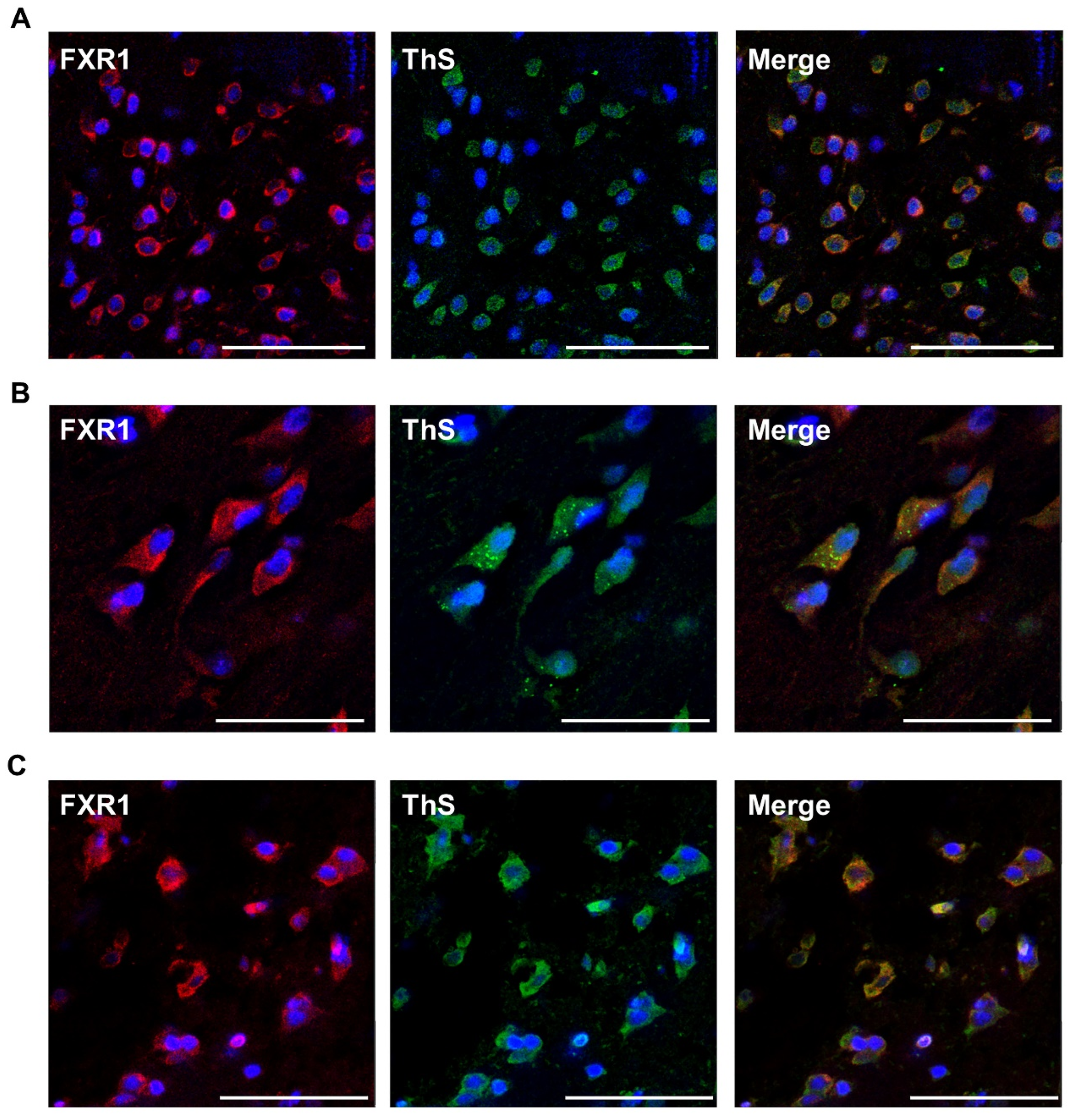
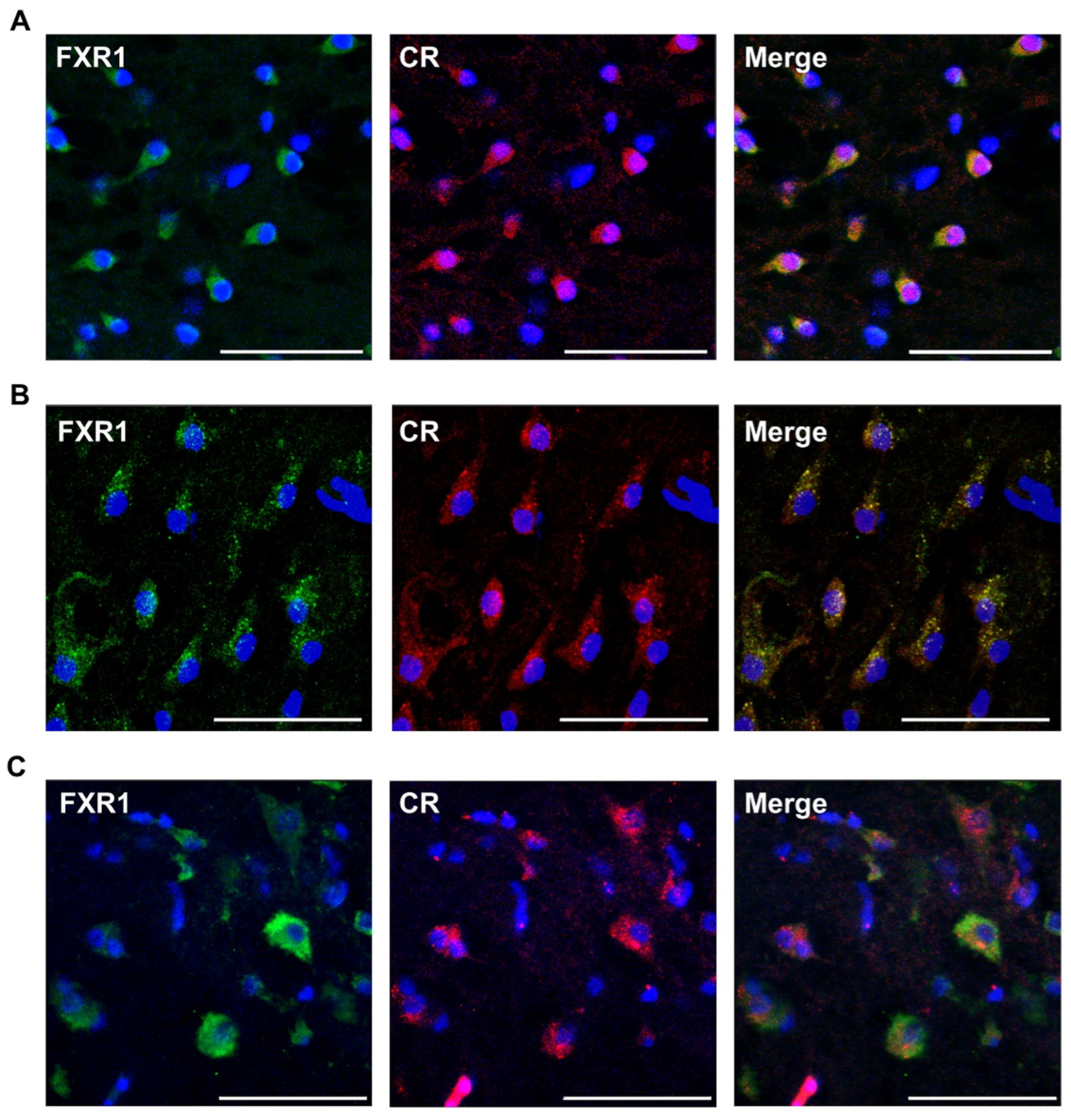
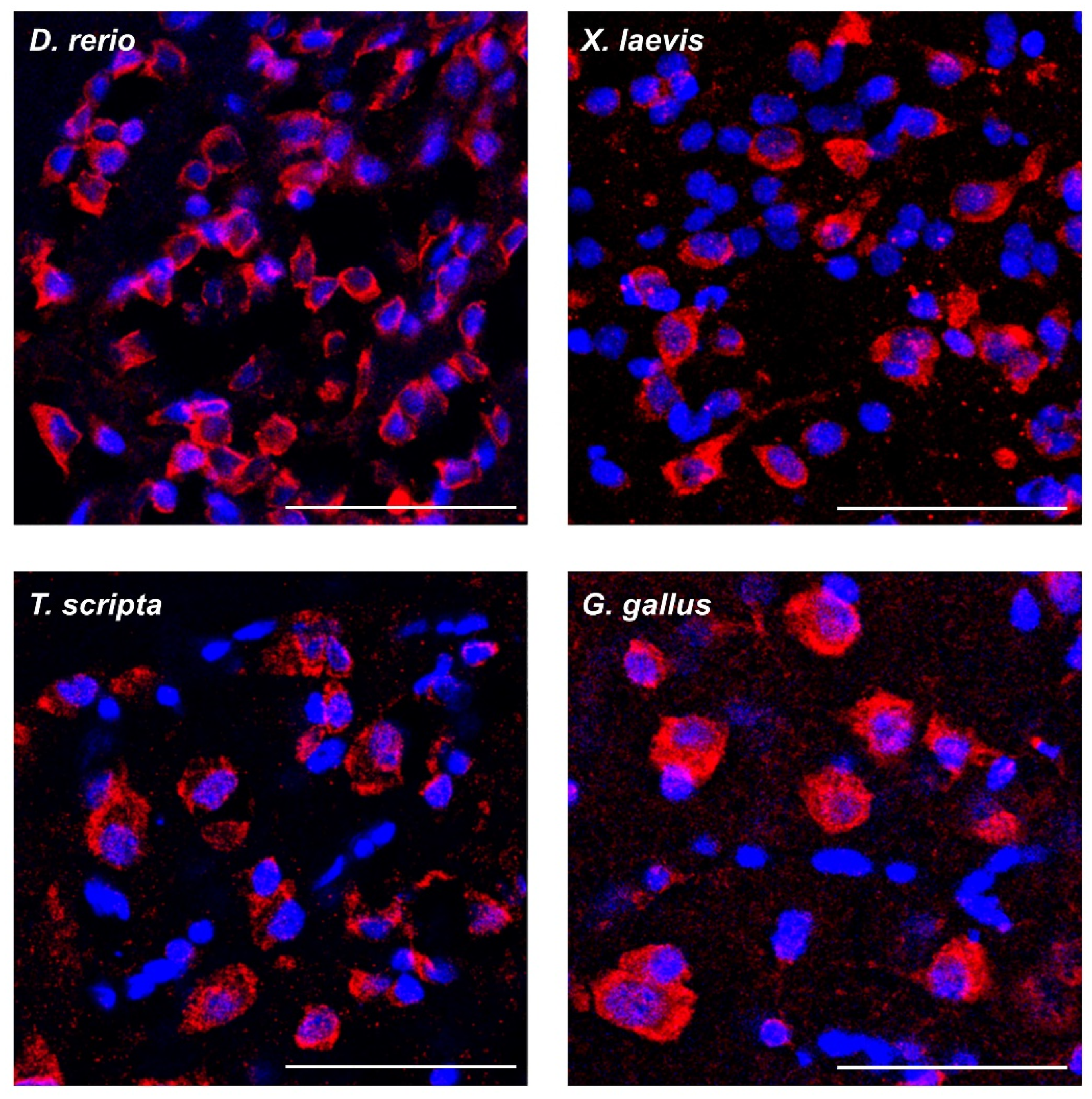
2.2. FXR1 Colocalizes with Amyloid-Specific Dyes Thioflavin S and Congo Red on Cryosections of the Brain of Clawed Frog, Red-Eared Turtle, and Chicken
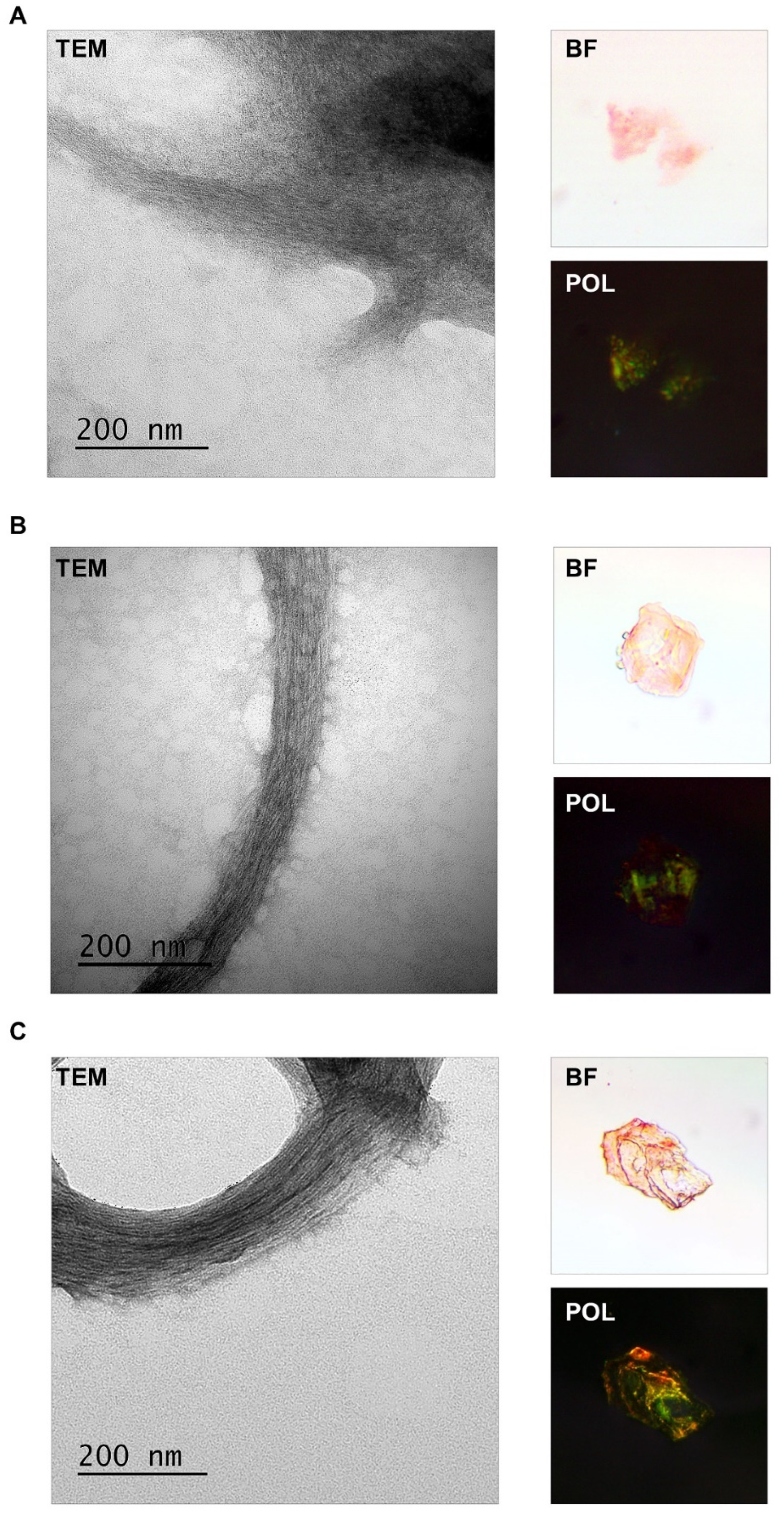
2.3. FXR1 Isolated from the Clawed Frog, Red-Eared Turtle, and Chicken Brain Is Detected as Fibrils That Stain with CR and Show Yellow-Green Birefringence
2.4. Functional Amyloid Fibrils Arose in the Brain at the Dawn of the Evolution of Jawed Vertebrates
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Preparation of the Brain Cryosections and Brain Homogenization
4.3. Protein Analysis
4.4. Immunoprecipitation
4.5. Electron Microscopy
4.6. CR Staining and Polarization Microscopy
4.7. Immunohistochemistry and Staining with Amyloid-Specific Dyes
4.8. Comparative Analysis of Sequences of FXR1 Protein in Different Vertebrate Species
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fowler, D.M.; Koulov, A.V.; Alory-Jost, C.; Marks, M.S.; Balch, W.E.; Kelly, J.W. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2006, 4, e6. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.K.; Wang, L.; Greenwald, J.; Riek, R. Structure-Activity Relationship of Amyloid Fibrils. FEBS Lett. 2009, 583, 2610–2617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erskine, E.; MacPhee, C.E.; Stanley-Wall, N.R. Functional Amyloid and Other Protein Fibers in the Biofilm Matrix. J. Mol. Biol. 2018, 430, 3642–3656. [Google Scholar] [CrossRef]
- Sopova, J.V.; Koshel, E.I.; Belashova, T.A.; Zadorsky, S.P.; Sergeeva, A.V.; Siniukova, V.A.; Shenfeld, A.A.; Velizhanina, M.E.; Volkov, K.V.; Nizhnikov, A.A.; et al. RNA-Binding Protein FXR1 Is Presented in Rat Brain in Amyloid Form. Sci. Rep. 2019, 9, 18983. [Google Scholar] [CrossRef] [Green Version]
- Hervas, R.; Rau, M.J.; Park, Y.; Zhang, W.; Murzin, A.G.; Fitzpatrick, J.A.J.; Scheres, S.H.W.; Si, K. Cryo-EM Structure of a Neuronal Functional Amyloid Implicated in Memory Persistence in Drosophila. Science 2020, 367, 1230–1234. [Google Scholar] [CrossRef]
- Maury, C.P.J. The Emerging Concept of Functional Amyloid. J. Intern. Med. 2009, 265, 329–334. [Google Scholar] [CrossRef]
- Mason, T.O.; Shimanovich, U. Fibrous Protein Self-Assembly in Biomimetic Materials. Adv. Mater. 2018, 30, 1706462. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Galkin, A.P. Functional Amyloids of Eukaryotes: Criteria, Classification, and Biological Significance. Curr. Genet. 2020, 66, 849–866. [Google Scholar] [CrossRef]
- Huot, M.E.; Bisson, N.; Moss, T.; Khandjian, E.W. Manipulating the Fragile X Mental Retardation Proteins in the Frog. Modeling Fragile X Syndr. 2012, 54, 165–179. [Google Scholar]
- Xu, X.-L.; Zong, R.; Li, Z.; Biswas, M.H.U.; Fang, Z.; Nelson, D.L.; Gao, F.-B. FXR1P but Not FMRP Regulates the Levels of Mammalian Brain-Specific MicroRNA-9 and MicroRNA-124. J. Neurosci. 2011, 31, 13705–13709. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Johnson, R.H.; Palanisamy, V. Fragile X-Related Protein Family: A Double-Edged Sword in Neurodevelopmental Disorders and Cancer. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Davidovic, L.; Durand, N.; Khalfallah, O.; Tabet, R.; Barbry, P.; Mari, B.; Sacconi, S.; Moine, H.; Bardoni, B. A Novel Role for the RNA-Binding Protein FXR1P in Myoblasts Cell-Cycle Progression by Modulating P21/Cdkn1a/Cip1/Waf1 MRNA Stability. PLoS Genet. 2013, 9, e1003367. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Li, Y.; Kadamberi, I.P.; Parashar, D.; Tsaih, S.W.; Gupta, P.; Geethadevi, A.; Chen, C.; Ghosh, C.; Sun, Y.; et al. RNA-Binding Protein FXR1 Drives CMYC Translation by Recruiting EIF4F Complex to the Translation Start Site. Cell Rep. 2021, 37, 109934. [Google Scholar] [CrossRef]
- Garnon, J.; Lachance, C.; Di Marco, S.; Hel, Z.; Marion, D.; Ruiz, M.C.; Newkirk, M.M.; Khandjian, E.W.; Radzioch, D. Fragile X-Related Protein FXR1P Regulates Proinflammatory Cytokine Tumor Necrosis Factor Expression at the Post-Transcriptional Level. J. Biol. Chem. 2005, 280, 5750–5763. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.; Nuro, E.; Jones, E.V.; Altimimi, H.F.; Farmer, W.T.; Gandin, V.; Hanna, E.; Zong, R.; Barbon, A.; Nelson, D.L.; et al. FXR1P Limits Long-Term Memory, Long-Lasting Synaptic Potentiation, and de Novo GluA2 Translation. Cell Rep. 2014, 9, 1402–1416. [Google Scholar] [CrossRef] [Green Version]
- Del’Guidice, T.; Latapy, C.; Rampino, A.; Khlghatyan, J.; Lemasson, M.; Gelao, B.; Quarto, T.; Rizzo, G.; Barbeau, A.; Lamarre, C.; et al. FXR1P Is a GSK3β Substrate Regulating Mood and Emotion Processing. Proc. Natl. Acad. Sci. USA 2015, 112, E4610–E4619. [Google Scholar] [CrossRef] [Green Version]
- Khandjian, E.W. Biology of the Fragile X Mental Retardation Protein, an RNA-Binding Protein. Biochem. Cell Biol. 1999, 77, 331–342. [Google Scholar] [CrossRef]
- Wolfe, L.S.; Calabrese, M.F.; Nath, A.; Blaho, D.V.; Miranker, A.D.; Xiong, Y. Protein-Induced Photophysical Changes to the Amyloid Indicator Dye Thioflavin, T. Proc. Natl. Acad. Sci. USA 2010, 107, 16863–16868. [Google Scholar] [CrossRef] [Green Version]
- Siniukova, V.A.; Sopova, J.V.; Galkina, S.A.; Galkin, A.P. Search for Functional Amyloid Structures in Chicken and Fruit Fly Female Reproductive Cells. Prion 2020, 14, 278–282. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Belashova, T.A.; Bondarev, S.A.; Velizhanina, M.E.; Barbitoff, Y.A.; Matveenko, A.G.; Valina, A.A.; Simanova, A.L.; Zhouravleva, G.A.; Galkin, A.P. Direct Proof of the Amyloid Nature of Yeast Prions [PSI+] and [PIN+] by the Method of Immunoprecipitation of Native Fibrils. FEMS Yeast Res. 2021, 21, foab046. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Head, E.; Sarsoza, F.; Saing, T.; Cotman, C.W.; Necula, M.; Margol, L.; Wu, J.; Breydo, L.; Thompson, J.L.; et al. Fibril Specific, Conformation Dependent Antibodies Recognize a Generic Epitope Common to Amyloid Fibrils and Fibrillar Oligomers That Is Absent in Prefibrillar Oligomers. Mol. Neurodegener. 2007, 2, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervas, R.; Li, L.; Majumdar, A.; Fernandez-Ramirez, M.D.C.; Unruh, J.R.; Slaughter, B.D.; Galera-Prat, A.; Santana, E.; Suzuki, M.; Nagai, Y.; et al. Molecular Basis of Orb2 Amyloidogenesis and Blockade of Memory Consolidation. PLoS Biol. 2016, 14, e1002361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakley, H.; Cole, S.L.; Logan, S.; Maus, E.; Shao, P.; Craft, J.; Guillozet-Bongaarts, A.; Ohno, M.; Disterhoft, J.; Van Eldik, L.; et al. Intraneuronal $β$-Amyloid Aggregates, Neurodegeneration, and Neuron Loss in Transgenic Mice with Five Familial Alzheimer’s Disease Mutations: Potential Factors in Amyloid Plaque Formation. J. Neurosci. 2006, 26, 10129–10140. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.B.; Znassi, N.; Château, M.T.; Kajava, A.V. A Structure-Based Approach to Predict Predisposition to Amyloidosis. Alzheimer’s Dement. 2015, 11, 681–690. [Google Scholar] [CrossRef]
- Emily, M.; Talvas, A.; Delamarche, C. MetAmyl: A METa-Predictor for AMYLoid Proteins. PLoS ONE 2013, 8, e79722. [Google Scholar] [CrossRef]
- Iadanza, M.G.; Jackson, M.P.; Hewitt, E.W.; Ranson, N.A.; Radford, S.E. A New Era for Understanding Amyloid Structures and Disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 755–773. [Google Scholar] [CrossRef]
- Schenck, A.; Bardoni, B.; Moro, A.; Bagni, C.; Mandel, J.-L. A Highly Conserved Protein Family Interacting with the Fragile X Mental Retardation Protein (FMRP) and Displaying Selective Interactions with FMRP-Related Proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. USA 2001, 98, 8844–8849. [Google Scholar] [CrossRef] [Green Version]
- Bechara, E.; Davidovic, L.; Melko, M.; Bensaid, M.; Tremblay, S.; Grosgeorge, J.; Khandjian, E.W.; Lalli, E.; Bardoni, B. Fragile X Related Protein 1 Isoforms Differentially Modulate the Affinity of Fragile X Mental Retardation Protein for G-Quartet RNA Structure. Nucleic Acids Res. 2007, 35, 299–306. [Google Scholar] [CrossRef]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velizhanina, M.E.; Galkin, A.P. Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates. Int. J. Mol. Sci. 2022, 23, 7997. https://doi.org/10.3390/ijms23147997
Velizhanina ME, Galkin AP. Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates. International Journal of Molecular Sciences. 2022; 23(14):7997. https://doi.org/10.3390/ijms23147997
Chicago/Turabian StyleVelizhanina, Maria E., and Alexey P. Galkin. 2022. "Amyloid Properties of the FXR1 Protein Are Conserved in Evolution of Vertebrates" International Journal of Molecular Sciences 23, no. 14: 7997. https://doi.org/10.3390/ijms23147997




