Characterization and Comparative Genomics Analysis of a New Bacteriophage BUCT610 against Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae
Abstract
1. Introduction
2. Results
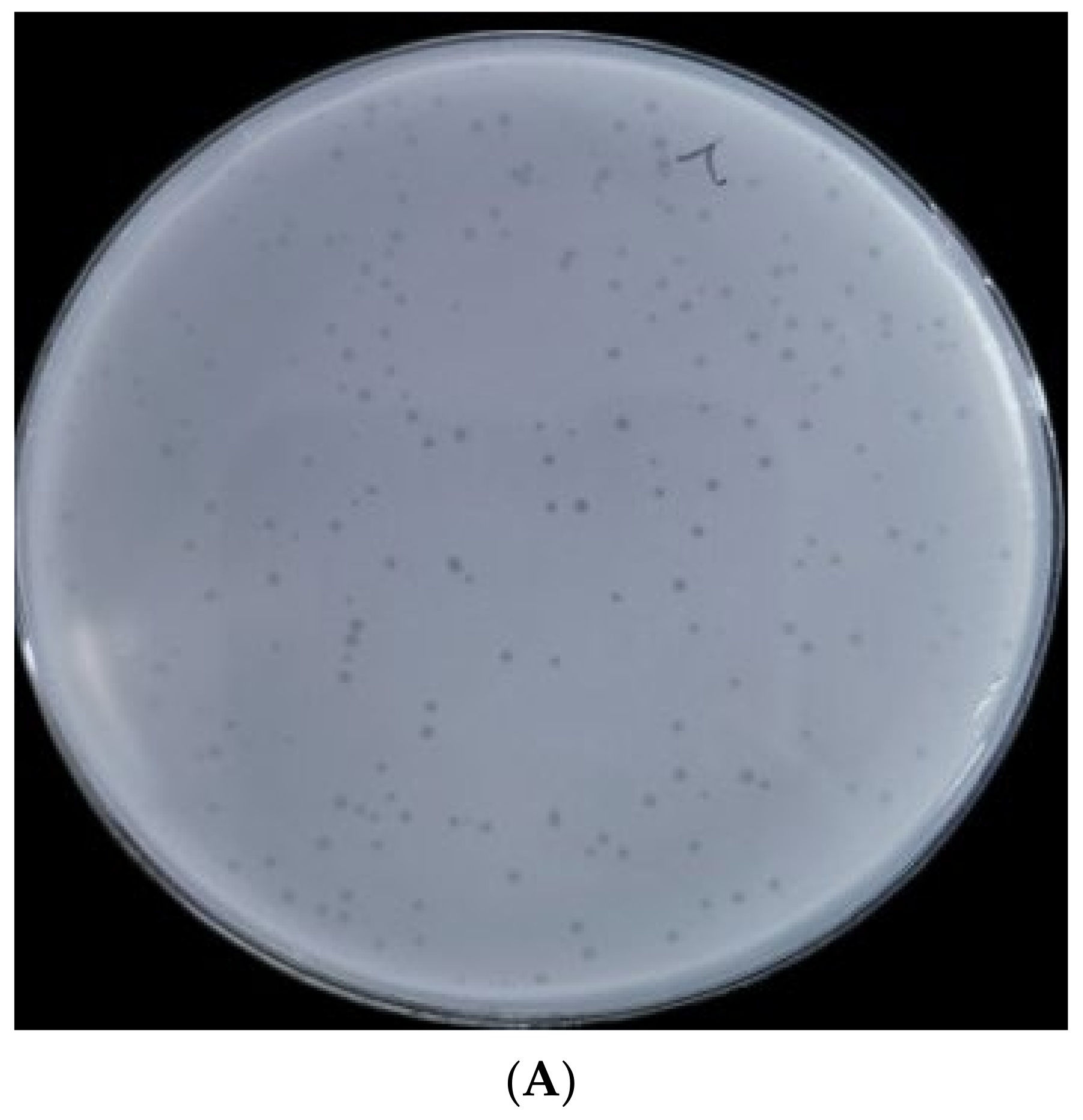
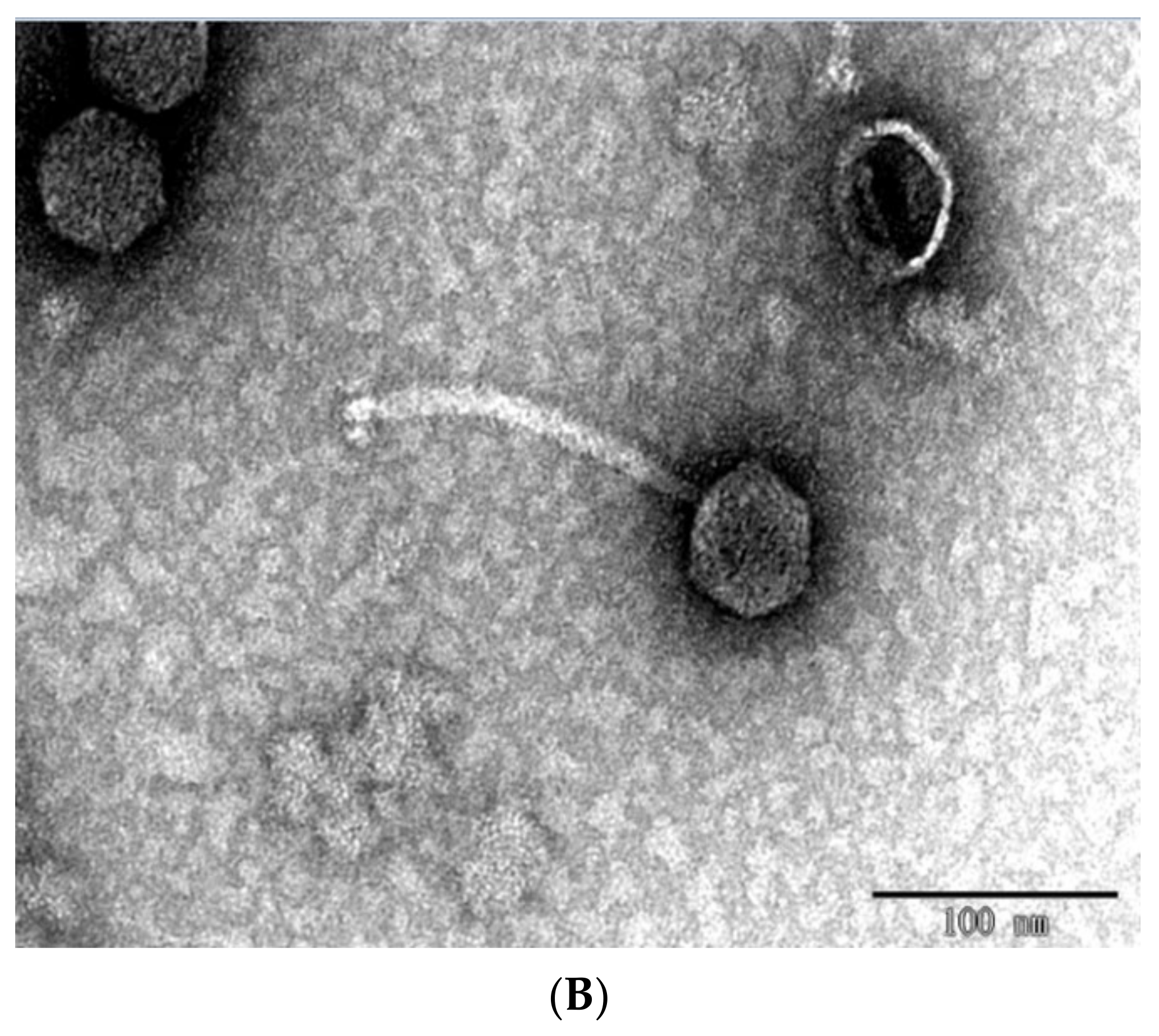
2.1. Morphological Characteristics and Lytic Rang of Phage BUCT610
2.2. Multilocus Sequence Typing and Capsule Type Identification of K. pneumoniae
2.3. Lifestyle of BUCT610
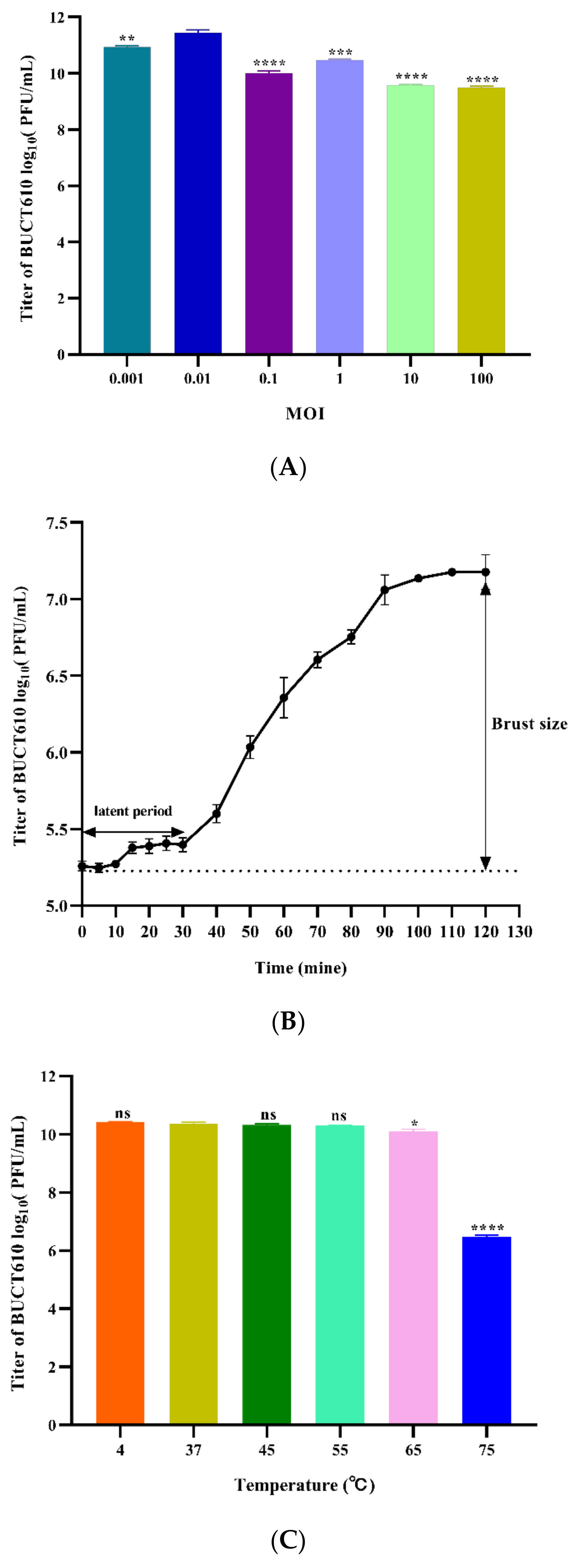
2.4. Physiological Characterization of Phage BUCT610
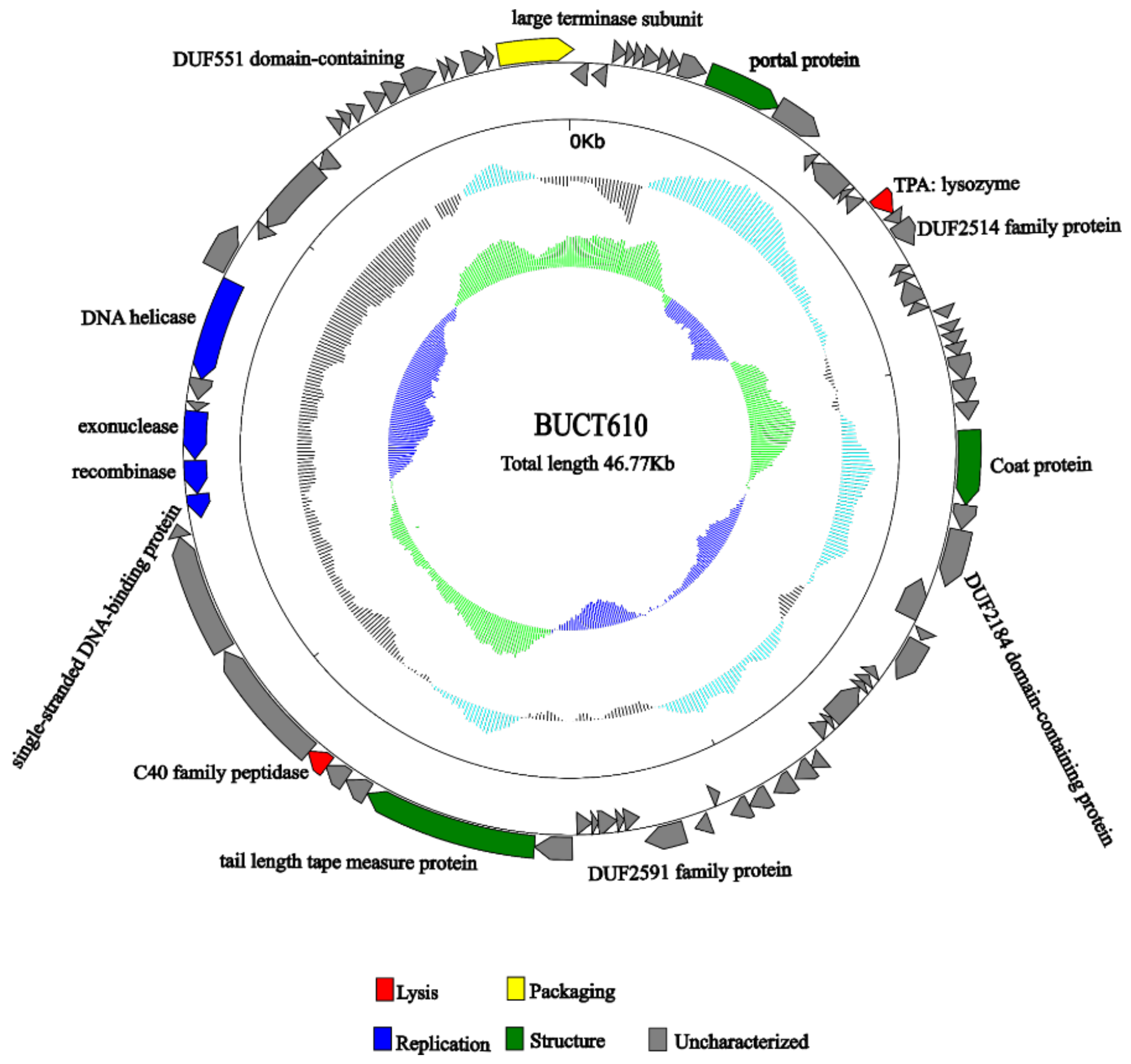
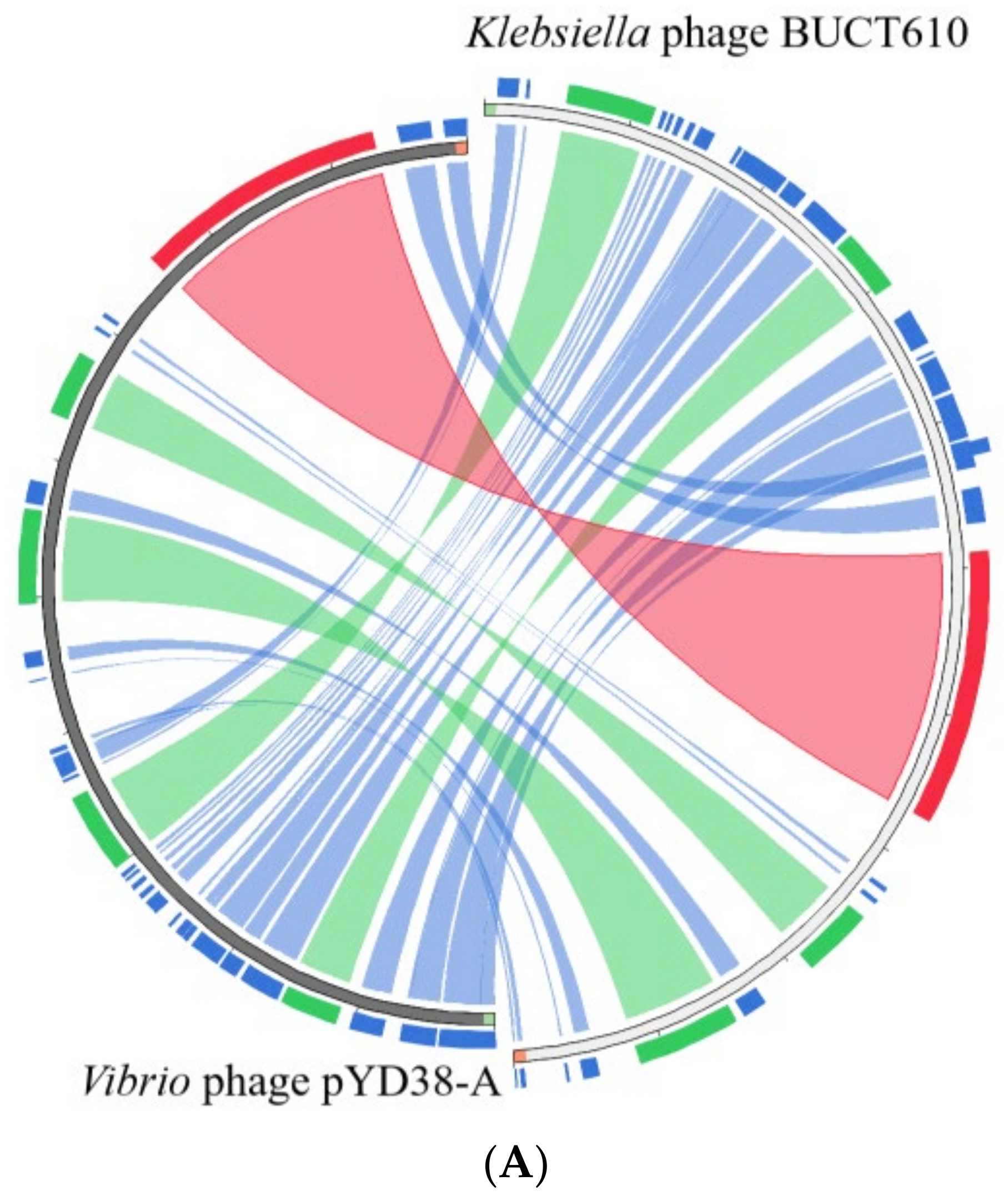
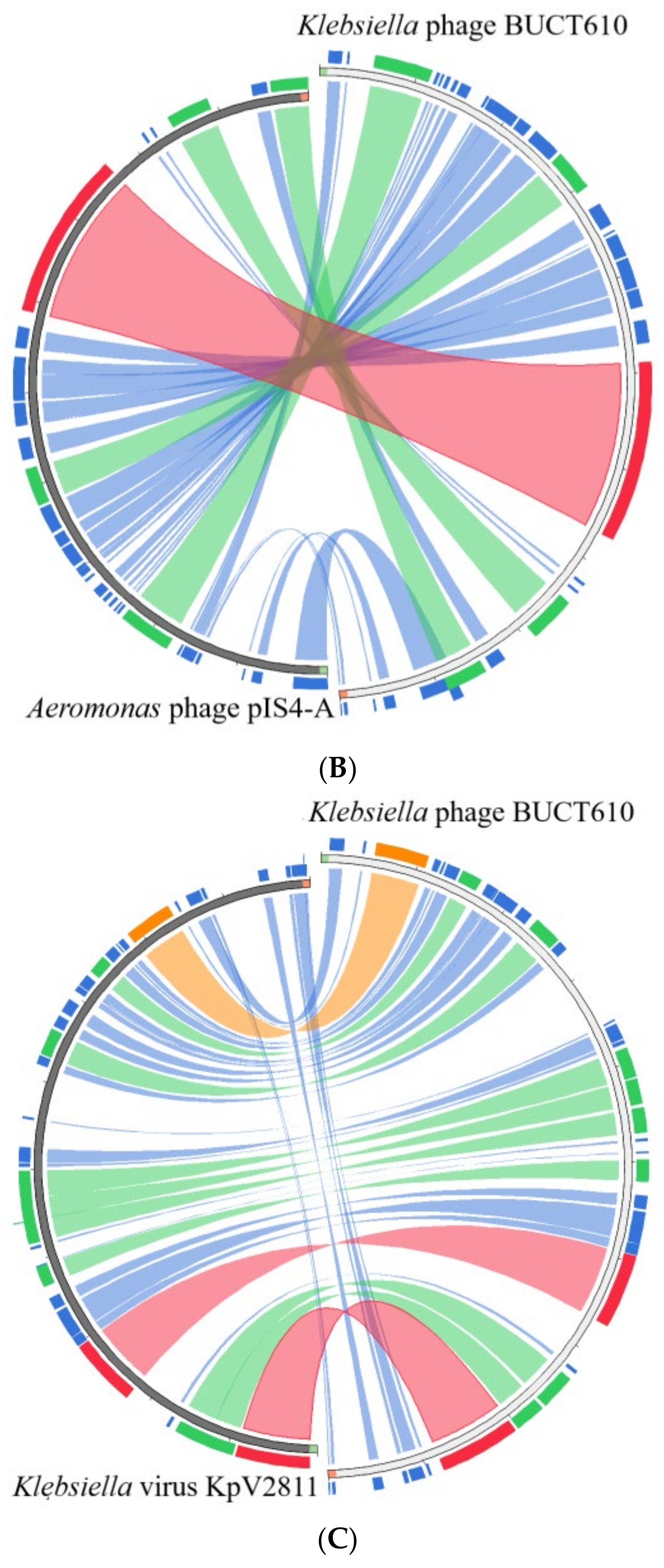
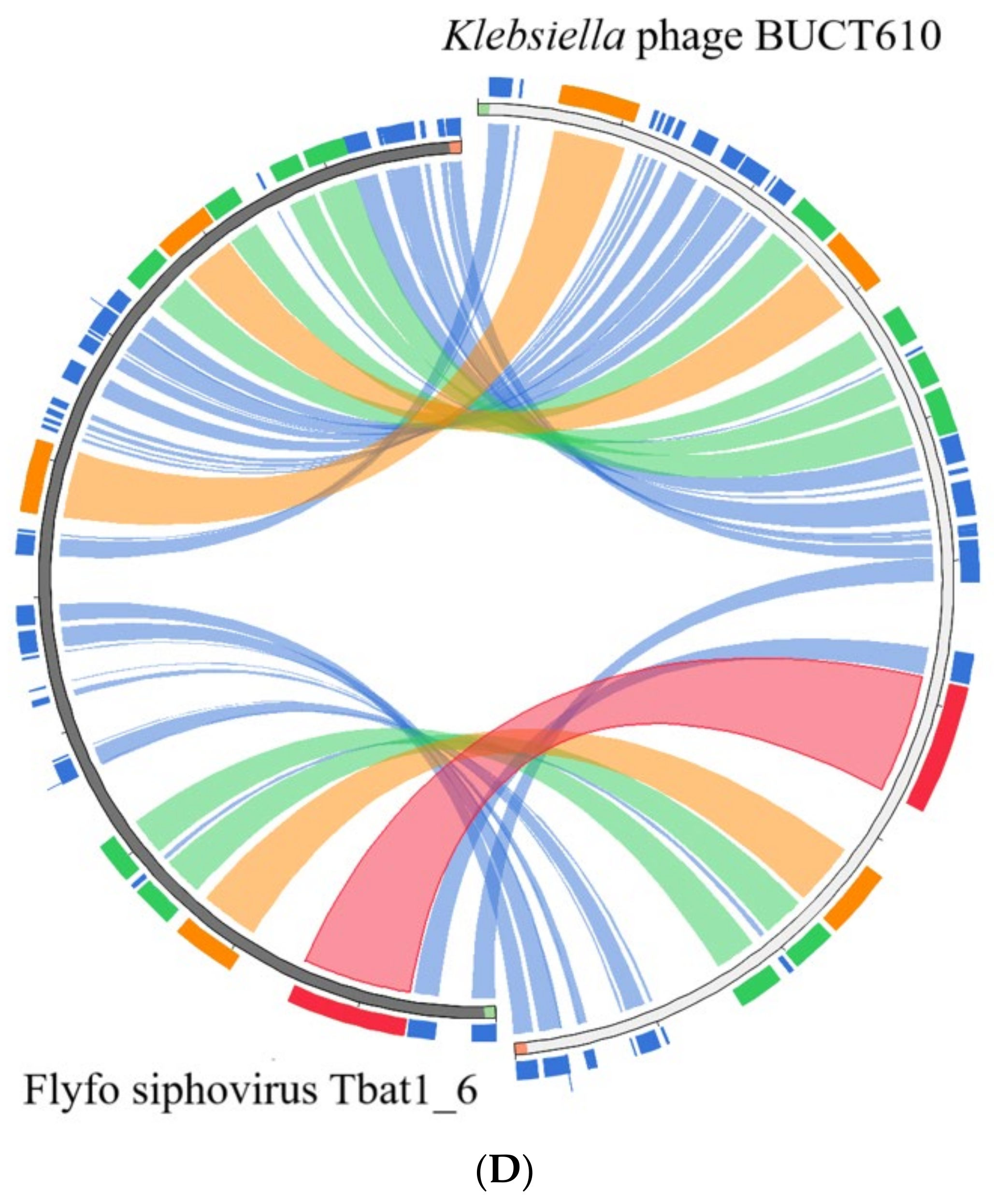
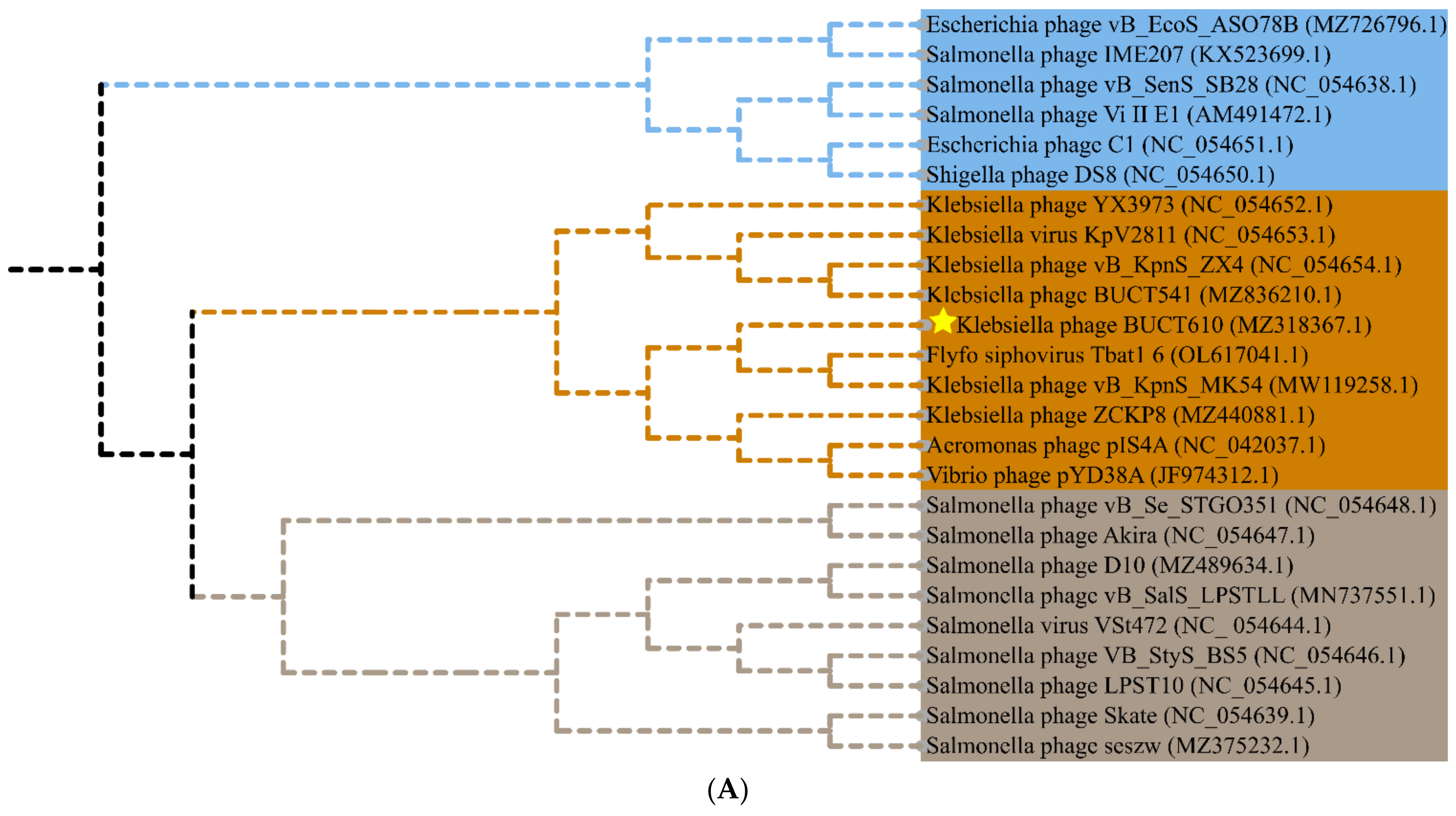
2.5. Annotations and Characteristics of the Complete Genome of BUCT610
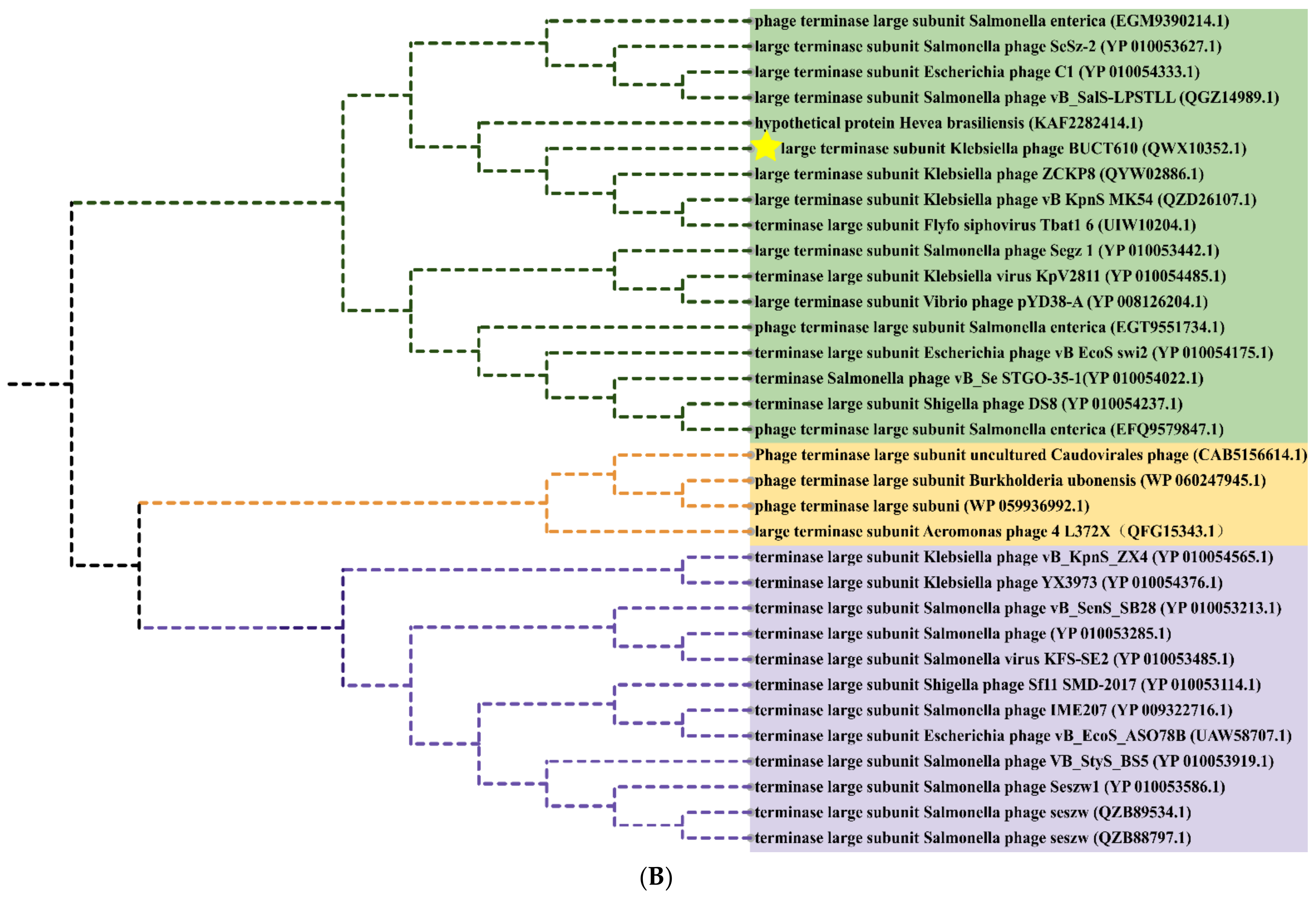
2.6. Replication-, Transcription-, and Structure-Related Genes of Phage BUCT610
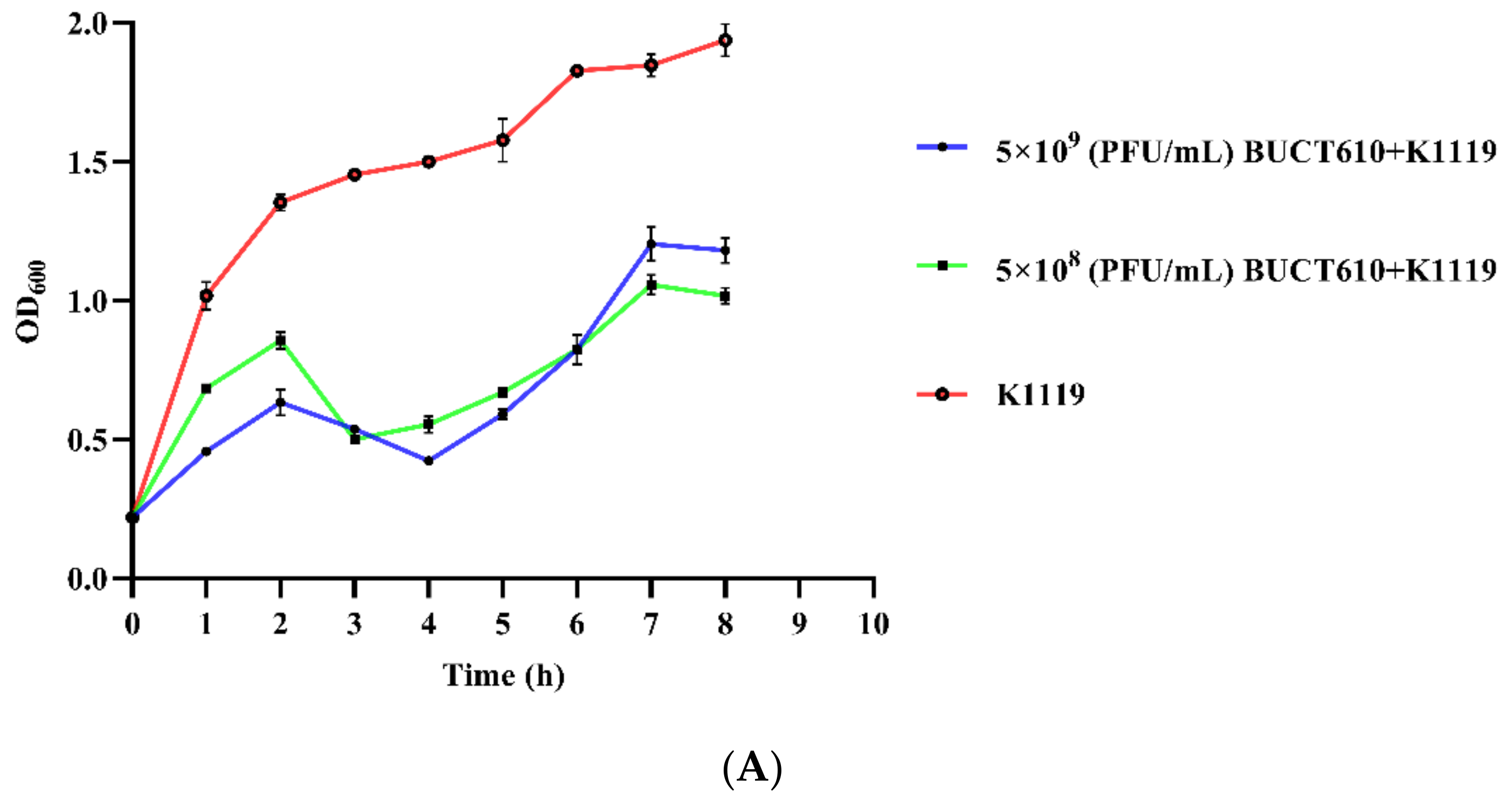
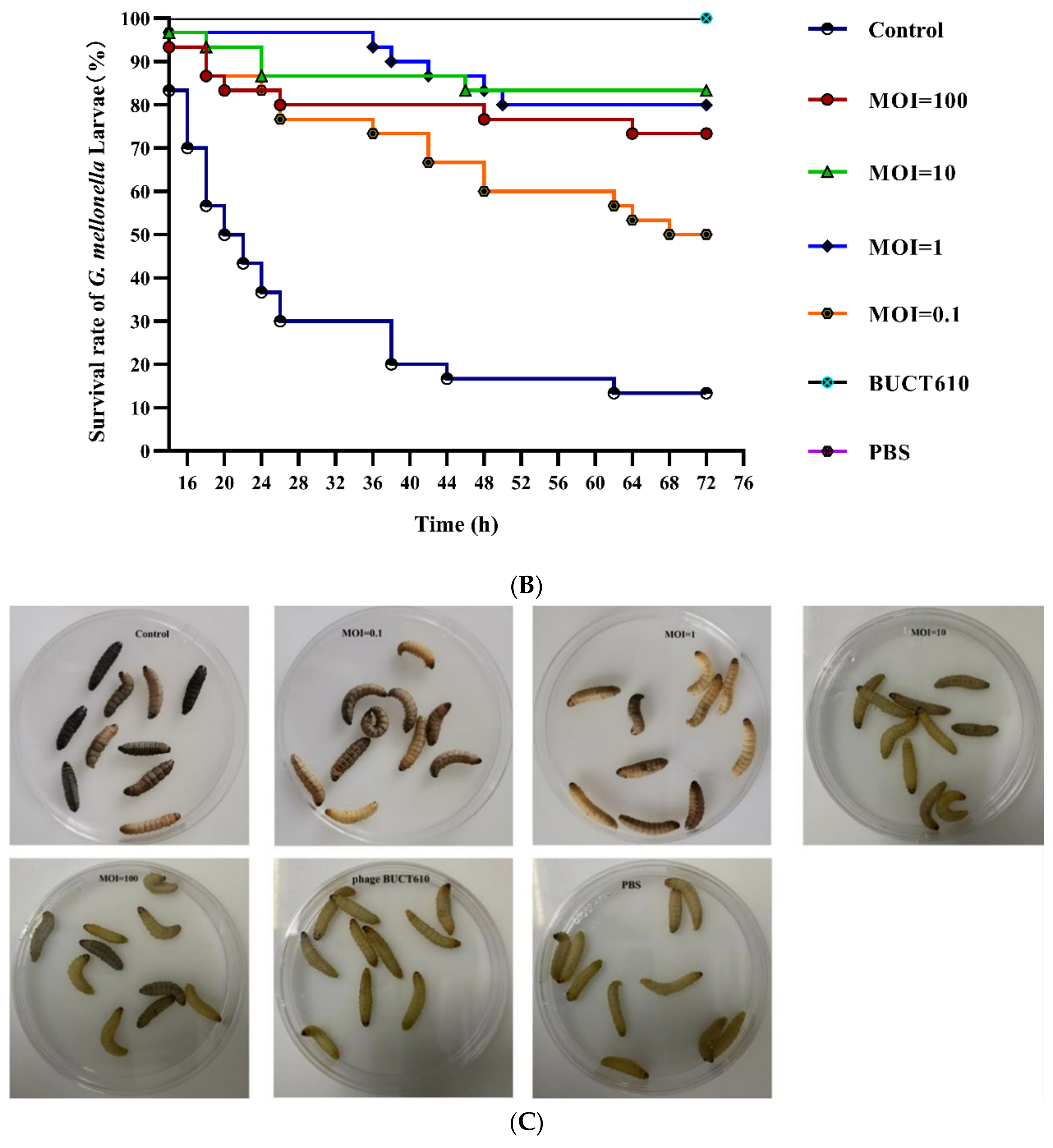
2.7. Assessment the Efficacy of BUCT610 against K1119 In Vitro and In Vivo
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain Cultures
4.2. Bacteriophage Isolation
4.3. Purification of Phage BUCT610
4.4. Electron Microscopy
4.5. Identified Lytic Range of Phage BUCT610
4.6. Multilocus Sequence Typing (MLST) and Capsule Type of K. pneumoniae
4.7. Lifestyle Identification of BUCT610
4.8. Optimal Multiplicity of Infection and One-Step Growth Curve of BUCT610
4.9. Stability Assessment of Phage BUCT610
4.10. Phage Sequencing and Bioinformatics Analysis
4.11. Assessment the Efficacy of BUCT610 against K. pneumoniae K1119 In Vitro
4.12. Therapeutic Effect of Phage BUCT610 in the G. mellonella Larvae
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gaiarsa, S.; Comandatore, F.; Gaibani, P.; Corbella, M.; Dalla Valle, C.; Epis, S.; Scaltriti, E.; Carretto, E.; Farina, C.; Labonia, M.; et al. Genomic epidemiology of Klebsiella pneumoniae in Italy and novel insights into the origin and global evolution of its resistance to carbapenem antibiotics. Antimicrob. Agents Chemother. 2015, 59, 389–396. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bengoechea, J.A.; Sa Pessoa, J. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019, 43, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and molecular perspectives. J. Intern. Med. 2020, 287, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef]
- Grillon, A.; Schramm, F.; Kleinberg, M.; Jehl, F. Comparative Activity of Ciprofloxacin, Levofloxacin and Moxifloxacin against Klebsiella pneumoniae, Pseudomonas aeruginosa and Stenotrophomonas maltophilia Assessed by Minimum Inhibitory Concentrations and Time-Kill Studies. PLoS ONE 2016, 11, e0156690. [Google Scholar] [CrossRef]
- Mikhail, S.; Singh, N.B.; Kebriaei, R.; Rice, S.A.; Stamper, K.C.; Castanheira, M.; Rybak, M.J. Evaluation of the Synergy of Ceftazidime-Avibactam in Combination with Meropenem, Amikacin, Aztreonam, Colistin, or Fosfomycin against Well-Characterized Multidrug-Resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e00779-19. [Google Scholar] [CrossRef]
- Motsch, J.; Murta de Oliveira, C.; Stus, V.; Köksal, I.; Lyulko, O.; Boucher, H.W.; Kaye, K.S.; File, T.M.; Brown, M.L.; Khan, I.; et al. RESTORE-IMI 1: A Multicenter, Randomized, Double-blind Trial Comparing Efficacy and Safety of Imipenem/Relebactam vs Colistin Plus Imipenem in Patients with Imipenem-nonsusceptible Bacterial Infections. Clin. Infect. Dis. 2020, 70, 1799–1808. [Google Scholar] [CrossRef]
- Zhen, X.; Stålsby Lundborg, C.; Sun, X.; Hu, X.; Dong, H. Clinical and Economic Impact of Third-Generation Cephalosporin-Resistant Infection or Colonization Caused by Escherichia coli and Klebsiella pneumoniae: A Multicenter Study in China. Int. J. Environ. Res. Public Health 2020, 17, 9285. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Łobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N.; et al. Phage Therapy: What Have We Learned. Viruses 2018, 10, 288. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J.; et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes. Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef]
- Cano, E.J.; Caflisch, K.M.; Bollyky, P.L.; Van Belleghem, J.D.; Patel, R.; Fackler, J.; Brownstein, M.J.; Horne, B.; Biswas, B.; Henry, M.; et al. Phage Therapy for Limb-threatening Prosthetic Knee Klebsiella pneumoniae Infection: Case Report and In Vitro Characterization of Anti-biofilm Activity. Clin. Infect. Dis. 2021, 73, e144–e151. [Google Scholar] [CrossRef]
- Eskenazi, A.; Lood, C.; Wubbolts, J.; Hites, M.; Balarjishvili, N.; Leshkasheli, L.; Askilashvili, L.; Kvachadze, L.; van Noort, V.; Wagemans, J.; et al. Combination of pre-adapted bacteriophage therapy and antibiotics for treatment of fracture-related infection due to pandrug-resistant Klebsiella pneumoniae. Nat. Commun. 2022, 13, 302. [Google Scholar] [CrossRef]
- Talebzadeh, H.; Mellali, H.; Solgi, H. Association of fluoroquinolone resistance and ESBL production in hypervirulent Klebsiella pneumoniae ST11 and ST893 in Iran. Acta Microbiol. Immunol. Hung. 2022, 135–143, online ahead of print. [Google Scholar] [CrossRef]
- Solgi, H.; Badmasti, F.; Giske, C.G.; Aghamohammad, S.; Shahcheraghi, F. Molecular epidemiology of NDM-1- and OXA-48-producing Klebsiella pneumoniae in an Iranian hospital: Clonal dissemination of ST11 and ST893. J. Antimicrob. Chemother. 2018, 73, 1517–1524. [Google Scholar] [CrossRef]
- Al-Anany, A.M.; Fatima, R.; Hynes, A.P. Temperate phage-antibiotic synergy eradicates bacteria through depletion of lysogens. Cell Rep. 2021, 35, 109172. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Smith, M. Phage-encoded Serine Integrases and Other Large Serine Recombinases. Microbiol. Spectr. 2015, 3, 253–272. [Google Scholar] [CrossRef]
- Murphy, K.C. Phage recombinases and their applications. Adv. Virus Res. 2012, 83, 367–414. [Google Scholar]
- Murphy, K.C. λ Recombination and Recombineering. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef]
- Keijzers, G.; Liu, D.; Rasmussen, L.J. Exonuclease 1 and its versatile roles in DNA repair. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 440–451. [Google Scholar] [CrossRef]
- Chongchai, A.; Waramit, S.; Wongwichai, T.; Kampangtip, J.; Phitak, T.; Kongtawelert, P.; Hajitou, A.; Suwan, K.; Pothacharoen, P. Targeting Human Osteoarthritic Chondrocytes with Ligand Directed Bacteriophage-Based Particles. Viruses 2021, 13, 2343. [Google Scholar] [CrossRef]
- Kumagai, I. Display of foreign protein on a coat protein of filamentous phage–phage antibody. Seikagaku J. Jpn. Biochem. Soc. 1993, 65, 1509–1512. [Google Scholar]
- Belcaid, M.; Bergeron, A.; Poisson, G. The evolution of the tape measure protein: Units, duplications and losses. BMC Bioinform. 2011, 12 (Suppl. S9), S10. [Google Scholar] [CrossRef]
- Mahony, J.; Alqarni, M.; Stockdale, S.; Spinelli, S.; Feyereisen, M.; Cambillau, C.; Sinderen, D.V. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci. Rep. 2016, 6, 36667. [Google Scholar] [CrossRef]
- Feiss, M.; Rao, V.B. The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 2012, 726, 489–509. [Google Scholar]
- Hwang, Y.; Feiss, M. Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage lambda terminase, inactivate the endonuclease activity of terminase. J. Mol. Biol. 1996, 261, 524–535. [Google Scholar] [CrossRef]
- Tomka, M.A.; Catalano, C.E. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage lambda. Biochemistry 1993, 32, 11992–11997. [Google Scholar] [CrossRef]
- Nemecek, D.; Gilcrease, E.B.; Kang, S.; Prevelige, P.E., Jr.; Casjens, S.; Thomas, G.J., Jr. Subunit conformations and assembly states of a DNA-translocating motor: The terminase of bacteriophage P22. J. Mol. Biol. 2007, 374, 817–836. [Google Scholar] [CrossRef]
- Gong, Q.; Wang, X.; Huang, H.; Sun, Y.; Qian, X.; Xue, F.; Ren, J.; Dai, J.; Tang, F. Novel Host Recognition Mechanism of the K1 Capsule-Specific Phage of Escherichia coli: Capsular Polysaccharide as the First Receptor and Lipopolysaccharide as the Secondary Receptor. J. Virol. 2021, 95, e0092021. [Google Scholar] [CrossRef]
- Chen, P.; Sun, H.; Ren, H.; Liu, W.; Li, G.; Zhang, C. LamB, OmpC, and the Core Lipopolysaccharide of Escherichia coli K-12 Function as Receptors of Bacteriophage Bp7. J. Virol. 2020, 94, e00325-20. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage Cocktail Development for Bacteriophage Therapy: Toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef] [PubMed]
- Mijbel Ali, B.; Gatea Kaabi, S.A.; Al-Bayati, M.A.; Musafer, H.K. A Novel Phage Cocktail Therapy of the Urinary Tract Infection in a Mouse Model. Arch. Razi Inst. 2021, 76, 1229–1236. [Google Scholar] [PubMed]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Jonge, P.A.; Meijenfeldt, F.; Rooijen, L.; Brouns, S.; Dutilh, B.E. Evolution of BACON Domain Tandem Repeats in crAssphage and Novel Gut Bacteriophage Lineages. Viruses 2019, 11, 1085. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B.N. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Han, K.; He, X.; Fan, H.; Song, L.; An, X.; Li, M.; Tong, Y. Characterization and genome analysis of a novel Stenotrophomonas maltophilia bacteriophage BUCT598 with extreme pH resistance. Virus Res. 2022, 314, 198751. [Google Scholar] [CrossRef]
- Jończyk-Matysiak, E.; Łodej, N.; Kula, D.; Owczarek, B.; Orwat, F.; Międzybrodzki, R.; Neuberg, J.; Bagińska, N.; Weber-Dąbrowska, B.; Górski, A. Factors determining phage stability/activity: Challenges in practical phage application. Expert. Rev. Anti Infect. Ther. 2019, 17, 583–606. [Google Scholar] [CrossRef]
- Kawahara, K. Variation, Modification and Engineering of Lipid A in Endotoxin of Gram-Negative Bacteria. Int. J. Mol. Sci. 2021, 22, 2281. [Google Scholar] [CrossRef]
- Wu, G.; Yi, Y.; Sun, J.; Li, M.; Qiu, L. No evidence for priming response in Galleria mellonella larvae exposed to toxin protein PirA2B2 from Photorhabdus luminescens TT01: An association with the inhibition of the host cellular immunity. Vaccine 2015, 33, 6307–6313. [Google Scholar] [CrossRef]
- Wu, G.; Yi, Y.; Lv, Y.; Li, M.; Wang, J.; Qiu, L. The lipopolysaccharide (LPS) of Photorhabdus luminescens TT01 can elicit dose- and time-dependent immune priming in Galleria mellonella larvae. J. Invertebr. Pathol. 2015, 127, 63–72. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Taha, O.A.; Connerton, P.L.; Connerton, I.F.; El-Shibiny, A. Bacteriophage ZCKP1: A Potential Treatment for Klebsiella pneumoniae Isolated from Diabetic Foot Patients. Front. Microbiol. 2018, 9, 2127. [Google Scholar] [CrossRef]
- Fangman, W.L. Separation of very large DNA molecules by gel electrophoresis. Nucleic Acids Res. 1978, 5, 653–665. [Google Scholar] [CrossRef]
- Serwer, P.; Wright, E.T. In-Gel Isolation and Characterization of Large (and Other) Phages. Viruses 2020, 12, 410. [Google Scholar] [CrossRef]
- Serwer, P.; Hunter, B.; Wright, E.T. Electron Microscopy of In-Plaque Phage T3 Assembly: Proposed Analogs of Neurodegenerative Disease Triggers. Pharmaceuticals 2020, 13, 18. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, R.; Hu, Y.; Liu, Y.; Wang, L.; An, X.; Song, L.; Shi, T.; Fan, H.; Tong, Y.; et al. Biological characteristics and genomic analysis of a Stenotrophomonas maltophilia phage vB_SmaS_BUCT548. Virus Genes. 2021, 57, 205–216. [Google Scholar] [CrossRef]
- Liu, Y.; Leung, S.; Huang, Y.; Guo, Y.; Jiang, N.; Li, P.; Chen, J.; Wang, R.; Bai, C.; Mi, Z.; et al. Identification of Two Depolymerases from Phage IME205 and Their Antivirulent Functions on K47 Capsule of Klebsiella pneumoniae. Front. Microbiol. 2020, 11, 218. [Google Scholar] [CrossRef]
- Sofy, A.R.; El-Dougdoug, N.K.; Refaey, E.E.; Dawoud, R.A.; Hmed, A.A. Characterization and Full Genome Sequence of Novel KPP-5 Lytic Phage against Klebsiella pneumoniae Responsible for Recalcitrant Infection. Biomedicines 2021, 9, 342. [Google Scholar] [CrossRef]
- Xing, S.; Zhang, X.; Sun, Q.; Wang, J.; Mi, Z.; Pei, G.; Huang, Y.; An, X.; Fu, K.; Zhou, L.; et al. Complete genome sequence of a novel, virulent Ahjdlikevirus bacteriophage that infects Enterococcus faecium. Arch. Virol. 2017, 162, 3843–3847. [Google Scholar] [CrossRef]
- Madaha, E.L.; Mienie, C.; Gonsu, H.K.; Bughe, R.N.; Fonkoua, M.C.; Mbacham, W.F.; Alayande, K.A.; Bezuidenhout, C.C.; Ateba, C.N. Whole-genome sequence of multi-drug resistant Pseudomonas aeruginosa strains UY1PSABAL and UY1PSABAL2 isolated from human broncho-alveolar lavage, Yaoundé, Cameroon. PLoS ONE 2020, 15, e0238390. [Google Scholar] [CrossRef]
- Xie, X.T.; Kropinski, A.M.; Tapscott, B.; Weese, J.S.; Turner, P.V. Prevalence of fecal viruses and bacteriophage in Canadian farmed mink (Neovison vison). Microbiologyopen 2019, 8, e00622. [Google Scholar] [CrossRef]
- Zoledowska, S.; Motyka-Pomagruk, A.; Sledz, W.; Mengoni, A.; Lojkowska, E. High genomic variability in the plant pathogenic bacterium Pectobacterium parmentieri deciphered from de novo assembled complete genomes. BMC Genom. 2018, 19, 751. [Google Scholar] [CrossRef]
- Marques-Bastos, S.; Coelho, M.; de Sousa Santos, I.N.; Farias, F.M.; Silva Francisco, M.; Albano, R.M.; Sales Alviano, C.; de Freire Bastos, M. Draft genome sequence of the producer strain of aureocin 4181, an antimicrobial peptide with antagonistic activity against multidrug-resistant staphylococci. J. Glob. Antimicrob. Resist. 2020, 23, 331–333. [Google Scholar] [CrossRef]
- Karunarathna, K.; Senathilake, N.; Mewan, K.M.; Weerasena, O.; Perera, S. In silico structural homology modelling of EST073 motif coding protein of tea Camellia sinensis (L.). J. Genet. Eng. Biotechnol. 2020, 18, 32. [Google Scholar] [CrossRef]
- Alam, T.I.; Rao, V.B. The ATPase domain of the large terminase protein, gp17, from bacteriophage T4 binds DNA: Implications to the DNA packaging mechanism. J. Mol. Biol. 2008, 376, 1272–1281. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Gupta, S.K.; Rolain, J.M. Reply to “Comparison of the web tools ARG-ANNOT and ResFinder for detection of resistance genes in bacteria”. Antimicrob. Agents Chemother. 2014, 58, 4987. [Google Scholar] [CrossRef][Green Version]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]

| Species | Strain | ST Type | Capsule Type | Origin | Sensitivity |
|---|---|---|---|---|---|
| K. pneumoniae | K1119 | ST893 | No match | 307 hospital | + |
| K. pneumoniae | K7 | ST23 | K1 | Aviation General Hospital | - |
| K. pneumoniae | 064 | ST23 | K1 | Aviation General Hospital | - |
| K. pneumoniae | 081 | ST23 | K1 | Aviation General Hospital | - |
| K. pneumoniae | 1241 | ST11 | K15 | 307 hospital | - |
| K. pneumoniae | 1291 | ST11 | K27 | 307 hospital | - |
| K. pneumoniae | 1299 | ST723 | N/A | 307 hospital | - |
| K. pneumoniae | 1300 | ST11 | K16 | 307 hospital | - |
| K. pneumoniae | 1301 | ST65 | K2 | 307 hospital | - |
| K. pneumoniae | 1307 | ST65 | K2 | 307 hospital | - |
| K. pneumoniae | 2012 | ST11 | K25 | Aviation General Hospital | - |
| K. pneumoniae | 2013 | ST15 | K19 | Aviation General Hospital | - |
| K. pneumoniae | 2015 | ST11 | K47 | Aviation General Hospital | - |
| K. pneumoniae | 2017 | ST15 | K19 | Aviation General Hospital | - |
| K. pneumoniae | 2019 | N/A | K47 | Aviation General Hospital | - |
| K. pneumoniae | 2021 | N/A | K64 | Aviation General Hospital | - |
| K. pneumoniae | 2068 | ST15 | K19 | Aviation General Hospital | - |
| K. pneumoniae | 2086 | ST11 | K64 | Aviation General Hospital | - |
| K. pneumoniae | 2752 | ST23 | K1 | Aviation General Hospital | - |
| K. pneumoniae | 2755 | ST23 | K1 | Aviation General Hospital | - |
| ORF | Strand | Start | Stop | Length (AA) | Putative Function | Best-Match BLASTp Result | Query Cover (%) | E-Values | Identity (%) | Accession | MW (kDa) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF1 | - | 352 | 20 | 110 | hypothetical protein | Klebsiella phage KMI7 | 98% | 7.00E-41 | 62.04% | QEG10317.1 | 13.14 |
| ORF2 | - | 729 | 439 | 96 | hypothetical protein | Klebsiella phage YX3973 | 100% | 2.00E-62 | 98.96% | YP_010054377.1 | 10.70 |
| ORF3 | + | 818 | 1057 | 79 | hypothetical protein | Klebsiella phage ZX4 | 100% | 3.00E-48 | 97.47% | YP_010054495.1 | 9.32 |
| ORF4 | + | 1054 | 1206 | 50 | hypothetical protein | Trabulsiella odontotermitis | 96% | 1.00E-06 | 50.00% | WP_158674504.1 | 5.67 |
| ORF5 | + | 1215 | 1394 | 59 | hypothetical protein | Salmonella phage SeSz-2 | 100% | 1.00E-14 | 61.02% | YP_010053632.1 | 6.84 |
| ORF6 | + | 1394 | 1690 | 98 | hypothetical protein | Pectinobacterium phage PEAT2 | 95% | 1.00E-37 | 63.83% | YP_009702196.1 | 11.68 |
| ORF7 | + | 1687 | 1875 | 62 | hypothetical protein | Escherichia coli | 100% | 4.00E-24 | 70.97% | MBB0589830.1 | 6.70 |
| ORF8 | + | 1875 | 2075 | 66 | hypothetical protein | Salmonella enterica subsp | 100% | 4.00E-07 | 34.85% | HAZ2983796.1 | 7.66 |
| ORF9 | + | 2085 | 2597 | 170 | hypothetical protein | Salmonella enterica | 97% | 6.00E-64 | 57.74% | EGT9551729.1 | 19.34 |
| ORF10 | + | 2646 | 4094 | 482 | portal protein | Klebsiella phage ZX4 | 100% | 0 | 97.30% | YP_010054497.1 | 54.12 |
| ORF11 | + | 4075 | 5019 | 314 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 0 | 97.13% | YP_010054477.1 | 35.56 |
| ORF12 | - | 5226 | 5032 | 64 | hypothetical protein | Klebsiella phage YX3973 | 100% | 5.00E-31 | 82.81% | YP_010054385.1 | 7.53 |
| ORF13 | - | 6049 | 5228 | 273 | hypothetical protein | Klebsiella phage YX3973 | 100% | 5.00E-91 | 56.12% | YP_010054386.1 | 30.50 |
| ORF14 | - | 6177 | 6046 | 43 | hypothetical protein | Klebsiella phage ZX4 | 100% | 3.00E-21 | 93.02% | YP_010054501.1 | 5.12 |
| ORF15 | - | 6467 | 6177 | 96 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 1.00E-57 | 94.79% | YP_010054472.1 | 9.95 |
| ORF16 | + | 6566 | 7003 | 145 | TPA: lysozyme | Salmonella enterica subsp | 100% | 3E-103 | 97.93% | HAZ2983869.1 | 15.70 |
| ORF17 | + | 7000 | 7245 | 81 | hypothetical protein | K. pneumoniae | 100% | 3.00E-51 | 100.00% | WP_142483241.1 | 9.06 |
| ORF18 | + | 7220 | 7717 | 165 | DUF2514 family protein | Klebsiella virus KpV2811 | 96% | 3.00E-91 | 93.71% | YP_010054469.1 | 18.13 |
| ORF19 | - | 8013 | 7864 | 49 | hypothetical protein | K. pneumoniae | 100% | 5.00E-25 | 95.92% | WP_185939371.1 | 5.64 |
| ORF20 | - | 8233 | 8021 | 70 | hypothetical protein | Klebsiella phage YX3973 | 100% | 1.00E-40 | 91.43% | YP_010054391.1 | 8.20 |
| ORF21 | - | 8700 | 8233 | 155 | hypothetical protein | Vibrio phage pYD38-A | 100% | 7.00E-91 | 86.16% | YP_008126189.1 | 17.50 |
| ORF22 | - | 8900 | 8700 | 66 | hypothetical protein | K. pneumoniae | 100% | 2.00E-40 | 100.00% | WP_142483236.1 | 8.02 |
| ORF23 | + | 8997 | 9212 | 71 | hypothetical protein | Klebsiella phage YX3973 | 100% | 2.00E-44 | 100.00% | YP_010054394.1 | 8.26 |
| ORF24 | + | 9296 | 9487 | 63 | hypothetical protein | K. pneumoniae | 100% | 1.00E-35 | 96.83% | WP_142483234.1 | 7.70 |
| ORF25 | + | 9484 | 9723 | 79 | hypothetical protein | Vibrio phage pYD38-A | 100% | 6.00E-35 | 89.87% | YP_008126184.1 | 9.62 |
| ORF26 | + | 9704 | 9946 | 80 | hypothetical protein | Salmonella phage SeSz-2 | 97% | 8.00E-28 | 66.67% | YP_010053654.1 | 8.97 |
| ORF27 | + | 9936 | 10,406 | 156 | hypothetical protein | Klebsiella phage YX3973 | 100% | 8E-113 | 100.00% | YP_010054398.1 | 17.67 |
| ORF28 | + | 10,394 | 10,819 | 141 | hypothetical protein | Shigella phage Sf11 SMD-2017 | 55% | 1.00E-39 | 80.77% | YP_010053088.1 | 16.52 |
| ORF29 | + | 10,816 | 11,160 | 167 | hypothetical protein | Klebsiella phage YX3973 | 68% | 8.00E-74 | 93.86% | YP_010054400.1 | 13.05 |
| ORF30 | + | 11,343 | 12,746 | 467 | hypothetical protein | Klebsiella phage YX3973 | 100% | 0 | 96.15% | YP_010054401.1 | 51.39 |
| ORF31 | + | 12,746 | 13,210 | 207 | hypothetical protein | Vibrio phage pYD38-A | 74% | 7E-103 | 97.40% | YP_008126178.1 | 15.64 |
| ORF32 | + | 13,222 | 14,310 | 362 | DUF2184 domain containing protein | K. pneumoniae | 100% | 0 | 99.45% | WP_142483227.1 | 40.05 |
| ORF33 | - | 15,123 | 14,326 | 265 | hypothetical protein | Salmonella enterica | 100% | 5E-116 | 60.97% | ECD2278621.1 | 31.84 |
| ORF34 | + | 15,215 | 15,409 | 64 | hypothetical protein | Vibrio phage pYD38-A | 100% | 6.00E-37 | 98.44% | YP_008126175.1 | 7.11 |
| ORF35 | + | 15,453 | 16,271 | 272 | hypothetical protein | K. pneumoniae | 94% | 1E-180 | 97.66% | WP_142483225.1 | 28.65 |
| ORF36 | - | 16,483 | 16,301 | 60 | hypothetical protein | Salmonella phage IME207 | 100% | 2.00E-28 | 83.33% | YP_009322759.1 | 6.76 |
| ORF37 | - | 16,682 | 16,509 | 57 | hypothetical protein | K. pneumoniae | 100% | 4.00E-33 | 96.49% | WP_185939376.1 | 6.71 |
| ORF38 | - | 16,819 | 16,682 | 45 | hypothetical protein | Bacteriophage sp. | 97% | 3.00E-15 | 68.18% | QHJ81118.1 | 5.27 |
| ORF39 | - | 17,634 | 16,819 | 271 | hypothetical protein | K. pneumoniae | 100% | 0 | 98.52% | WP_142483222.1 | 29.94 |
| ORF40 | - | 17,758 | 17,642 | 38 | hypothetical protein | Klebsiella phage ZX4 | 100% | 5.00E-14 | 92.11% | YP_010054525.1 | 4.42 |
| ORF41 | - | 18,048 | 17,755 | 97 | hypothetical protein | Vibrio phage pYD38-A | 100% | 6.00E-22 | 53.61% | YP_008126169.1 | 10.02 |
| ORF42 | + | 18,279 | 18,566 | 95 | hypothetical protein | Klebsiella phage YX3973 | 100% | 5.00E-62 | 100.00% | YP_010054411.1 | 10.16 |
| ORF43 | + | 18,544 | 18,936 | 130 | hypothetical protein | Klebsiella phage YX3973 | 100% | 9.00E-89 | 97.69% | YP_010054412.1 | 13.88 |
| ORF44 | + | 18,974 | 19,399 | 141 | hypothetical protein | Vibrio phage pYD38-A | 80% | 8.00E-79 | 100.00% | YP_008126166.1 | 15.18 |
| ORF45 | + | 19,504 | 19,920 | 138 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 5.00E-94 | 98.55% | YP_010054440.1 | 14.99 |
| ORF46 | + | 19,917 | 20,300 | 127 | hypothetical protein | Vibrio phage pYD38-A | 100% | 6.00E-89 | 100.00% | YP_008126240.1 | 14.47 |
| ORF47 | - | 20,509 | 20,324 | 61 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 1.00E-35 | 95.08% | YP_010054438.1 | 7.06 |
| ORF48 | + | 20,713 | 21,015 | 100 | hypothetical protein | Pectobacterium polonicum | 39% | 9.00E-09 | 82.05% | WP_137741933.1 | 11.10 |
| ORF49 | + | 21,219 | 21,974 | 251 | hypothetical protein | K. pneumoniae | 99% | 2e-178 | 99.60% | WP_142483279.1 | 26.54 |
| ORF50 | - | 22,298 | 21,996 | 100 | hypothetical protein | Salmonella enterica | 100% | 5.00E-55 | 81.00% | EAQ4012616.1 | 12.12 |
| ORF51 | - | 22,450 | 22,298 | 50 | hypothetical protein | Klebsiella phage ZX4 | 100% | 1.00E-25 | 94.00% | YP_010054538.1 | 5.74 |
| ORF52 | - | 22,817 | 22,440 | 125 | DUF2591 family protein | K. pneumoniae | 96% | 2.00E-44 | 63.11% | WP_116961239.1 | 13.94 |
| ORF53 | - | 22,948 | 22,814 | 44 | hypothetical protein | Klebsiella phage ZX4 | 95% | 3.00E-17 | 88.10% | YP_010054539.1 | 5.15 |
| ORF54 | - | 23,244 | 22,945 | 99 | hypothetical protein | Vibrio phage pYD38-A | 100% | 4.00E-60 | 92.93% | YP_008126235.1 | 10.99 |
| ORF55 | + | 23,338 | 24,045 | 235 | hypothetical protein | Klebsiella phage ZX4 | 100% | 7E-171 | 97.45% | YP_010054540.1 | 26.55 |
| ORF56 | + | 24,045 | 27,338 | 1097 | tail length tape measure protein | Vibrio phage pYD38-A | 100% | 0 | 96.08% | YP_008126233.1 | 114.54 |
| ORF57 | + | 27,338 | 27,808 | 156 | hypothetical protein | K. pneumoniae | 100% | 3E-112 | 100.00% | WP_142483272.1 | 17.99 |
| ORF58 | + | 27,808 | 28,278 | 156 | hypothetical protein | Klebsiella phage YX3973 | 100% | 1E-109 | 98.72% | YP_010054357.1 | 17.93 |
| ORF59 | + | 28,241 | 28,699 | 152 | C40 family peptidase | K. pneumoniae | 100% | 2E-107 | 98.03% | WP_142483270.1 | 17.40 |
| ORF60 | + | 28,671 | 31,127 | 818 | hypothetical protein | Vibrio phage pYD38-A | 100% | 0 | 99.02% | YP_008126229.1 | 89.86 |
| ORF61 | + | 31,166 | 33,430 | 754 | hypothetical protein | K. pneumoniae | 95% | 0 | 66.02% | WP_191974704.1 | 80.67 |
| ORF62 | + | 33,427 | 33,672 | 81 | hypothetical protein | K. pneumoniae | 96% | 8.00E-19 | 47.06% | WP_211775938.1 | 9.06 |
| ORF63 | - | 34,175 | 33,702 | 157 | single-strandedDNA-binding protein | Vibrio phage pYD38-A | 100% | 9.00E-48 | 52.10% | YP_008126226.1 | 17.65 |
| ORF64 | - | 34,844 | 34,185 | 219 | recombinase | Klebsiella phage ZX4 | 100% | 1E-154 | 98.17% | YP_010054548.1 | 23.76 |
| ORF65 | - | 35,824 | 34,868 | 318 | exonuclease | Klebsiella virus KpV2811 | 100% | 0 | 97.80% | YP_010054422.1 | 36.00 |
| ORF66 | - | 36,012 | 35,824 | 62 | hypothetical protein | Vibrio phage pYD38-A | 100% | 4.00E-37 | 98.39% | YP_008126223.1 | 7.13 |
| ORF67 | - | 36,475 | 36,059 | 138 | hypothetical protein | Vibrio phage pYD38-A | 100% | 5.00E-95 | 94.93% | YP_008126222.1 | 15.74 |
| ORF68 | - | 38,504 | 36,462 | 680 | DNA helicase | Klebsiella virus KpV2811 | 100% | 0 | 95.15% | YP_010054420.1 | 76.95 |
| ORF69 | + | 38,579 | 39,472 | 297 | hypothetical protein | K. pneumoniae | 100% | 0 | 93.94% | WP_142483294.1 | 33.36 |
| ORF70 | - | 39,783 | 39,487 | 98 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 7.00E-62 | 90.82% | YP_010054418.1 | 11.46 |
| ORF71 | - | 41,336 | 39,783 | 517 | hypothetical protein | Klebsiella phage ZX4 | 100% | 0 | 99.81% | YP_010054556.1 | 57.52 |
| ORF72 | - | 41,707 | 41,333 | 124 | hypothetical protein | Aeromonas phage pIS4-A | 87% | 6.00E-59 | 87.04% | YP_009614674.1 | 13.87 |
| ORF73 | + | 42,064 | 42,285 | 73 | hypothetical protein | Vibrio phage pYD38-A | 100% | 4.00E-40 | 90.41% | YP_008126215.1 | 8.05 |
| ORF74 | + | 42,282 | 42,467 | 61 | hypothetical protein | Vibrio phage pYD38-A | 100% | 3.00E-36 | 96.72% | YP_008126214.1 | 6.76 |
| ORF75 | + | 42,495 | 42,761 | 88 | hypothetical protein | Klebsiella virus KpV2811 | 100% | 4.00E-55 | 98.86% | YP_010054492.1 | 10.03 |
| ORF76 | + | 42,873 | 43,220 | 115 | hypothetical protein | Klebsiella | 99% | 9.00E-50 | 74.56% | WP_004151294.1 | 12.92 |
| ORF77 | + | 43,217 | 43,615 | 132 | hypothetical protein | K. pneumoniae | 100% | 2.00E-94 | 99.24% | WP_117033037.1 | 15.28 |
| ORF78 | + | 43,618 | 44,235 | 205 | DUF551 domain-containing | Escherichia coli | 59% | 2.00E-60 | 81.60% | EFM1806727.1 | 22.84 |
| ORF79 | + | 44,328 | 44,486 | 52 | hypothetical protein | Klebsiella phage YX3973 | 100% | 9.00E-29 | 92.31% | YP_010054372.1 | 6.06 |
| ORF80 | + | 44,486 | 44,680 | 64 | hypothetical protein | Klebsiella phage YX3973 | 100% | 4.00E-38 | 95.31% | YP_010054373.1 | 7.10 |
| ORF81 | + | 44,784 | 45,203 | 139 | hypothetical protein | K. pneumoniae | 99% | 5.00E-92 | 99.28% | WP_142483254.1 | 15.55 |
| ORF82 | + | 45,196 | 45,351 | 51 | hypothetical protein | NO hit | - | - | - | - | 5.58 |
| ORF83 | + | 45,341 | 46,762 | 473 | large terminase subunit | Salmonella phage SeSz-2 | 100% | 0 | 89.64% | YP_010053627.1 | 54.99 |
| PCR Primers Information | |
|---|---|
| Gene Name | Sequence |
| rpoB | F: Vic3oF: GTTTTCCCAGTCACGACGTTGTAGGCGAAATGGCWGAGAACCA R: Vic2oR: TTGTGAGCGGATAACAATTTCGAGTCTTCGAAGTTGTAACC |
| gapA | F: gapA173oF: GTTTTCCCAGTCACGACGTTGTATGAAATATGACTCCACTCACGG R: gapA181oR: TTGTGAGCGGATAACAATTTCCTTCAGAAGCGGCTTTGATGGCTT |
| mdh | F: mdh130oF: GTTTTCCCAGTCACGACGTTGTA CCCAACTCGCTTCAGGTTCAG R: mdh867oR: TTGTGAGCGGATAACAATTTCCCGTTTTTCCCCAGCAGCAG |
| pgi | F: pgi1FoF: GTTTTCCCAGTCACGACGTTGTAGAGAAAAACCTGCCTGTACTGCTGGC R: pgi1RoR: TTGTGAGCGGATAACAATTTCCGCGCCACGCTTTATAGCGGTTAAT |
| phoE | F: phoE604.1oF: GTTTTCCCAGTCACGACGTTGTAACCTACCGCAACACCGACTTCTTCGG R: phoE604.2oR: TTGTGAGCGGATAACAATTTCTGATCAGAACTGGTAGGTGAT |
| infB | F: infB1FoF: GTTTTCCCAGTCACGACGTTGTACTCGCTGCTGGACTATATTCG R: infB1RoR: TTGTGAGCGGATAACAATTTC CGCTTTCAGCTCAAGAACTTC |
| tonB | F: tonB1FoF: GTTTTCCCAGTCACGACGTTGTACTTTATACCTCGGTACATCAGGTT R: tonB2RoR: TTGTGAGCGGATAACAATTTCATTCGCCGGCTGRGCRGAGAG |
| universal sequencing primers | F: GTTTTCCCAGTCACGACGTTGTA R: TTGTGAGCGGATAACAATTTC |
| Wzi | F: GTG CCGCGAGCGCTT TCTATCTTGGTATTCC R: GAGAGCCACTGGTTCCAGAATTACCGC |
| qPCR primers information | |
| recombinase | F: CCGCTACTCTATTGCGTCTATG R: GTTTGCTGCTGAACGAATGAG |
| conserved hypothetical protein | F: CCCGGTAATGCGTCAGAATA R: TGCCGGATGGTCTGTAATTT |
| large terminase subunit | F: GTCGCAATGGTTGAGGTTTATG R: TGCCGGATGGTCTGTAATTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, M.; Han, P.; Zhang, G.; Liu, Y.; Li, Y.; Li, F.; Li, M.; An, X.; Song, L.; Chen, Y.; et al. Characterization and Comparative Genomics Analysis of a New Bacteriophage BUCT610 against Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae. Int. J. Mol. Sci. 2022, 23, 8040. https://doi.org/10.3390/ijms23148040
Pu M, Han P, Zhang G, Liu Y, Li Y, Li F, Li M, An X, Song L, Chen Y, et al. Characterization and Comparative Genomics Analysis of a New Bacteriophage BUCT610 against Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae. International Journal of Molecular Sciences. 2022; 23(14):8040. https://doi.org/10.3390/ijms23148040
Chicago/Turabian StylePu, Mingfang, Pengjun Han, Guangye Zhang, Yucong Liu, Yahao Li, Fei Li, Mengzhe Li, Xiaoping An, Lihua Song, Yiming Chen, and et al. 2022. "Characterization and Comparative Genomics Analysis of a New Bacteriophage BUCT610 against Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae" International Journal of Molecular Sciences 23, no. 14: 8040. https://doi.org/10.3390/ijms23148040
APA StylePu, M., Han, P., Zhang, G., Liu, Y., Li, Y., Li, F., Li, M., An, X., Song, L., Chen, Y., Fan, H., & Tong, Y. (2022). Characterization and Comparative Genomics Analysis of a New Bacteriophage BUCT610 against Klebsiella pneumoniae and Efficacy Assessment in Galleria mellonella Larvae. International Journal of Molecular Sciences, 23(14), 8040. https://doi.org/10.3390/ijms23148040






