Isolated Variable Domains of an Antibody Can Assemble on Blood Coagulation Factor VIII into a Functional Fv-like Complex
Abstract
1. Introduction
2. Results
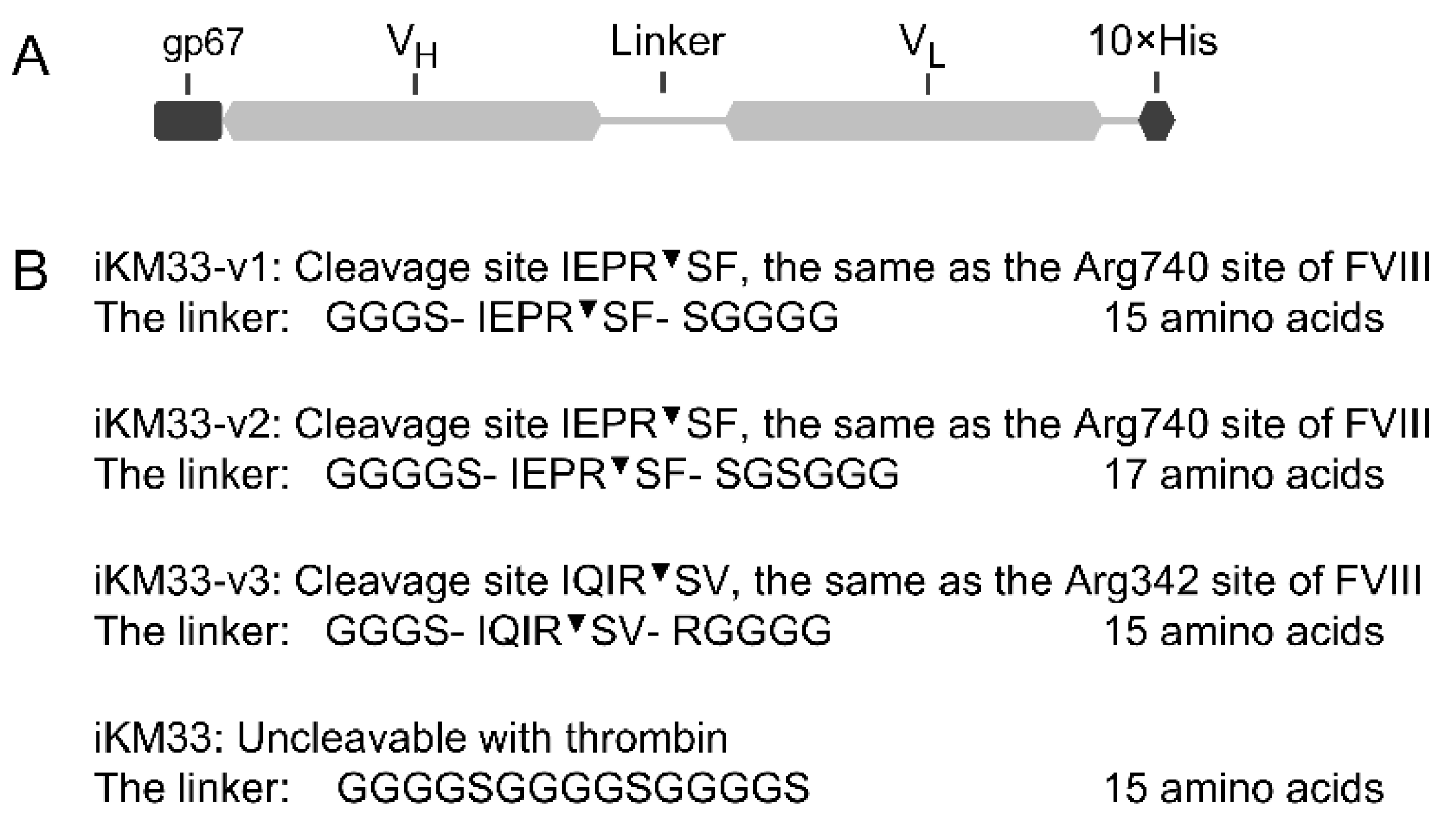
2.1. Design and Expression of iKM33 Variants with Thrombin Cleavage Site
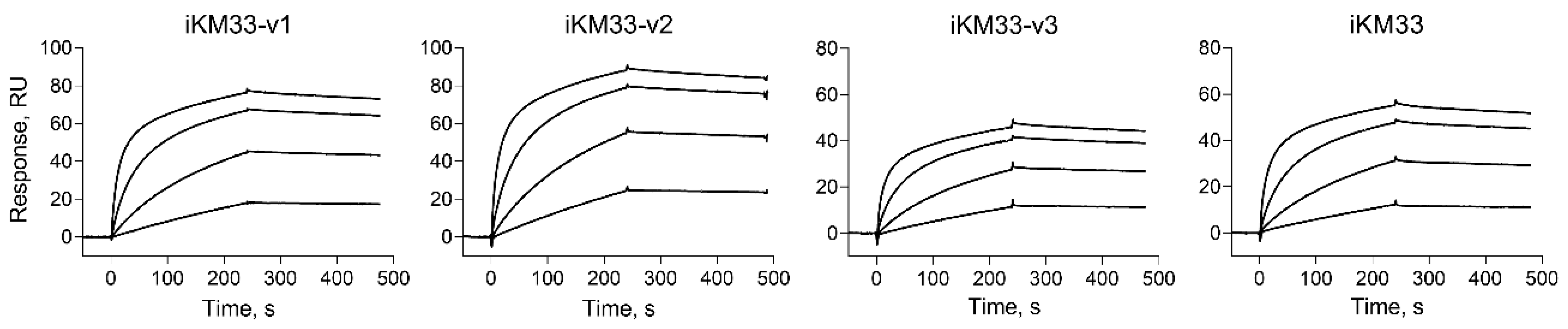
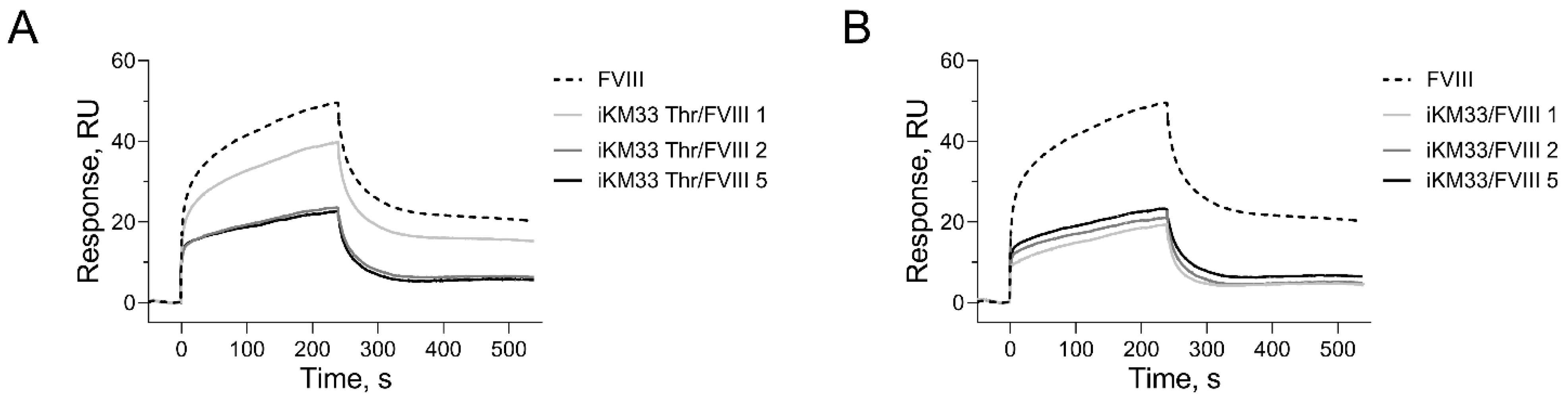
2.2. Testing iKM33 Variants for Binding with FVIII
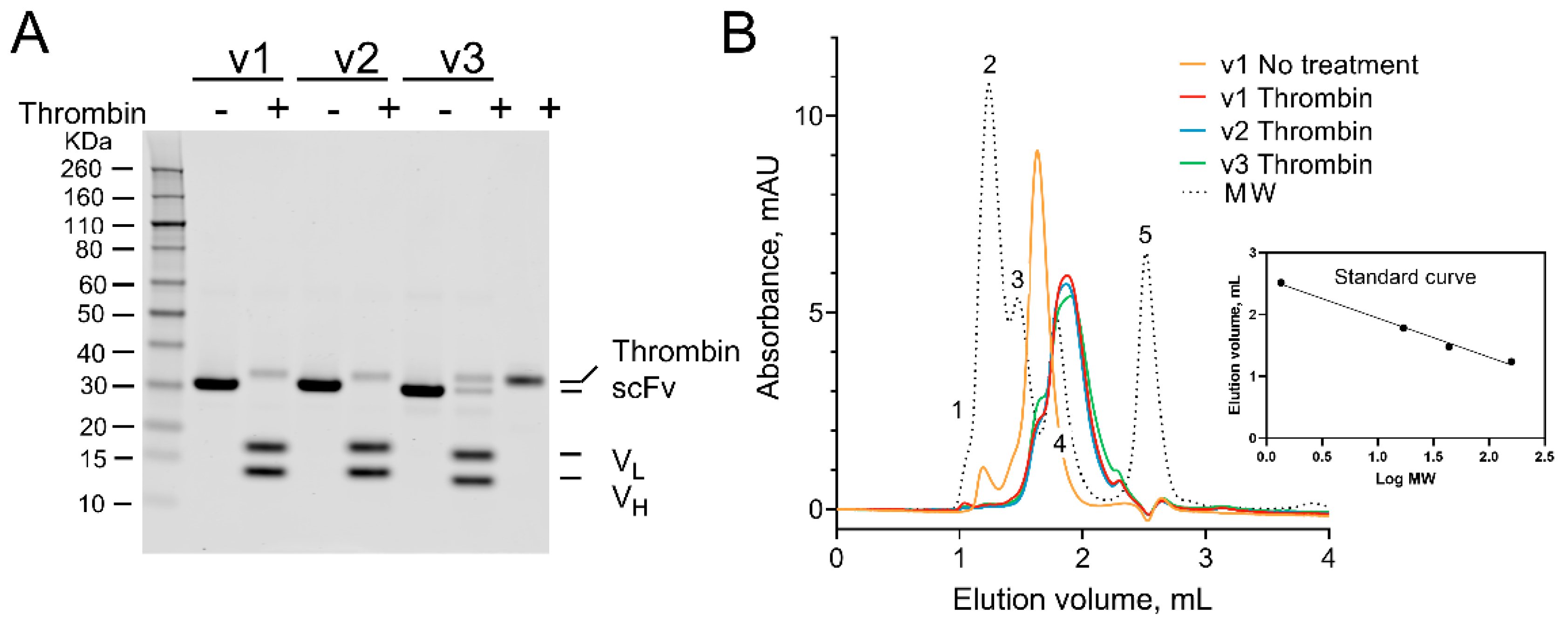
2.3. Testing iKM33 Variants for Thrombin Cleavage
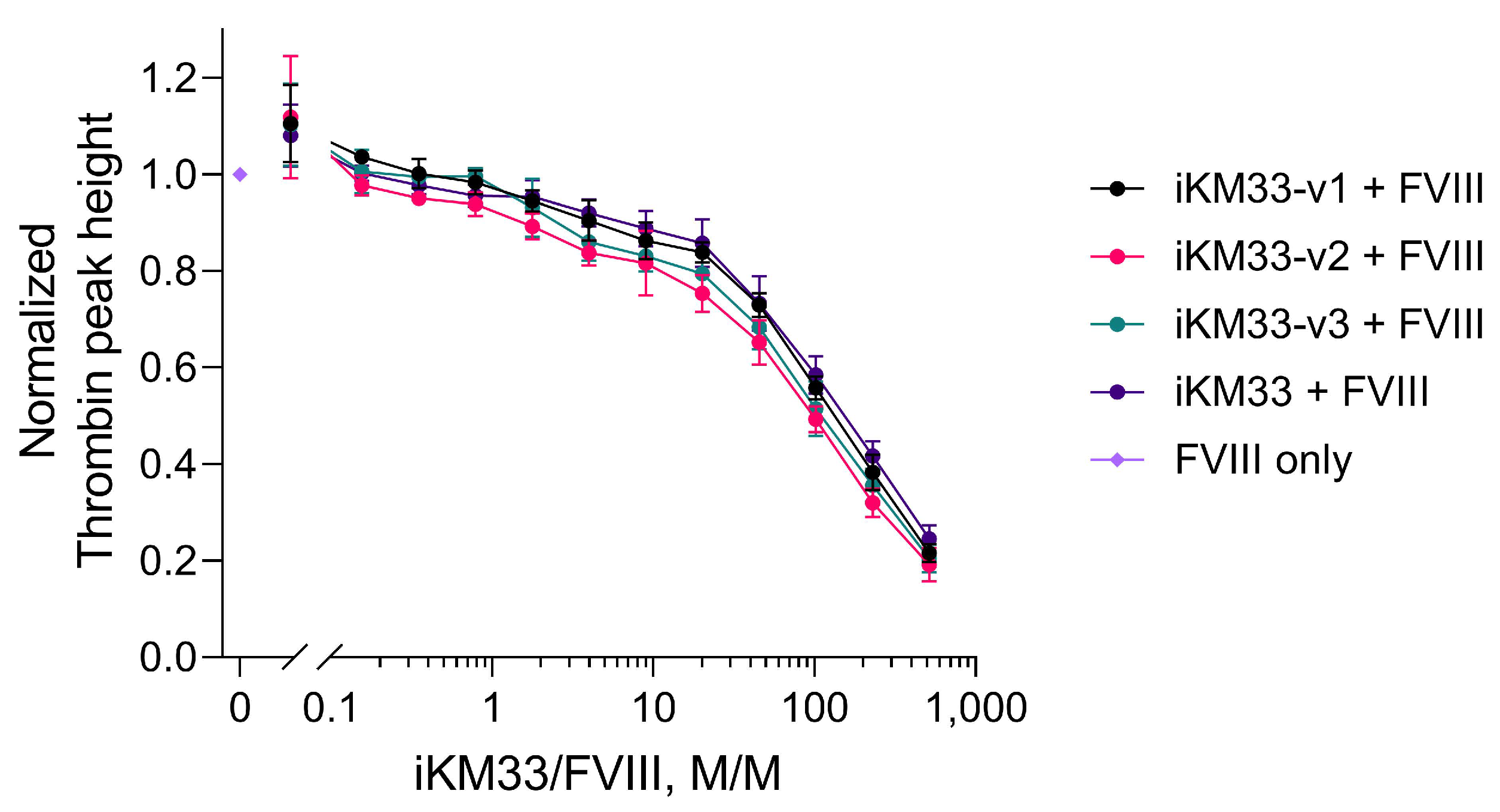
2.4. Effect of iKM33 Variants on FVIII Activity
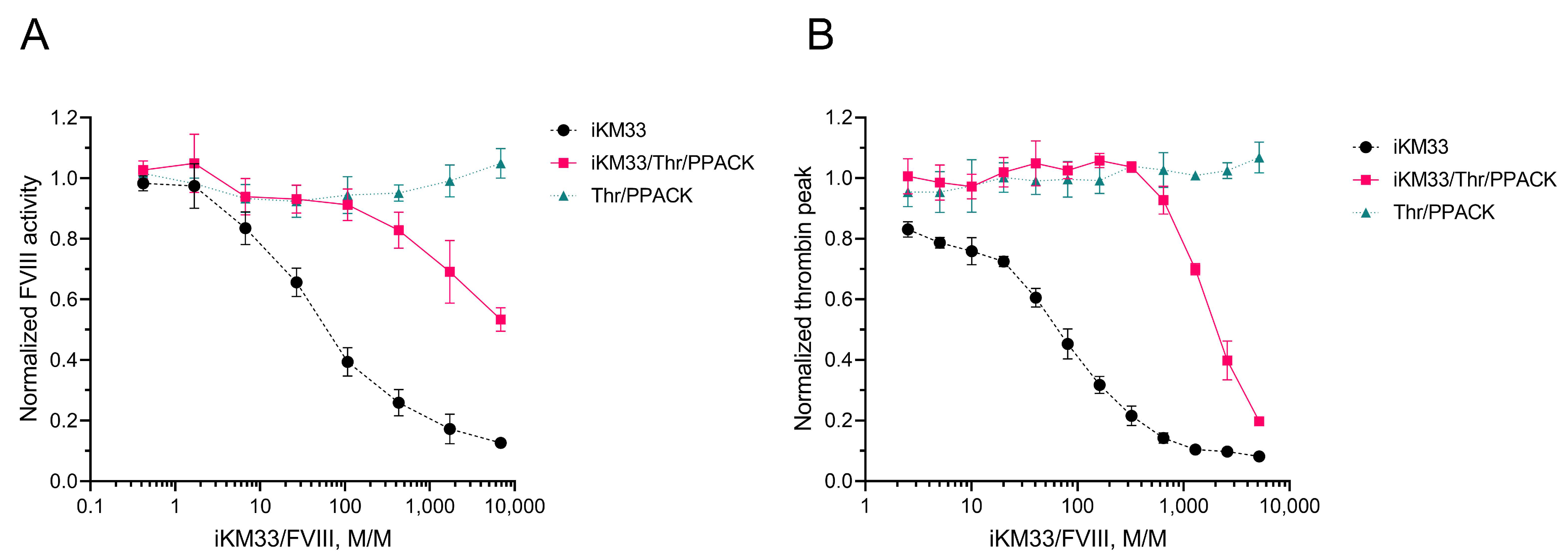
2.5. Effect of Thrombin-Cleaved iKM33-v2 on FVIII Activity
2.6. Effect of Thrombin-Cleaved iKM33-v2 on Binding FVIII to LRP1
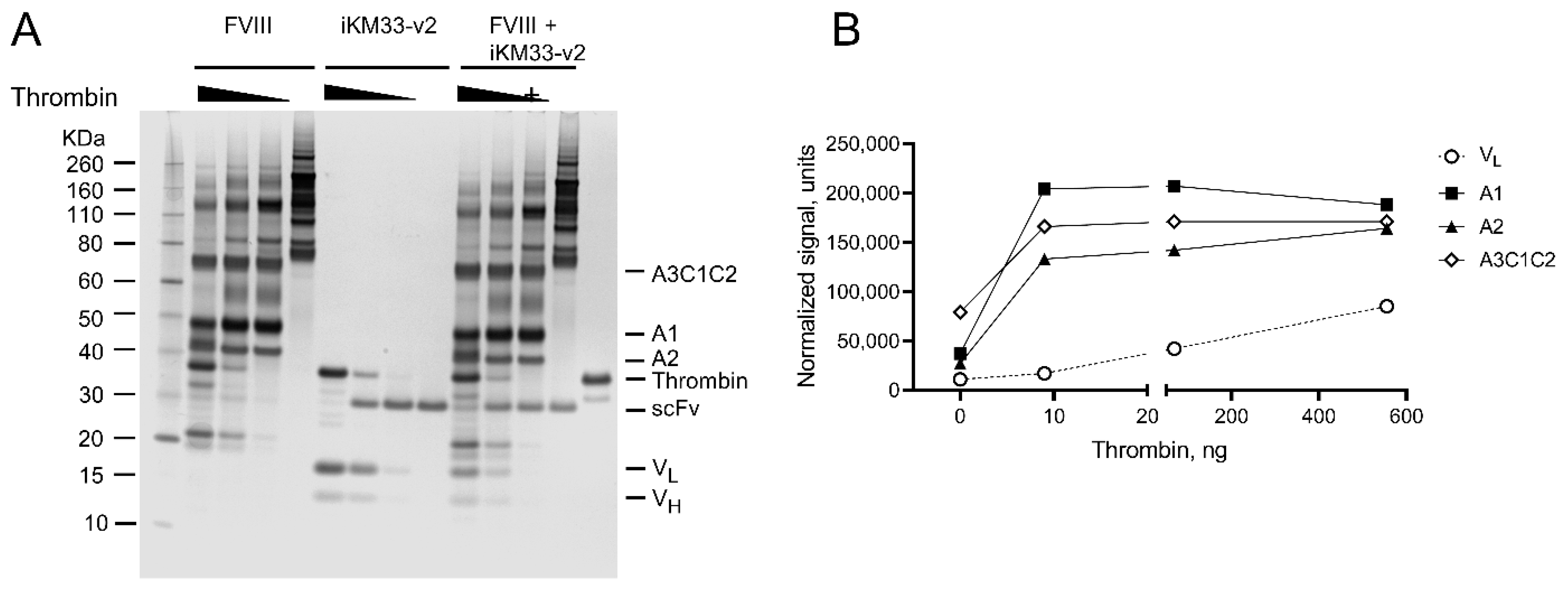
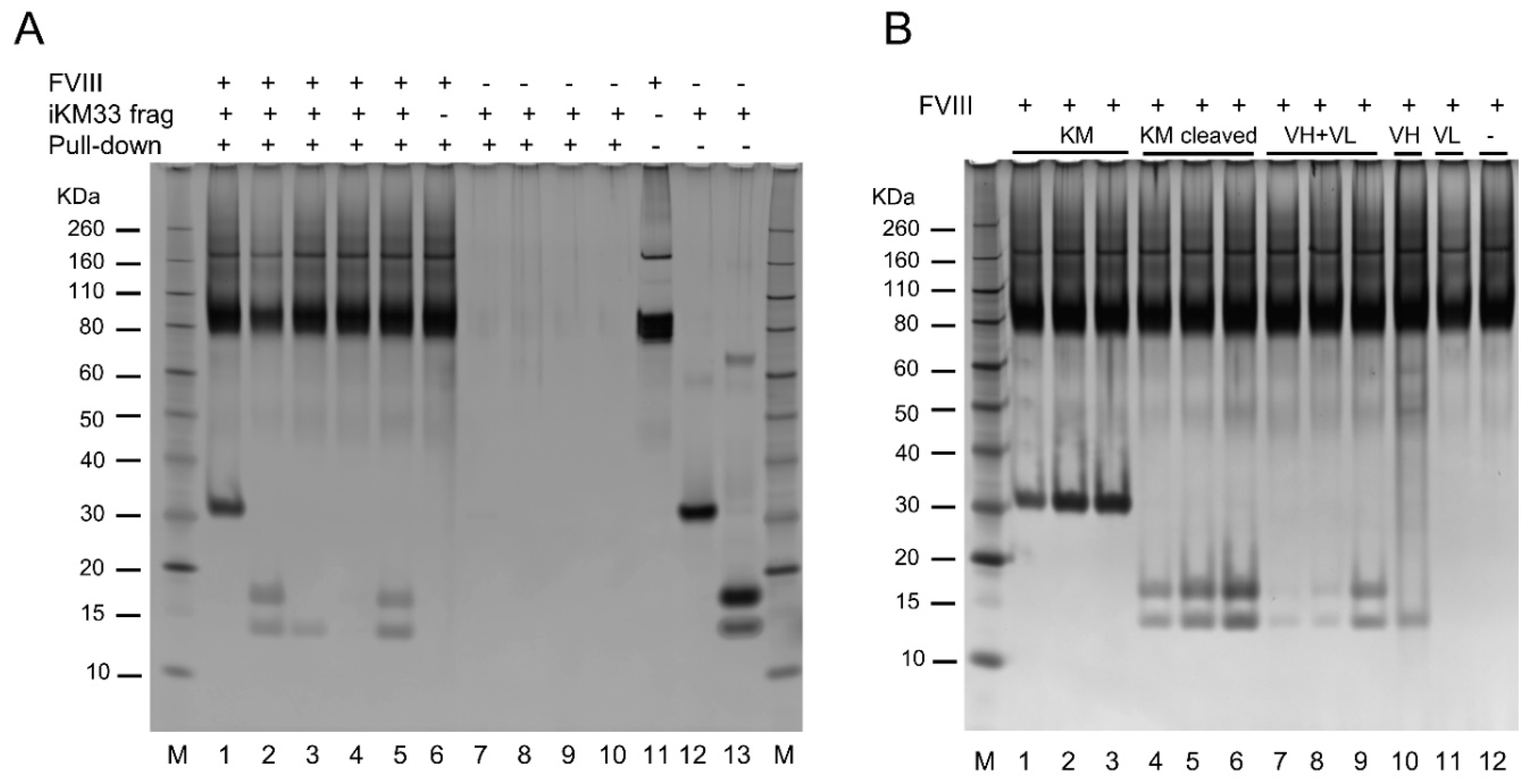
2.7. Testing Interaction between FVIII and Isolated iKM33 Subunits
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Generation of Vectors for Protein Expression
4.3. Protein Expression and Purification
4.4. Cleavage of scFv Variants by Thrombin and Isolation of Individual VH and VL Subunits
4.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
4.6. Size-Exclusion Chromatography Assay
4.7. Surface Plasmon Resonance Assay
4.8. Factor VIII Activity Assays
4.9. Pull-Down Assay with FVIII and iKM33 Fragments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APTT | activated partial thromboplastin time |
| BDD | B-domain deleted FVIII |
| CS | chromogenic substrate |
| EHL | extended plasma half-life |
| Fv | variable fragment of immunoglobulin |
| FVIII | coagulation factor VIII |
| FVIIIa | activated coagulation factor VIII |
| FXa | activated coagulation factor X |
| LDLR | low-density lipoprotein receptor |
| LRP1 | low-density lipoprotein receptor-related protein 1 |
| PPACK | Phe-Pro-Arg-chloromethylketone |
| MW | molecular weight |
| scFv | single-chain variable fragment |
| SDS-PAGE | sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SE-FPLC | size-exclusion fast protein chromatography |
| SPR | surface plasmon resonance |
| TG | thrombin generation |
| VWF | von Willebrand factor |
| VH and VL | heavy and light chains of immunoglobulin variable fragment |
References
- Glockshuber, R.; Malia, M.; Pfitzinger, I.; Pluckthun, A. A comparison of strategies to stabilize immunoglobulin Fv-fragments. Biochemistry 1990, 29, 1362–1367. [Google Scholar] [CrossRef]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 242, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotny, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R.; et al. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [PubMed]
- Colcher, D.; Pavlinkova, G.; Beresford, G.; Booth, B.J.; Batra, S.K. Single-chain antibodies in pancreatic cancer. Ann. N. Y. Acad. Sci. 1999, 880, 263–280. [Google Scholar] [CrossRef]
- Jain, M.; Kamal, N.; Batra, S.K. Engineering antibodies for clinical applications. Trends Biotechnol. 2007, 25, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Weisser, N.E.; Hall, J.C. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol. Adv. 2009, 27, 502–520. [Google Scholar] [CrossRef]
- Calvez, T.; Chambost, H.; Claeyssens-Donadel, S.; d’Oiron, R.; Goulet, V.; Guillet, B.; Heritier, V.; Milien, V.; Rothschild, C.; Roussel-Robert, V.; et al. Recombinant factor VIII products and inhibitor development in previously untreated boys with severe hemophilia A. Blood 2014, 124, 3398–3408. [Google Scholar] [CrossRef]
- van den Brink, E.N.; Turenhout, E.A.; Bovenschen, N.; Heijnen, B.G.; Mertens, K.; Peters, M.; Voorberg, J. Multiple VH genes are used to assemble human antibodies directed toward the A3-C1 domains of factor VIII. Blood 2001, 97, 966–972. [Google Scholar] [CrossRef]
- Fay, P.J. Activation of factor VIII and mechanisms of cofactor action. Blood Rev. 2004, 18, 1–15. [Google Scholar] [CrossRef]
- Butenas, S.; van‘t Veer, C.; Mann, K.G. Evaluation of the initiation phase of blood coagulation using ultrasensitive assays for serine proteases. J. Biol. Chem. 1997, 272, 21527–21533. [Google Scholar] [CrossRef]
- Newell, J.L.; Fay, P.J. Proteolysis at Arg740 facilitates subsequent bond cleavages during thrombin-catalyzed activation of factor VIII. J. Biol. Chem. 2007, 282, 25367–25375. [Google Scholar] [CrossRef]
- Hill-Eubanks, D.C.; Lollar, P. von Willebrand factor is a cofactor for thrombin-catalyzed cleavage of the factor VIII light chain. J. Biol. Chem. 1990, 265, 17854–17858. [Google Scholar] [CrossRef]
- Newell-Caito, J.L.; Griffiths, A.E.; Fay, P.J. P3-P3’ residues flanking scissile bonds in factor VIII modulate rates of substrate cleavage and procofactor activation by thrombin. Biochemistry 2012, 51, 3451–3459. [Google Scholar] [CrossRef]
- Walenga, J.M. Normal Hemostasis. In Rodak’s Hematology: Clinical Principles and Applications, 6th ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 626–649. [Google Scholar] [CrossRef]
- Davie, E.W.; Kulman, J.D. An overview of the structure and function of thrombin. Semin. Thromb. Hemost. 2006, 32 (Suppl. 1), 3–15. [Google Scholar] [CrossRef]
- Borissoff, J.I.; Joosen, I.A.; Versteylen, M.O.; Spronk, H.M.; ten Cate, H.; Hofstra, L. Accelerated in vivo thrombin formation independently predicts the presence and severity of CT angiographic coronary atherosclerosis. JACC Cardiovasc. Imaging 2012, 5, 1201–1210. [Google Scholar] [CrossRef]
- Jaberi, N.; Soleimani, A.; Pashirzad, M.; Abdeahad, H.; Mohammadi, F.; Khoshakhlagh, M.; Khazaei, M.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Role of thrombin in the pathogenesis of atherosclerosis. J Cell Biochem. 2019, 120, 4757–4765. [Google Scholar] [CrossRef]
- Lee, D.Y.; Park, K.W.; Jin, B.K. Thrombin induces neurodegeneration and microglial activation in the cortex in vivo and in vitro: Proteolytic and non-proteolytic actions. Biochem. Biophys. Res. Commun. 2006, 346, 727–738. [Google Scholar] [CrossRef]
- Iannucci, J.; Renehan, W.; Grammas, P. Thrombin, a Mediator of Coagulation, Inflammation, and Neurotoxicity at the Neurovascular Interface: Implications for Alzheimer’s Disease. Front. Neurosci. 2020, 14, 762. [Google Scholar] [CrossRef]
- Huntington, J.A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005, 3, 1861–1872. [Google Scholar] [CrossRef]
- Gallwitz, M.; Enoksson, M.; Thorpe, M.; Hellman, L. The extended cleavage specificity of human thrombin. PLoS ONE 2012, 7, e31756. [Google Scholar] [CrossRef]
- Bloem, E.; van den Biggelaar, M.; Wroblewska, A.; Voorberg, J.; Faber, J.H.; Kjalke, M.; Stennicke, H.R.; Mertens, K.; Meijer, A.B. Factor VIII C1 domain spikes 2092-2093 and 2158-2159 comprise regions that modulate cofactor function and cellular uptake. J. Biol. Chem. 2013, 288, 29670–29679. [Google Scholar] [CrossRef]
- Wroblewska, A.; van Haren, S.D.; Herczenik, E.; Kaijen, P.; Ruminska, A.; Jin, S.Y.; Zheng, X.L.; van den Biggelaar, M.; ten Brinke, A.; Meijer, A.B.; et al. Modification of an exposed loop in the C1 domain reduces immune responses to factor VIII in hemophilia A mice. Blood 2012, 119, 5294–5300. [Google Scholar] [CrossRef]
- Meems, H.; Meijer, A.B.; Cullinan, D.B.; Mertens, K.; Gilbert, G.E. Factor VIII C1 domain residues Lys 2092 and Phe 2093 contribute to membrane binding and cofactor activity. Blood 2009, 114, 3938–3946. [Google Scholar] [CrossRef][Green Version]
- Lu, J.; Pipe, S.W.; Miao, H.; Jacquemin, M.; Gilbert, G.E. A membrane-interactive surface on the factor VIII C1 domain cooperates with the C2 domain for cofactor function. Blood 2011, 117, 3181–3189. [Google Scholar] [CrossRef]
- Meems, H.; van den Biggelaar, M.; Rondaij, M.; van der Zwaan, C.; Mertens, K.; Meijer, A.B. C1 domain residues Lys 2092 and Phe 2093 are of major importance for the endocytic uptake of coagulation factor VIII. Int. J. Biochem. Cell Biol. 2011, 43, 1114–1121. [Google Scholar] [CrossRef]
- van den Biggelaar, M.; Madsen, J.J.; Faber, J.H.; Zuurveld, M.G.; van der Zwaan, C.; Olsen, O.H.; Stennicke, H.R.; Mertens, K.; Meijer, A.B. Factor VIII Interacts with the Endocytic Receptor Low-density Lipoprotein Receptor-related Protein 1 via an Extended Surface Comprising “Hot-Spot” Lysine Residues. J. Biol. Chem. 2015, 290, 16463–16476. [Google Scholar] [CrossRef]
- Przeradzka, M.A.; Freato, N.; Boon-Spijker, M.; van Galen, J.; van der Zwaan, C.; Mertens, K.; van den Biggelaar, M.; Meijer, A.B. Unique surface-exposed hydrophobic residues in the C1 domain of factor VIII contribute to cofactor function and von Willebrand factor binding. J. Thromb. Haemost. 2020, 18, 364–372. [Google Scholar] [CrossRef]
- Herczenik, E.; van Haren, S.D.; Wroblewska, A.; Kaijen, P.; van den Biggelaar, M.; Meijer, A.B.; Martinez-Pomares, L.; ten Brinke, A.; Voorberg, J. Uptake of blood coagulation factor VIII by dendritic cells is mediated via its C1 domain. J. Allergy Clin. Immunol. 2012, 129, 501–509.e5. [Google Scholar] [CrossRef]
- Kurasawa, J.H.; Shestopal, S.A.; Karnaukhova, E.; Struble, E.B.; Lee, T.K.; Sarafanov, A.G. Mapping the binding region on the low density lipoprotein receptor for blood coagulation factor VIII. J. Biol. Chem. 2013, 288, 22033–22041. [Google Scholar] [CrossRef]
- Peyvandi, F.; Mannucci, P.M.; Garagiola, I.; El-Beshlawy, A.; Elalfy, M.; Ramanan, V.; Eshghi, P.; Hanagavadi, S.; Varadarajan, R.; Karimi, M.; et al. A Randomized Trial of Factor VIII and Neutralizing Antibodies in Hemophilia A. N. Engl. J. Med. 2016, 374, 2054–2064. [Google Scholar] [CrossRef]
- Mertens, K.; Bovenschen, A.; Voorberg, J.; Rieger, M.; Scheiflinger, F. Antagonists of Factor VIII Interaction with Low-Density Lipoprotein Receptor-Related Protein. Patent No. US20080219983A1. US20080219983A1. 2008. Available online: https://patents.google.com/patent/US20080219983A1/en?oq=20080219983 (accessed on 1 July 2022).
- Mannucci, P.M. Benefits and limitations of extended plasma half-life factor VIII products in hemophilia A. Expert Opin. Investig. Drugs 2020, 29, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Kurasawa, J.H.; Shestopal, S.A.; Jha, N.K.; Ovanesov, M.V.; Lee, T.K.; Sarafanov, A.G. Insect cell-based expression and characterization of a single-chain variable antibody fragment directed against blood coagulation factor VIII. Protein Expr. Purif. 2013, 88, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Kettner, C.; Shaw, E. Inactivation of trypsin-like enzymes with peptides of arginine chloromethyl ketone. Methods Enzymol. 1981, 80 Pt C, 826–842. [Google Scholar] [CrossRef]
- Bode, W.; Mayr, I.; Baumann, U.; Huber, R.; Stone, S.R.; Hofsteenge, J. The refined 1.9 A crystal structure of human alpha-thrombin: Interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989, 8, 3467–3475. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, M.A.; Patel, H.; Rouse, J.C.; Marzilli, L.A.; Weston, S.B.; Sharpe, P.J. Defining ‘full-length’ recombinant factor VIII: A comparative structural analysis. Haemophilia 2007, 13, 30–37. [Google Scholar] [CrossRef]
- D’Amici, G.M.; Timperio, A.M.; Gevi, F.; Grazzini, G.; Zolla, L. Recombinant clotting factor VIII concentrates: Heterogeneity and high-purity evaluation. Electrophoresis 2010, 31, 2730–2739. [Google Scholar] [CrossRef]
- Ueda, H.; Tsumoto, K.; Kubota, K.; Suzuki, E.; Nagamune, T.; Nishimura, H.; Schueler, P.A.; Winter, G.; Kumagai, I.; Mohoney, W.C. Open sandwich ELISA: A novel immunoassay based on the interchain interaction of antibody variable region. Nat. Biotechnol. 1996, 14, 1714–1718. [Google Scholar] [CrossRef]
- Ihara, M.; Suzuki, T.; Kobayashi, N.; Goto, J.; Ueda, H. Open-sandwich enzyme immunoassay for one-step noncompetitive detection of corticosteroid 11-deoxycortisol. Anal. Chem. 2009, 81, 8298–8304. [Google Scholar] [CrossRef]
- Michnick, D.A.; Pittman, D.D.; Wise, R.J.; Kaufman, R.J. Identification of individual tyrosine sulfation sites within factor VIII required for optimal activity and efficient thrombin cleavage. J. Biol. Chem. 1994, 269, 20095–20102. [Google Scholar] [CrossRef]
- Choi, K.Y.; Swierczewska, M.; Lee, S.; Chen, X. Protease-activated drug development. Theranostics 2012, 2, 156–178. [Google Scholar] [CrossRef]
- Poreba, M. Protease-activated prodrugs: Strategies, challenges, and future directions. FEBS J. 2020, 287, 1936–1969. [Google Scholar] [CrossRef]
- Gabriel, D.; Lange, N.; Chobaz-Peclat, V.; Zuluaga, M.F.; Gurny, R.; van den Bergh, H.; Busso, N. Thrombin-sensitive dual fluorescence imaging and therapeutic agent for detection and treatment of synovial inflammation in murine rheumatoid arthritis. J. Control Release 2012, 163, 178–186. [Google Scholar] [CrossRef]
- Kwon, S.P.; Jeon, S.; Lee, S.H.; Yoon, H.Y.; Ryu, J.H.; Choi, D.; Kim, J.Y.; Kim, J.; Park, J.H.; Kim, D.E.; et al. Thrombin-activatable fluorescent peptide incorporated gold nanoparticles for dual optical/computed tomography thrombus imaging. Biomaterials 2018, 150, 125–136. [Google Scholar] [CrossRef]
- Lux, J.; Vezeridis, A.M.; Hoyt, K.; Adams, S.R.; Armstrong, A.M.; Sirsi, S.R.; Mattrey, R.F. Thrombin-Activatable Microbubbles as Potential Ultrasound Contrast Agents for the Detection of Acute Thrombosis. ACS Appl. Mater. Interfaces 2017, 9, 37587–37596. [Google Scholar] [CrossRef] [PubMed]
- Muczynski, V.; Verhenne, S.; Casari, C.; Cherel, G.; Panicot-Dubois, L.; Gueguen, P.; Trossaert, M.; Dubois, C.; Lenting, P.J.; Denis, C.V.; et al. A Thrombin-Activatable Factor X Variant Corrects Hemostasis in a Mouse Model for Hemophilia A. Thromb. Haemost. 2019, 119, 1981–1993. [Google Scholar] [CrossRef]
- Konkle, B.A.; Shapiro, A.D.; Quon, D.V.; Staber, J.M.; Kulkarni, R.; Ragni, M.V.; Chhabra, E.S.; Poloskey, S.; Rice, K.; Katragadda, S.; et al. BIVV001 Fusion Protein as Factor VIII Replacement Therapy for Hemophilia A. N. Engl. J. Med. 2020, 383, 1018–1027. [Google Scholar] [CrossRef]
- Selvin, P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef]
- Hwang, H.; Myong, S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014, 43, 1221–1229. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Paek, S.H.; Lee, S.H.; Cho, J.H.; Kim, Y.S. Development of rapid one-step immunochromatographic assay. Methods 2000, 22, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.; Tang, M.; Zhao, J.; Zhu, X. Nanobody: A promising toolkit for molecular imaging and disease therapy. EJNMMI Res. 2021, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Hamann, M.V.; Beschorner, N.; Vu, X.K.; Hauber, I.; Lange, U.C.; Traenkle, B.; Kaiser, P.D.; Foth, D.; Schneider, C.; Büning, H.; et al. Improved targeting of human CD4+ T cells by nanobody-modified AAV2 gene therapy vectors. PLoS ONE 2021, 16, e0261269. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C. Nanobody approval gives domain antibodies a boost. Nat. Rev. Drug Discov. 2019, 18, 485–487. [Google Scholar] [CrossRef]
- Peyron, I.; Kizlik-Masson, C.; Dubois, M.D.; Atsou, S.; Ferrière, S.; Denis, C.V.; Lenting, P.J.; Casari, C.; Christophe, O.D. Camelid-derived single-chain antibodies in hemostasis: Mechanistic, diagnostic, and therapeutic applications. Res. Pract. Thromb. Haemost. 2020, 4, 1087–1110. [Google Scholar] [CrossRef]
- Silva-Pilipich, N.; Smerdou, C.; Vanrell, L. A Small Virus to Deliver Small Antibodies: New Targeted Therapies Based on AAV Delivery of Nanobodies. Microorganisms 2021, 9, 1956. [Google Scholar] [CrossRef]
- Nilvebrant, J.; Ereño-Orbea, J.; Gorelik, M.; Julian, M.C.; Tessier, P.M.; Julien, J.P.; Sidhu, S.S. Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains. J. Mol. Biol. 2021, 433, 167241. [Google Scholar] [CrossRef]
- Sarafanov, A.; Saenko, E. High-throughput optimization of protein expression in the baculovirus system based on determination of relative expression efficiency of viral stocks. Anal. Biochem. 2004, 328, 98–100. [Google Scholar] [CrossRef]
- Marakasova, E.; Olivares, P.; Karnaukhova, E.; Chun, H.; Hernandez, N.E.; Kurasawa, J.H.; Hassink, G.U.; Shestopal, S.A.; Strickland, D.K.; Sarafanov, A.G. Molecular chaperone RAP interacts with LRP1 in a dynamic bivalent mode and enhances folding of ligand-binding regions of other LDLR family receptors. J. Biol. Chem. 2021, 297, 100842. [Google Scholar] [CrossRef]
- Jha, N.K.; Shestopal, S.A.; Gourley, M.J.; Woodle, S.A.; Liang, Y.; Sarafanov, A.G.; Weinstein, M.; Ovanesov, M.V. Optimization of the thrombin generation test components to measure potency of factor VIII concentrates. Haemophilia 2016, 22, 780–789. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shestopal, S.A.; Parunov, L.A.; Olivares, P.; Chun, H.; Ovanesov, M.V.; Pettersson, J.R.; Sarafanov, A.G. Isolated Variable Domains of an Antibody Can Assemble on Blood Coagulation Factor VIII into a Functional Fv-like Complex. Int. J. Mol. Sci. 2022, 23, 8134. https://doi.org/10.3390/ijms23158134
Shestopal SA, Parunov LA, Olivares P, Chun H, Ovanesov MV, Pettersson JR, Sarafanov AG. Isolated Variable Domains of an Antibody Can Assemble on Blood Coagulation Factor VIII into a Functional Fv-like Complex. International Journal of Molecular Sciences. 2022; 23(15):8134. https://doi.org/10.3390/ijms23158134
Chicago/Turabian StyleShestopal, Svetlana A., Leonid A. Parunov, Philip Olivares, Haarin Chun, Mikhail V. Ovanesov, John R. Pettersson, and Andrey G. Sarafanov. 2022. "Isolated Variable Domains of an Antibody Can Assemble on Blood Coagulation Factor VIII into a Functional Fv-like Complex" International Journal of Molecular Sciences 23, no. 15: 8134. https://doi.org/10.3390/ijms23158134
APA StyleShestopal, S. A., Parunov, L. A., Olivares, P., Chun, H., Ovanesov, M. V., Pettersson, J. R., & Sarafanov, A. G. (2022). Isolated Variable Domains of an Antibody Can Assemble on Blood Coagulation Factor VIII into a Functional Fv-like Complex. International Journal of Molecular Sciences, 23(15), 8134. https://doi.org/10.3390/ijms23158134





