Comparative Transcriptome Analysis Reveals That WSSV IE1 Protein Plays a Crucial Role in DNA Replication Control
Abstract
:1. Introduction
2. Results
2.1. De Novo Assembly and Unigene Annotation
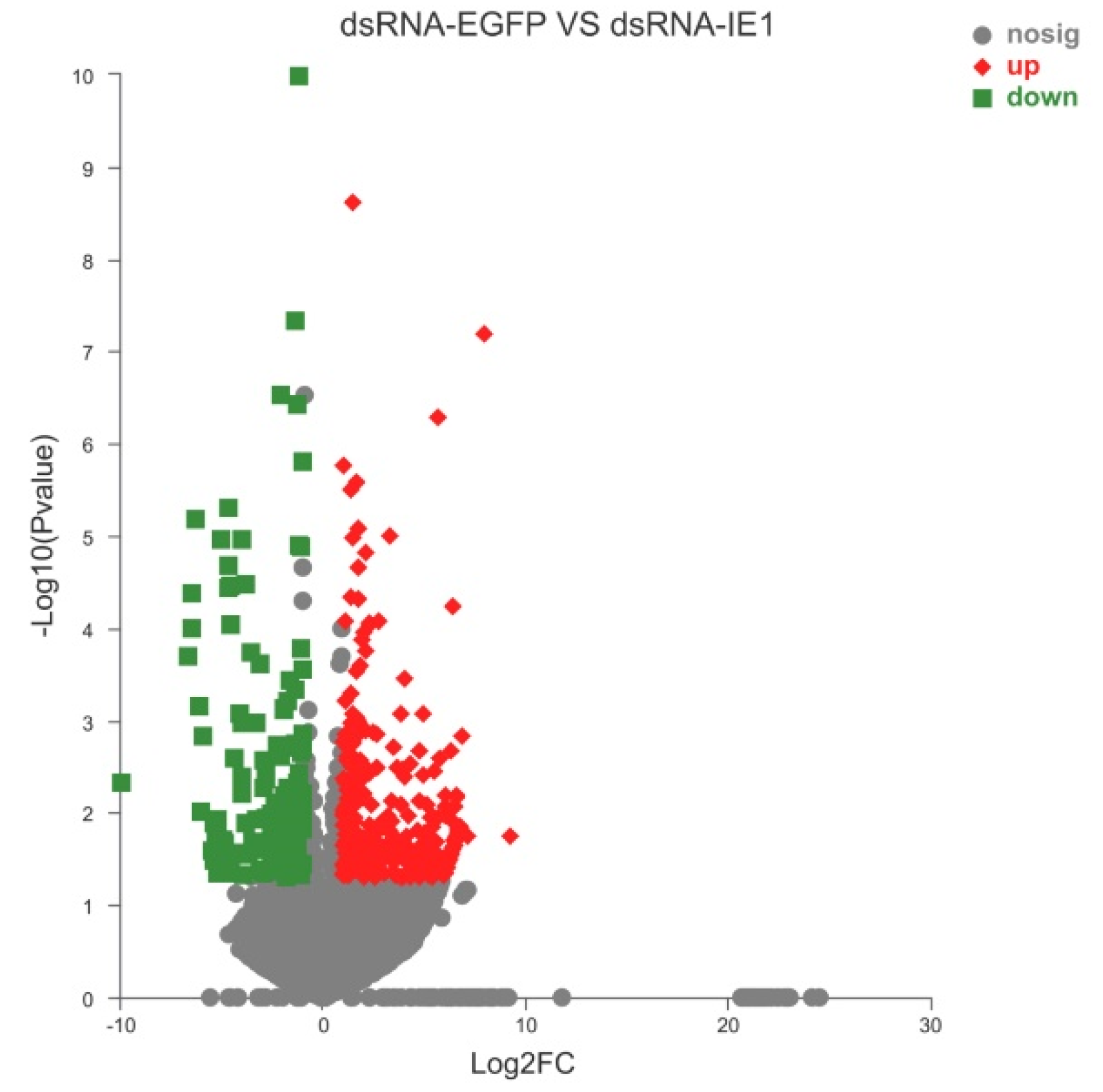
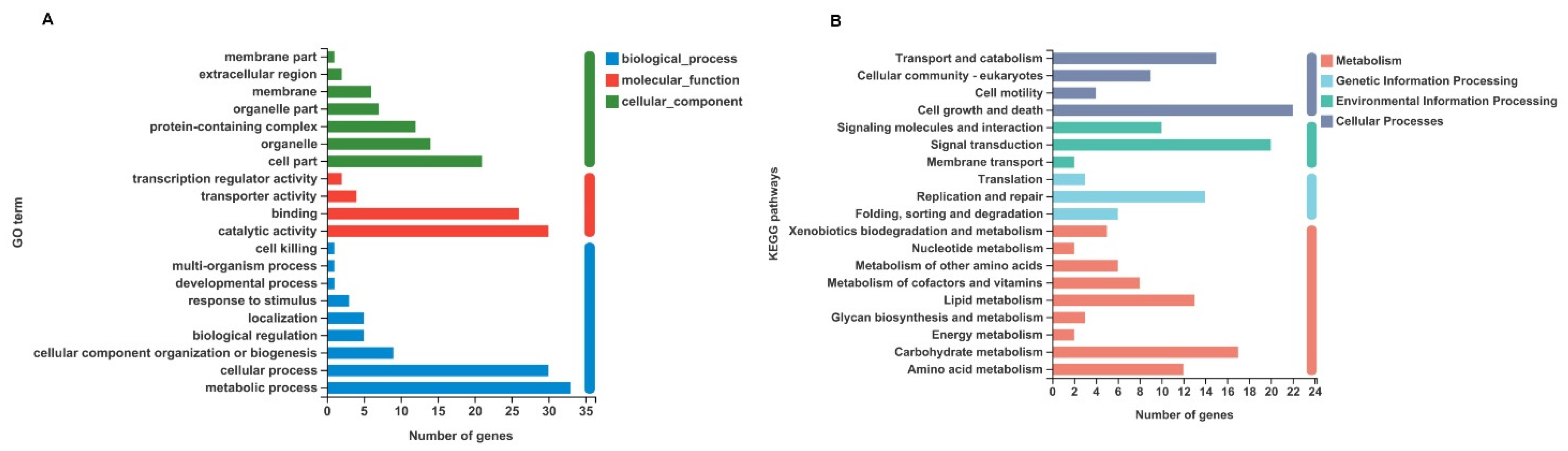
2.2. Identification and Functional Classification Analysis of DEGs
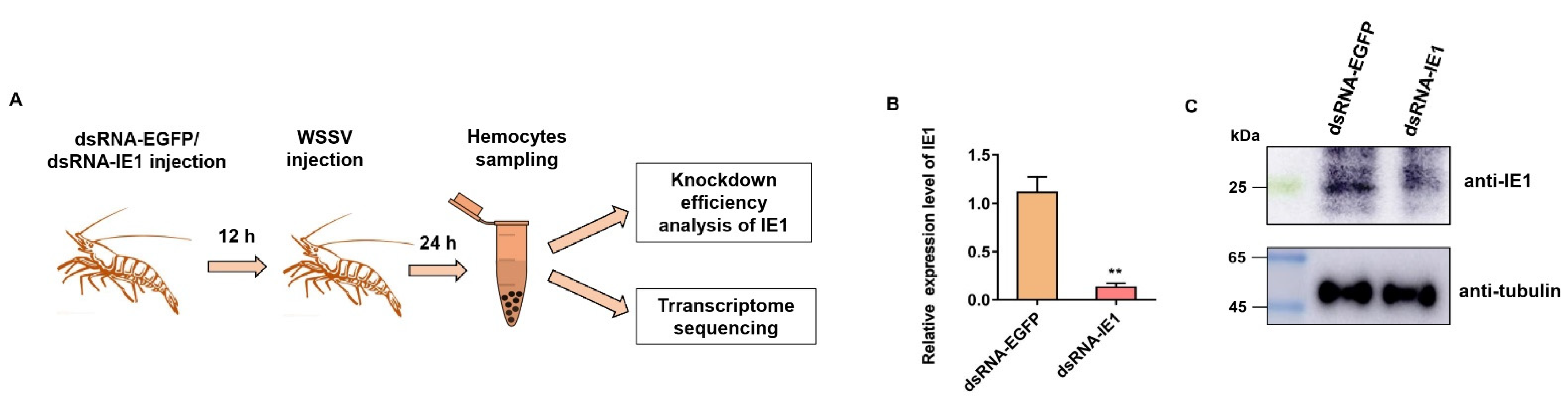
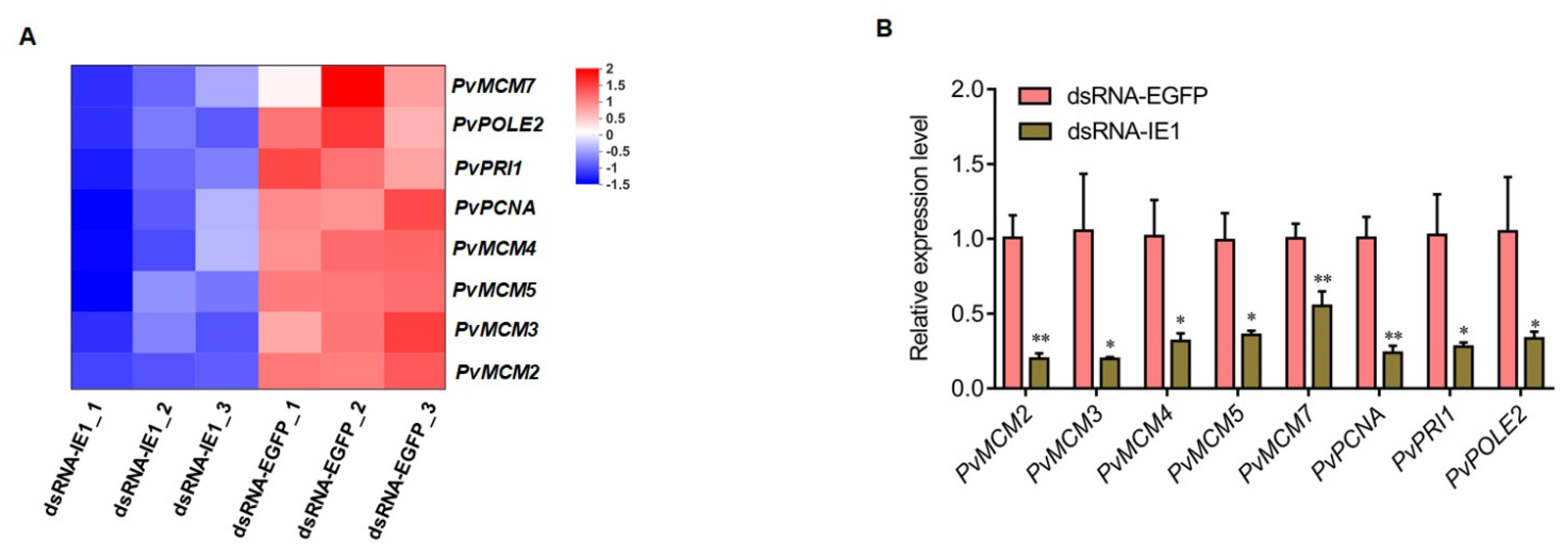
2.3. Validation of RNA-Seq Results by qPCR Analysis
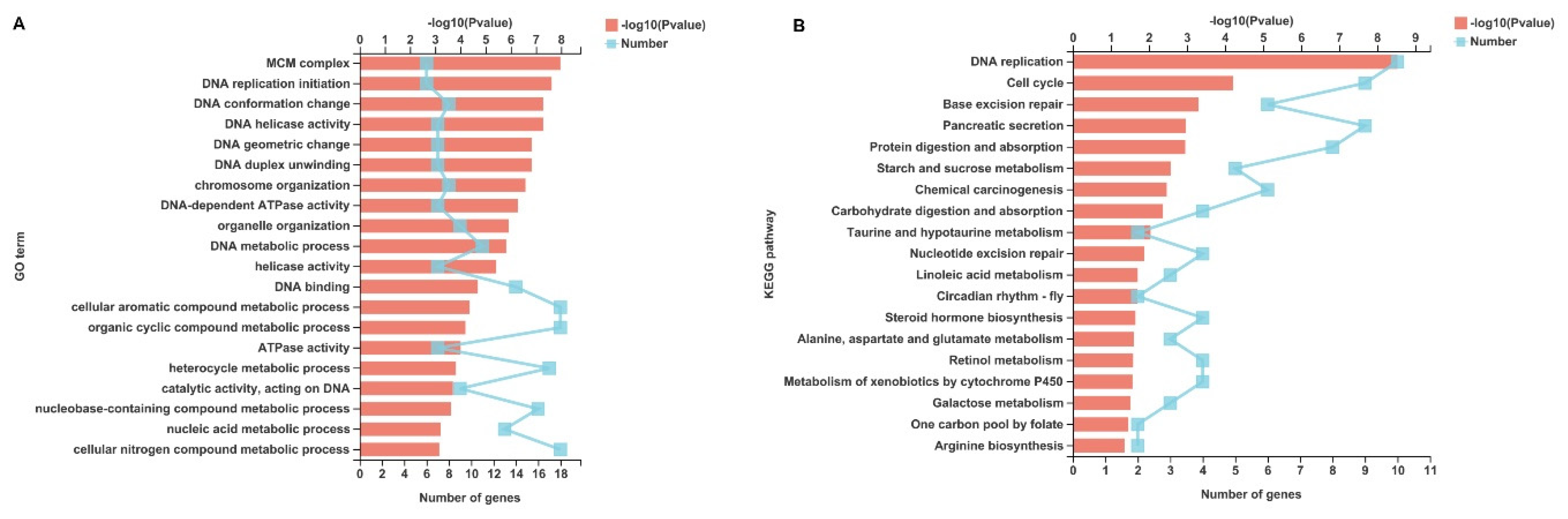
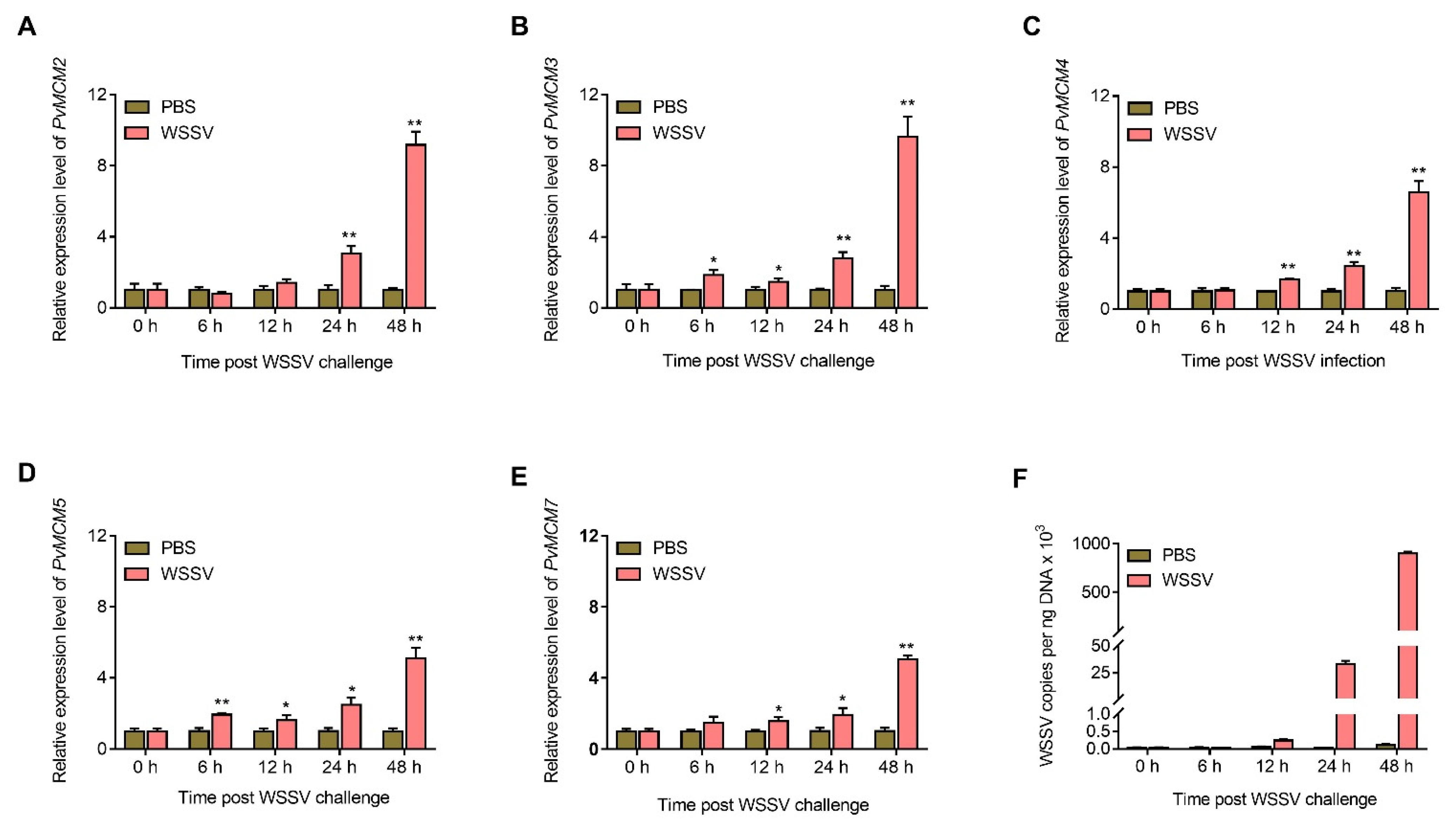
2.4. P. Vannamei MCM Complex Genes Are Upregulated after WSSV Challenge
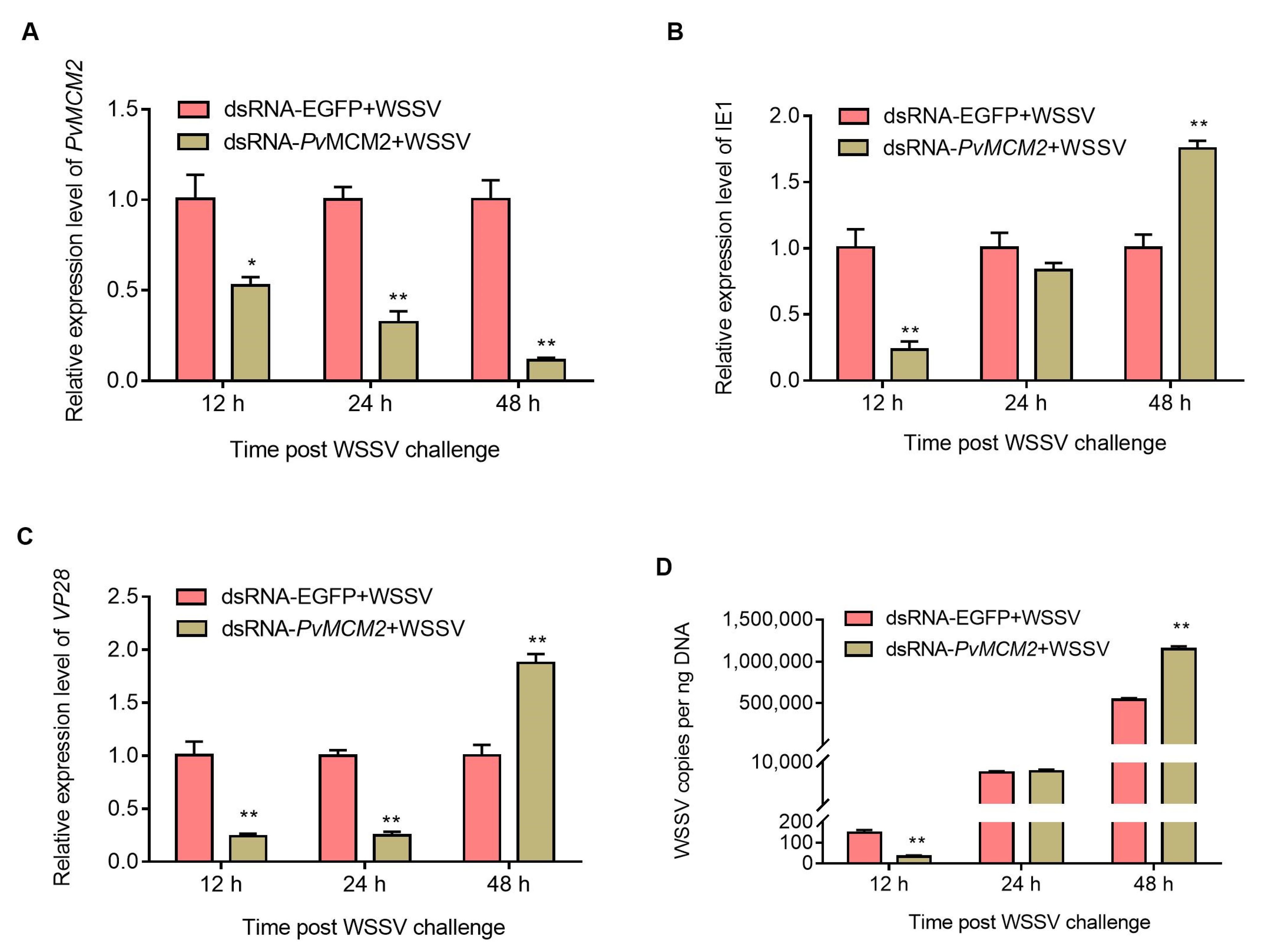
2.5. P. vannamei MCM Complex Enhances WSSV Replication at the Early Infection Stage
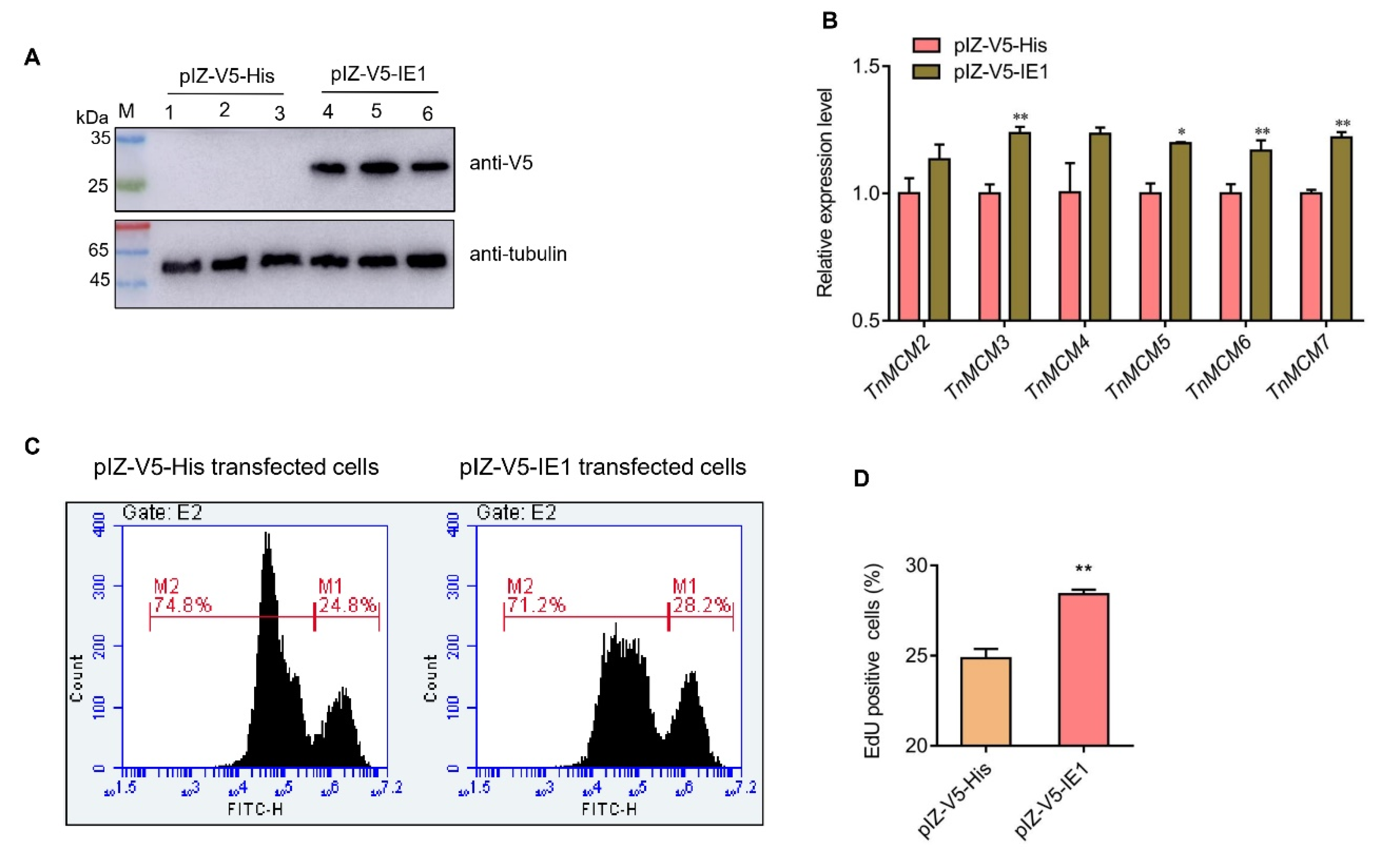
2.6. IE1 Promotes the Cellular DNA Synthesis In Vitro
3. Discussion
4. Materials and Methods
4.1. Shrimp Culture and Virus
4.2. RNAi-Mediated Knockdown of IE1 and Sample Collection
4.3. RNA Extraction, cDNA Library Construction, and Transcriptome Sequencing
4.4. Reads Mapping, De Novo Assembly, and Unigene Annotation
4.5. Identification and Functional Classification Analysis of Differential Expression Genes
4.6. Validation of DEGs by Real-Time Quantitative PCR (qPCR) Analysis
4.7. Expression Analysis of P. vannamei MCM Complex Genes after WSSV Infection
4.8. Detection of Viral Genes’ Expression and Viral Loads after PvMCM2 Knockdown
4.9. Plasmid Transfection and Analysis of MCM Complex Genes’ Expression and DNA Synthesis
4.10. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leu, J.H.; Yang, F.; Zhang, X.; Xu, X.; Kou, G.H.; Lo, C.F. Whispovirus. Curr. Top. Microbiol. Immunol. 2009, 328, 197–227. [Google Scholar] [PubMed]
- Escobedo-Bonilla, C.M.; Alday-Sanz, V.; Wille, M.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2008, 31, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; He, J.; Lin, X.H.; Li, Q.; Pan, D.; Zhang, X.B.; Xu, X. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 2001, 75, 11811–11820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.X.; Xu, L.M.; Yang, F. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 2006, 80, 10615–10623. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.C.; Xu, J.Y.; Liu, H.P. Cellular entry of white spot syndrome virus and antiviral immunity mediated by cellular receptors in crustaceans. Fish Shellfish Immunol. 2019, 93, 580–588. [Google Scholar] [CrossRef]
- Verbruggen, B.; Bickley, L.K.; van Aerle, R.; Bateman, K.S.; Stentiford, G.D.; Santos, E.M.; Tyler, C.R. Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses 2016, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Li, C.Z.; Weng, S.P.; He, J.G. WSSV-host interaction: Host response and immune evasion. Fish Shellfish Immunol. 2019, 84, 558–571. [Google Scholar] [CrossRef]
- Xu, D.D.; Liu, W.F.; Alvarez, A.; Huang, T.Z. Cellular immune responses against viral pathogens in shrimp. Dev. Comp. Immunol. 2014, 47, 287–297. [Google Scholar] [CrossRef]
- Marks, H.; Vorst, O.; van Houwelingen, A.; van Hulten, M.C.W.; Vlak, J.M. Gene-expression profiling of white spot syndrome virus in vivo. J. Gen. Virol. 2005, 86 Pt 7, 2081–2100. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Dybas, J.M.; Hughes, J.; Weitzman, M.D.; Boutell, C. The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection. Virus Res. 2020, 285, 198015. [Google Scholar] [CrossRef]
- Martinez, F.P.; Cosme, R.S.C.; Tang, Q.Y. Murine cytomegalovirus major immediate-early protein 3 interacts with cellular and viral proteins in viral DNA replication compartments and is important for early gene activation. J. Gen. Virol. 2010, 91, 2664–2676. [Google Scholar] [CrossRef] [PubMed]
- Torres, L.; Tang, Q. Immediate-Early (IE) gene regulation of cytomegalovirus: IE1- and pp71-mediated viral strategies against cellular defenses. Virol. Sin. 2014, 29, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Li, M.Y.; Ke, W.; Ji, Y.C.; Bian, X.F.; Yan, X.M. Identification of the immediate-early genes of white spot syndrome virus. Virology 2009, 385, 267–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, F.Y.; Huang, H.; Xu, L.M.; Li, F.; Yang, F. Identification of three immediate-early genes of white spot syndrome virus. Arch. Virol. 2011, 156, 1611–1614. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Chang, Y.S.; Wang, C.H.; Kou, G.H.; Lo, C.F. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 2005, 334, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.J.; Chang, Y.S.; Wang, A.H.; Kou, G.H.; Lo, C.F. White spot syndrome virus annexes a shrimp STAT to enhance expression of the immediate-early gene ie1. J. Virol. 2007, 81, 1461–1471. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.D.; Zhao, L.; Zhang, H.Q.; Xu, X.P.; Jia, X.T.; Chen, Y.H.; Wang, P.H.; Weng, S.P.; Yu, X.Q.; Yin, Z.X.; et al. Shrimp NF- κB binds to the immediate-early gene ie1 promoter of white spot syndrome virus and upregulates its activity. Virology 2010, 406, 176–180. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Li, H.Y.; Weng, S.P.; Li, C.Z.; He, J.G. White spot syndrome virus establishes a novel IE1/JNK/c-Jun positive feedback loop to drive replication. iScience 2020, 23, 100752. [Google Scholar] [CrossRef] [Green Version]
- Yao, D.F.; Ruan, L.W.; Lu, H.S.; Shi, H.; Xu, X. Shrimp STAT was hijacked by white spot syndrome virus immediate-early protein IE1 involved in modulation of viral genes. Fish Shellfish Immunol. 2016, 59, 268–275. [Google Scholar] [CrossRef]
- Chang, L.K.; Huang, P.H.; Shen, W.T.; Yang, S.H.; Liu, W.J.; Lo, C.F. Role of Penaeus monodon Kruppel-like factor (PmKLF) in infection by white spot syndrome virus. Dev. Comp. Immunol. 2012, 36, 121–129. [Google Scholar] [CrossRef]
- Huang, P.H.; Huang, T.Y.; Cai, P.S.; Chang, L.K. Role of Litopenaeus vannamei Yin Yang 1 in the regulation of the white spot syndrome virus immediate early gene ie1. J. Virol. 2017, 91, e02314-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.J.; Chang, Y.S.; Wang, H.C.; Leu, J.H.; Kou, G.H.; Lo, C.F. Transactivation, dimerization, and DNA-binding activity of white spot syndrome virus immediate-early protein IE1. J. Virol. 2008, 82, 11362–11373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, X.Z.; Bian, X.F.; Ji, Y.C.; Yan, X.M.; Yang, F.; Li, F. White spot syndrome virus IE1 and WSV056 modulate the G1/S transition by binding to the host retinoblastoma protein. J. Virol. 2013, 87, 12576–12582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.Z.; Ruan, L.W.; Zhou, C.Z.; Shi, H.; Xu, X. Characterization and function of a beta-catenin homolog from Litopenaeus vannamei in WSSV infection. Dev. Comp. Immunol. 2017, 76, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L.W.; Sun, J.Z.; Zhou, C.Z.; Shi, H.; Lei, K.Y. Cloning, identification and function analysis of a Chibby homolog from Litopenaeus vannamei. Fish Shellfish Immunol. 2018, 78, 114–120. [Google Scholar] [CrossRef]
- Wang, C.Q.; Wei, M.H.; Wu, G.C.; He, L.X.; Zhu, J.H.; Aweya, J.J.; Chen, X.L.; Zhao, Y.Z.; Zhang, Y.L.; Yao, D.F. Proteomics analysis reveals a critical role for the WSSV immediate-early protein IE1 in modulating the host prophenoloxidase system. Virulence 2022, 13, 936–948. [Google Scholar] [CrossRef]
- Maiorano, D.; Lutzmann, M.; Mechali, M. MCM proteins and DNA replication. Curr. Opin. Cell Biol. 2006, 18, 130–136. [Google Scholar] [CrossRef]
- Chen, L.L.; Wang, H.C.; Huang, C.J.; Peng, S.E.; Chen, Y.G.; Lin, S.J.; Chen, W.Y.; Dai, C.F.; Yu, H.T.; Wang, C.H.; et al. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology 2002, 301, 136–147. [Google Scholar] [CrossRef] [Green Version]
- Zang, K.; Li, F.H.; Ma, Q.J. The dUTPase of white spot syndrome virus assembles its active sites in a noncanonical manner. J. Biol. Chem. 2018, 293, 1088–1099. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, B.; Xu, H.Z.; Todorov, I.; Dutta, A.; Yates, J.L. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 2001, 98, 10085–10089. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, A.; Nagata, K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. Embo J. 2007, 26, 4566–4575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabral, P.; Uppal, T.; Rossetto, C.C.; Verma, S.C. Minichromosome maintenance proteins cooperate with LANA during the G1/S phase of the cell cycle to support viral DNA replication. J. Virol. 2019, 93, e02256-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhu, F. Minichromosome maintenance protein 7 regulates phagocytosis in kuruma shrimp Marsupenaeus japonicas against white spot syndrome virus. Fish Shellfish Immunol. 2016, 55, 293–303. [Google Scholar] [CrossRef]
- Zhu, F.; Qian, X.Y.; Wang, Z.Y. Molecular characterization of minichromosome maintenance protein (MCM7) in Scylla paramamosain and its role in white spot syndrome virus and Vibrio alginolyticus infection. Fish Shellfish Immunol. 2018, 83, 104–114. [Google Scholar] [CrossRef]
- Liu, W.J.; Chang, Y.S.; Huang, W.T.; Chen, I.T.; Wang, K.C.H.C.; Kou, G.H.; Lo, C.F. Penaeus monodon TATA box-binding protein interacts with the white spot syndrome virus transactivator IE1 and promotes its transcriptional activity. J. Virol. 2011, 85, 6535–6547. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, K.; Iwanaga, R.; Nakamura, M.; Ikeda, M.; Yabuta, N.; Tsuruga, H.; Nojima, H. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene 1999, 18, 2299–2309. [Google Scholar] [CrossRef] [Green Version]
- Bruemmer, D.; Yin, F.; Liu, J.; Kiyono, T.; Fleck, E.; Van Herle, A.J.; Law, R.E. Rapamycin inhibits E2F-dependent expression of minichromosome maintenance proteins in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2003, 303, 251–258. [Google Scholar] [CrossRef]
- Xie, X.X.; Li, H.Y.; Xu, L.M.; Yang, F. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 2005, 108, 63–67. [Google Scholar] [CrossRef]
- Zhou, Q.; Qi, Y.P.; Yang, F. Application of spectrophotometry to evaluate the concentration of purified white spot syndrome virus. J. Virol. Methods 2007, 146, 288–292. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.X.; Aweya, J.J.; Wang, F.; Yao, D.F.; Zhong, M.Q.; Chen, J.H.; Li, S.K.; Zhang, Y.L. Litopenaeus vannamei attenuates white spot syndrome virus replication by specific antiviral peptides generated from hemocyanin. Dev. Comp. Immunol. 2019, 91, 50–61. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wu, G.; Wang, C.; Zhang, H.; Zhu, J.; Zhang, Y.; Lin, Z.; Yao, D. Comparative Transcriptome Analysis Reveals That WSSV IE1 Protein Plays a Crucial Role in DNA Replication Control. Int. J. Mol. Sci. 2022, 23, 8176. https://doi.org/10.3390/ijms23158176
Chen Y, Wu G, Wang C, Zhang H, Zhu J, Zhang Y, Lin Z, Yao D. Comparative Transcriptome Analysis Reveals That WSSV IE1 Protein Plays a Crucial Role in DNA Replication Control. International Journal of Molecular Sciences. 2022; 23(15):8176. https://doi.org/10.3390/ijms23158176
Chicago/Turabian StyleChen, Yixi, Gaochun Wu, Chuanqi Wang, Huimin Zhang, Jinghua Zhu, Yueling Zhang, Zhongyang Lin, and Defu Yao. 2022. "Comparative Transcriptome Analysis Reveals That WSSV IE1 Protein Plays a Crucial Role in DNA Replication Control" International Journal of Molecular Sciences 23, no. 15: 8176. https://doi.org/10.3390/ijms23158176





