Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis
Abstract
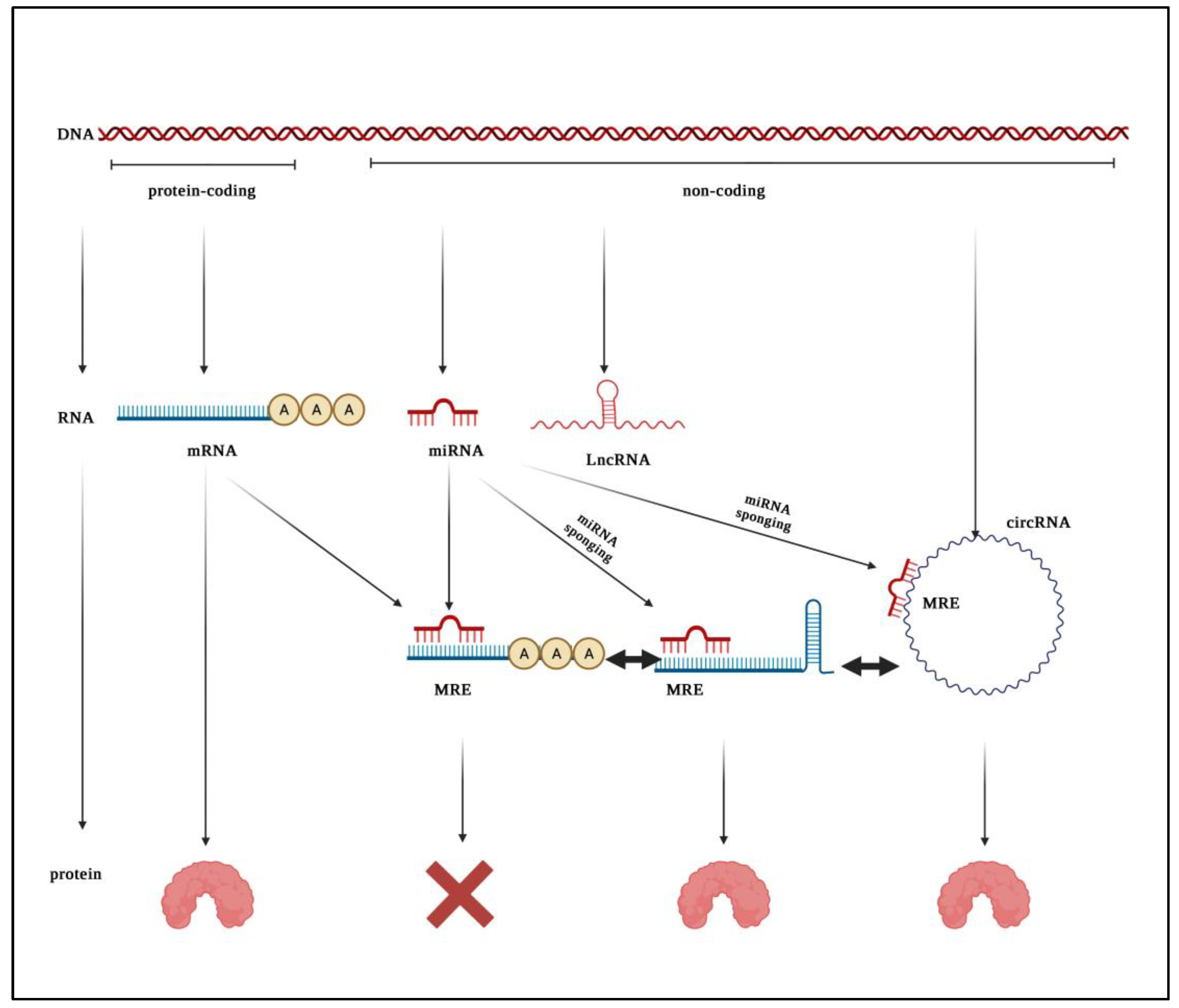
:1. Introduction
2. Oncogenic and Tumor Suppressor Roles of Exosomal Non-Coding RNAs in BC
2.1. Roles of Exosomal ncRNAs in BC Cell Growth and Proliferation
2.2. Roles of Exosomal ncRNAs in BC Metastasis
2.3. Roles of Exosomal ncRNAs in Immunoregulation and Cellular Polarization
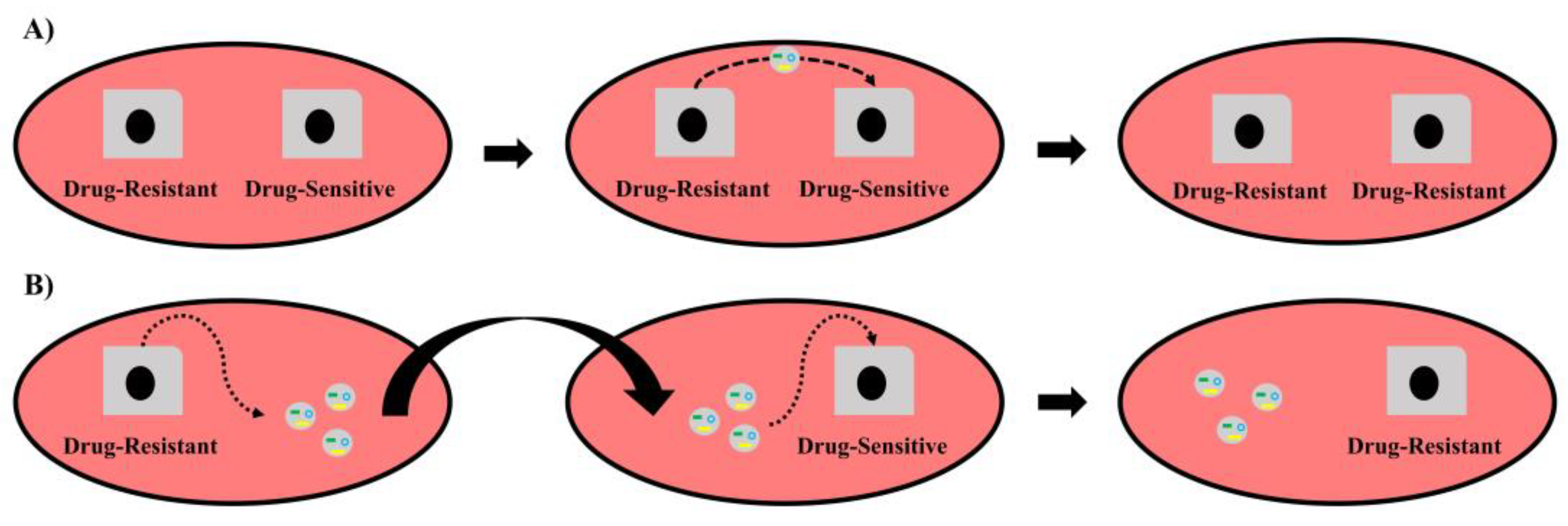
2.4. Roles of Exosomal ncRNAs in BC-Drug Sensitivity/Resistance
| Immunotherapy | |||||
|---|---|---|---|---|---|
| Exosomal ncRNA(s) | Drug | Role | Mechanism | Clinical Evidence and Status | Reference |
| miR-1246,miR-155 | Trastuzumab | Enhancement of trastuzumab resistance | N/A | yes, upregulated | [85] |
| miR-567 | Trastuzumab | Enhancement of trastuzumab sensitivity | Inhibiting autophagy via ATG5 suppression | yes, downregulated | [86] |
| SNHG14 | Trastuzumab | Enhancement of trastuzumab resistance | N/A | yes, upregulated | [77] |
| AGAP2-AS1 | Trastuzumab | Enhancement of trastuzumab resistance | N/A | no | [73] |
| AFAP1-AS1 | Trastuzumab | Enhancement of trastuzumab resistance | Promoting ERBB2 translation via AUF1 binding | yes, upregulated | [72] |
| Hormonal Therapy | |||||
| Exosomal ncRNA(s) | Drug | Role | Mechanism | Reference | |
| miR-221, miR-222 | Tamoxifen | Enhancement of tamoxifen resistance | Negatively regulating p27 and ERα | no | [81] |
| miR-205 | Tamoxifen | Enhancement of tamoxifen resistance | Inhibiting apoptosis via E2F1 downregulation | no | [68] |
| miR-181a-2 | Tamoxifen | Enhancement of tamoxifen resistance | Downregulating ERα and activating PI3K/AKT signaling | no | [87] |
| UCA1 | Tamoxifen | Enhancement of tamoxifen resistance | N/A | no | [80] |
| HOTAIR | Tamoxifen | Enhancement of tamoxifen resistance | N/A | yes, upregulated | [75] |
| circ_UBE2D2, miR-200a-3p | Tamoxifen | Enhancement of tamoxifen resistance | miR-200a-3p sponging, leading to alterations in cell viability, EMT, and ERα status | no | [71] |
| Chemotherapy | |||||
| Exosomal ncRNA(s) | Drug | Role | Mechanism | Clinical Evidence | Reference |
| multiple miRNAs | Docetaxel | Enhancement of docetaxel resistance | N/A | no | [82] |
| miR-23b | Docetaxel | Enhancement of docetaxel resistance | Inducing metastatic breast cancer cell dormancy via suppressing MARCKS | yes, upregulated | [84] |
| miR-134 | Cisplatin | Enhancement of cisplatin sensitivity | Negatively regulating STAT5B, Hsp90, and Bcl-2 | yes, downregulated | [88] |
| miR-222 | Adriamycin | Enhancement of adriamycin resistance | N/A | no | [79] |
| miR-222/223 | Carboplatin | Enhancement of carboplatin resistance | N/A | no | [83] |
| miR-1246 | Docetaxel, Epirubicin, Gemcitabine | Enhancement of docetaxel, epirubicin, and gemcitabine resistance | Negatively regulating Cyclin-G2 | yes, upregulated | [32] |
| miR-126a | Doxorubicin | Enhancement of doxorubicin resistance | Inducing IL-13+ Th2 cells, promoting angiogenesis, and enhancing cell viability via S100A8/A9 upregulation | no | [78] |
| miR-155 | Doxorubicin, Paclitaxel | Enhancement of doxorubicin, and paclitaxel resistance | N/A | no | [76] |
| miR-423-5p | Cisplatin | Enhancement of cisplatin resistance | N/A | no | [74] |
| miR-378a-3p, miR-378d | Doxorubicin, Paclitaxel (neoadjuvant) | Enhancement of doxorubicin and paclitaxel resistance | Activation of WNT and NOTCH stemness pathways via DKK3 and NUMB suppression. | yes, upregulated | [69] |
| HOTAIR | Neoadjuvant chemotherapy | Enhancement of chemoresistance | N/A | yes, upregulated | [75] |
| H19 | Doxorubicin | Enhancement of doxorubicin resistance | N/A | yes, upregulated | [70] |
3. Diagnostic, Prognostic, and Predictive Biomarker Potential of Exosomal ncRNAs
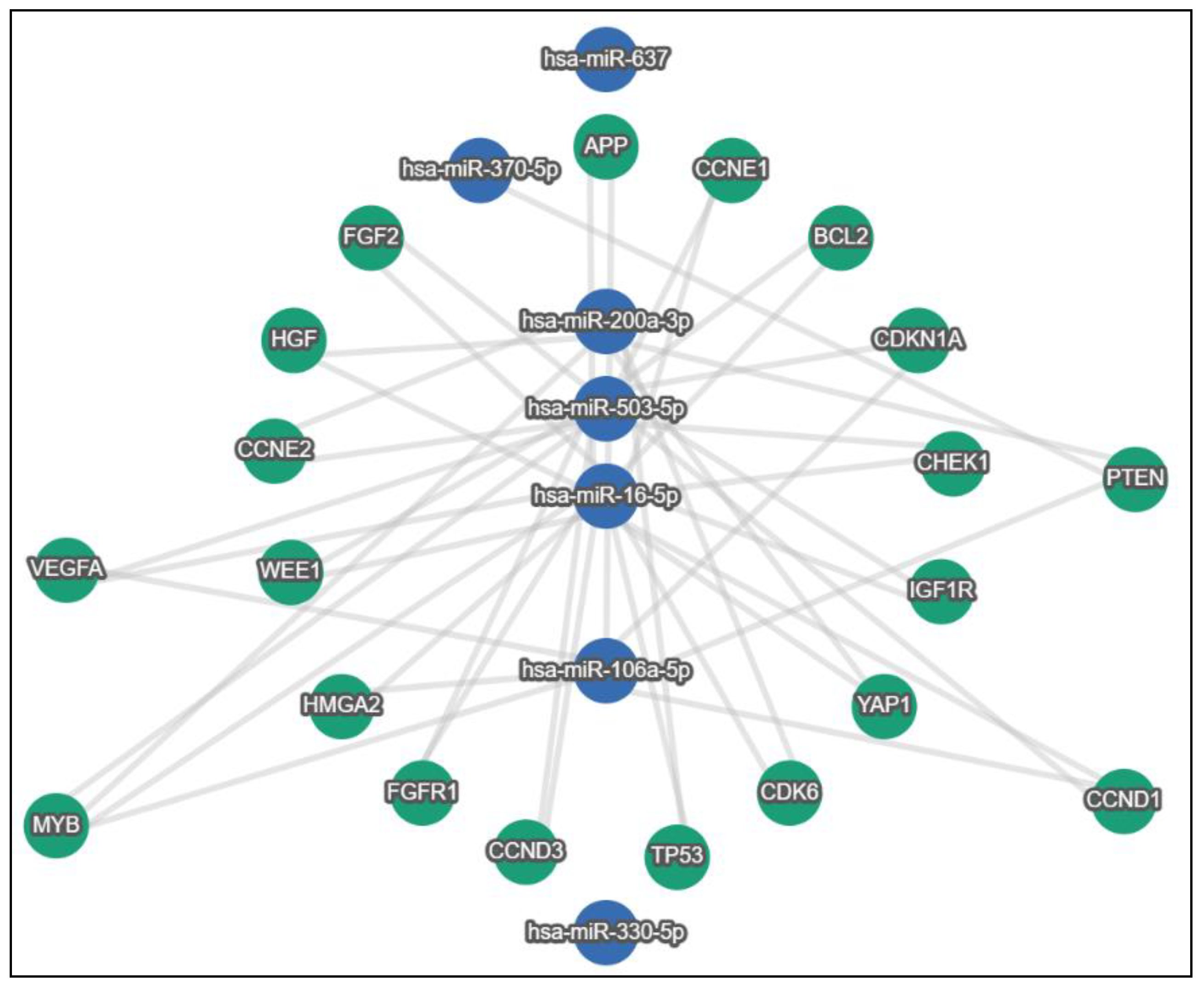
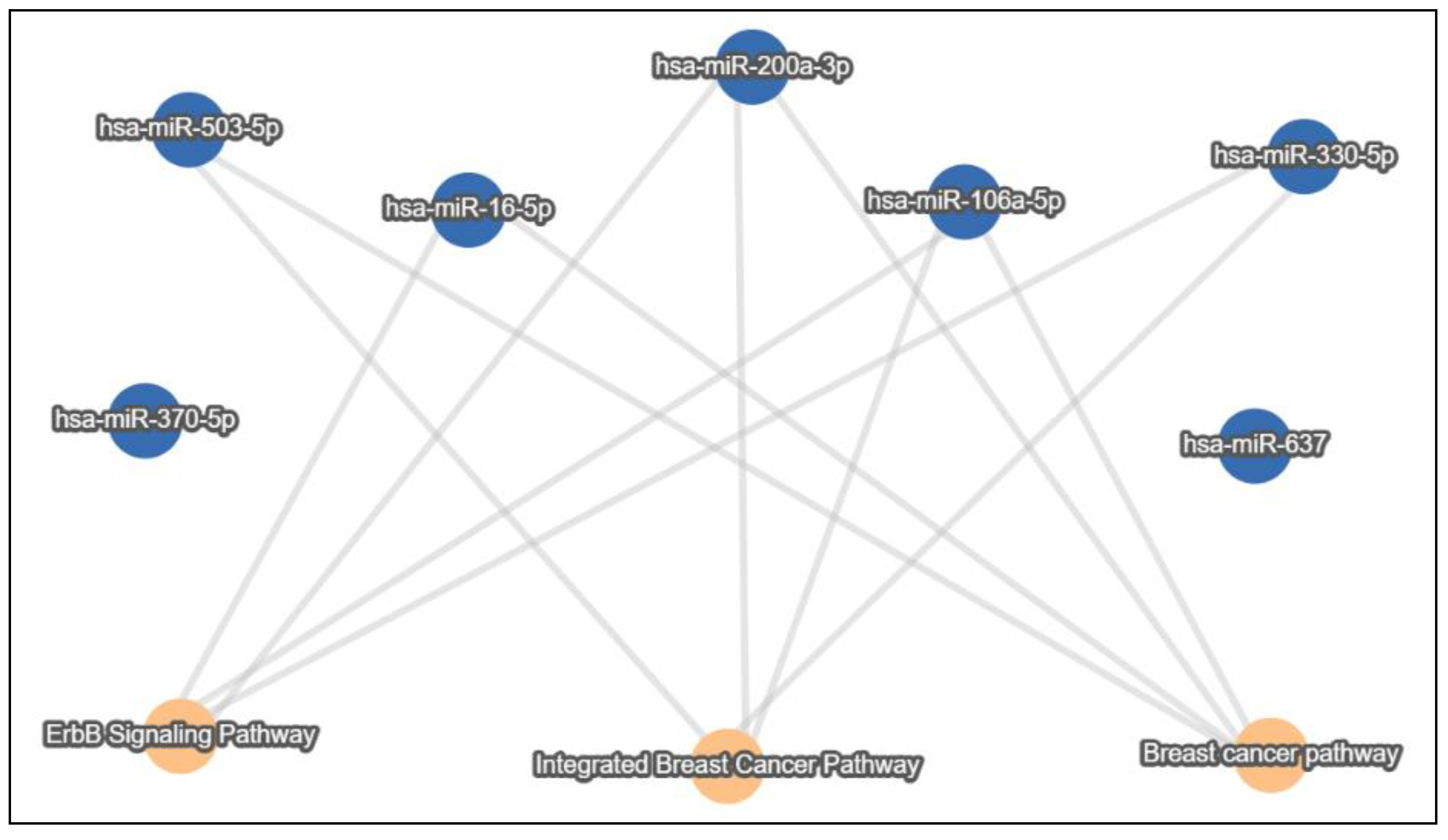
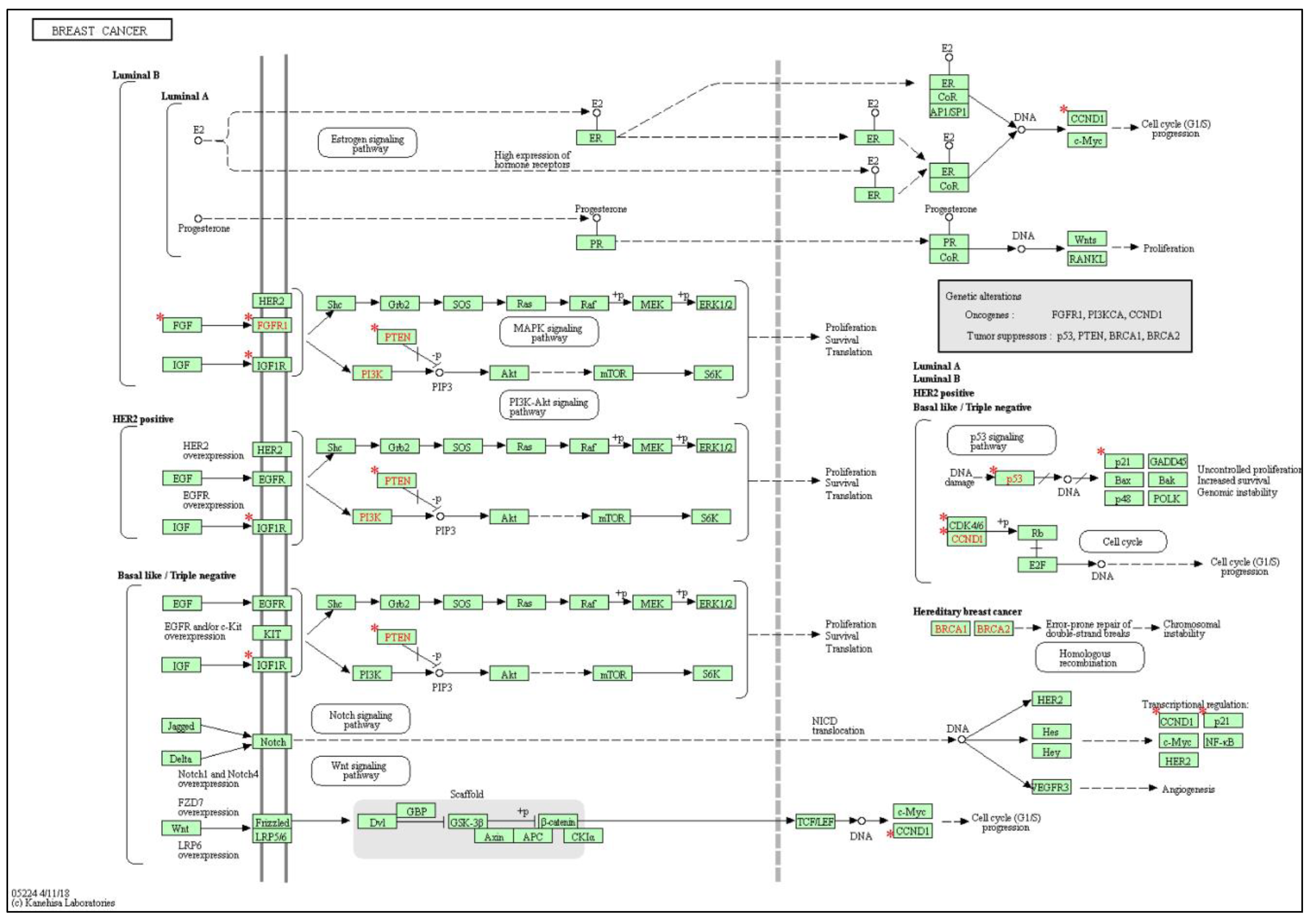
4. In Silico Analysis of lnc/circRNA-Sponged miRNAs’ Experimentally Validated Target Genes and Pathways in the BC Exosomal Axis
5. Search Strategy
6. Challenges
7. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Momenimovahed, Z.; Salehiniya, H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. Early diagnosis of breast cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef] [Green Version]
- Wong, G.L.; Abu Jalboush, S.; Lo, H.-W. Exosomal MicroRNAs and organotropism in breast cancer metastasis. Cancers 2020, 12, 1827. [Google Scholar] [CrossRef]
- Nahand, J.S.; Vandchali, N.R.; Darabi, H.; Doroudian, M.; Banafshe, H.R.; Moghoofei, M.; Babaei, F.; Salmaninejad, A.; Mirzaei, H. Exosomal microRNAs: Novel players in cervical cancer. Epigenomics 2020, 12, 1651–1660. [Google Scholar] [CrossRef]
- Zhou, R.; Chen, K.K.; Zhang, J.; Xiao, B.; Huang, Z.; Ju, C.; Sun, J.; Zhang, F.; Lv, X.-B.; Huang, G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Dixon, C.L.; Sheller-Miller, S.; Saade, G.R.; Fortunato, S.J.; Lai, A.; Palma, C.; Guanzon, D.; Salomon, C.; Menon, R. Amniotic fluid exosome proteomic profile exhibits unique pathways of term and preterm labor. Endocrinology 2018, 159, 2229–2240. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Z.; Bedi, B.; Sadikot, R.T. Bronchoalveolar lavage exosomes in lipopolysaccharide-induced septic lung injury. J. Vis. Exp. 2018, 135, e57737. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, Y.; Xiao, K.; Xiang, S.; Li, Z.; Weng, X. Emerging role of exosomes in the joint diseases. Cell. Physiol. Biochem. 2018, 47, 2008–2017. [Google Scholar] [CrossRef]
- Yoon, S.B.; Chang, J.H. Extracellular vesicles in bile: A game changer in the diagnosis of indeterminate biliary stenoses? Hepatobiliary Surg. Nutr. 2017, 6, 408. [Google Scholar] [CrossRef] [Green Version]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 47. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Gezer, U.; Özgür, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef]
- Li, Z.; Yanfang, W.; Li, J.; Jiang, P.; Peng, T.; Chen, K.; Zhao, X.; Zhang, Y.; Zhen, P.; Zhu, J.; et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018, 432, 237–250. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sang, Y.; Song, X.; Zhang, D.; Wang, L.; Zhao, W.; Liang, Y.; Zhang, N.; Yang, Q. Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 2021, 11, 3932–3947. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Hao, M.; Yeo, S.K.; Guan, J.L. FAK signaling in cancer-associated fibroblasts promotes breast cancer cell migration and metastasis by exosomal miRNAs-mediated intercellular communication. Oncogene 2020, 39, 2539–2549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, Y.; Du, J.; Lin, D.; Li, F. MiR-3613-3p from carcinoma-associated fibroblasts exosomes promoted breast cancer cell proliferation and metastasis by regulating SOCS2 expression. IUBMB Life 2020, 72, 1705–1714. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Liu, W.; Li, X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth through the Metabolic Reprogramming. Appl. Biochem. Biotechnol. 2020, 191, 1084–1099. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Gil Kim, B.; Jang, Y.; Kang, S.; Lee, J.H.; Cho, N.H. The stromal loss of miR-4516 promotes the FOSL1-dependent proliferation and malignancy of triple negative breast cancer. Cancer Lett. 2019, 469, 256–265. [Google Scholar] [CrossRef]
- Dou, D.; Ren, X.; Han, M.; Xu, X.; Ge, X.; Gu, Y.; Wang, X. Cancer-Associated Fibroblasts-Derived Exosomes Suppress Immune Cell Function in Breast Cancer via the miR-92/PD-L1 Pathway. Front. Immunol. 2020, 11, 2026. [Google Scholar] [CrossRef]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.H.; Miller, P.; Garcia-Contreras, M.; Ao, Z.; Machlin, L.; Issa, E.; El-Ashry, D. Hierarchical paracrine interaction of breast cancer associated fibroblasts with cancer cells via hMAPK-microRNAs to drive ER-negative breast cancer phenotype. Cancer Biol. Ther. 2015, 16, 1671–1681. [Google Scholar] [CrossRef] [Green Version]
- Sheykhhasan, M.; Kalhor, N.; Sheikholeslami, A.; Dolati, M.; Amini, E.; Fazaeli, H. Exosomes of Mesenchymal Stem Cells as a Proper Vehicle for Transfecting miR-145 into the Breast Cancer Cell Line and Its Effect on Metastasis. BioMed Res. Int. 2021, 2021, 5516078. [Google Scholar] [CrossRef]
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.-H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef] [Green Version]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef]
- Jung, K.O.; Youn, H.; Lee, C.-H.; Kang, K.W.; Chung, J.-K. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget 2017, 8, 9899–9910. [Google Scholar] [CrossRef] [Green Version]
- Xing, F.; Liu, Y.; Wu, S.-Y.; Wu, K.; Sharma, S.; Mo, Y.-Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2021, 78, 4316–4330, Erratum in Cancer Res. 2021, 81, 5582. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Liu, H.; Jiang, J.; Yang, Y.; Wang, W.; Jia, Z. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging 2021, 13, 18978–18992. [Google Scholar] [CrossRef]
- Chen, T.; Wang, X.; Li, C.; Zhang, H.; Liu, Y.; Han, D.; Li, Y.; Li, Z.; Luo, D.; Zhang, N.; et al. CircHIF1A regulated by FUS accelerates triple-negative breast cancer progression by modulating NFIB expression and translocation. Oncogene 2021, 40, 2756–2771. [Google Scholar] [CrossRef]
- Wang, X.; Sun, C.; Huang, X.; Li, J.; Fu, Z.; Li, W.; Yin, Y. The advancing roles of exosomes in breast cancer. Front. Cell Dev. Biol. 2021, 9, 2924. [Google Scholar] [CrossRef]
- Shen, S.; Song, Y.; Zhao, B.; Xu, Y.; Ren, X.; Zhou, Y.; Sun, Q. Cancer-derived exosomal miR-7641 promotes breast cancer progression and metastasis. Cell Commun. Signal. 2021, 19, 20. [Google Scholar] [CrossRef]
- Wang, B.; Mao, J.H.; Wang, B.Y.; Wang, L.X.; Wen, H.Y.; Xu, L.J.; Fue, J.-X.; Yang, H. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020, 489, 87–99. [Google Scholar] [CrossRef]
- Singh, R.; Pochampally, R.; Watabe, K.; Lu, Z.; Mo, Y.-Y. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 2014, 13, 256. [Google Scholar] [CrossRef] [Green Version]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-S.; Ma, S.; Dou, H.; Liu, F.; Zhang, S.-Y.; Jiang, C.; Xiao, M.; Huang, Y.-X. Breast cancer-derived exosomes regulate cell invasion and metastasis in breast cancer via miR-146a to activate cancer associated fibroblasts in tumor microenvironment. Exp. Cell Res. 2020, 391, 111983. [Google Scholar] [CrossRef]
- Gorczynski, R.M.; Zhu, F.; Chen, Z.; Kos, O.; Khatri, I. A comparison of serum miRNAs influencing metastatic growth of EMT6 vs 4THM tumor cells in wild-type and CD200R1KO mice. Breast Cancer Res. Treat. 2017, 162, 255–266. [Google Scholar] [CrossRef]
- Baroni, S.; Romero-Cordoba, S.L.; Plantamura, I.; Dugo, M.; D’Ippolito, E.; Cataldo, A.; Cosentino, G.; Angeloni, V.; Rossini, A.; Daidone, M.G.; et al. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016, 7, e2312. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Na-Er, A.; Xu, Y.-Y.; Liu, Y.-H.; Gan, Y.-J. Upregulation of serum exosomal SUMO1P3 predicts unfavorable prognosis in triple negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 154–160. [Google Scholar]
- Du, J.; Fan, J.-J.; Dong, C.; Li, H.-T.; Ma, B.-L. Inhibition effect of exosomes-mediated Let-7a on the development and metastasis of triple negative breast cancer by down-regulating the expression of c-Myc. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5301–5314. [Google Scholar]
- Park, E.J.; Jung, H.J.; Choi, H.J.; Jang, H.J.; Park, H.J.; Nejsum, L.N.; Kwon, T.H. Exosomes co-expressing AQP5-targeting miRNAs and IL-4 receptor-binding peptide inhibit the migration of human breast cancer cells. Faseb J. 2020, 34, 3379–3398. [Google Scholar] [CrossRef]
- Wei, Y.; Li, M.; Cui, S.; Wang, D.; Zhang, C.-Y.; Zen, K.; Li, L. Shikonin inhibits the proliferation of human breast cancer cells by reducing tumor-derived exosomes. Molecules 2016, 21, 777. [Google Scholar] [CrossRef] [PubMed]
- Le Pape, F.; Vargas, G.; Clézardin, P. The role of osteoclasts in breast cancer bone metastasis. J. Bone Oncol. 2016, 5, 93–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Feng, J.; Lyu, F.; Xing, F.; Sharma, S.; Liu, Y.; Wu, S.-Y.; Zhao, D.; Tyagi, A.; Deshpande, R.P.; et al. Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nat. Commun. 2021, 12, 5196. [Google Scholar] [CrossRef]
- Xun, J.; Du, L.; Gao, R.; Shen, L.; Wang, D.; Kang, L.; Chen, C.; Zhang, Z.; Zhang, Y.; Yue, S.; et al. Cancer-derived exosomal miR-138-5p modulates polarization of tumor-associated macrophages through inhibition of KDM6B. Theranostics 2021, 11, 6847–6859. [Google Scholar] [CrossRef]
- Feng, T.; Zhang, P.; Sun, Y.; Wang, Y.; Tong, J.; Dai, H.; Hua, Z. High throughput sequencing identifies breast cancer-secreted exosomal LncRNAs initiating pulmonary pre-metastatic niche formation. Gene 2019, 710, 258–264. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, L.; Li, L.; Cao, Y. Exosomes Derived from Brain Metastatic Breast Cancer Cells Destroy the Blood-Brain Barrier by Carrying lncRNA GS1-600G8.5. Biomed. Res. Int. 2020, 2020, 7461727. [Google Scholar] [CrossRef]
- Yang, S.J.; Wang, D.D.; Zhong, S.L.; Chen, W.Q.; Wang, F.L.; Zhang, J.; Xu, W.-X.; Xu, D.; Zhang, Q.; Li, J.; et al. Tumor-derived exosomal circPSMA1 facilitates the tumorigenesis, metastasis, and migration in triple-negative breast cancer (TNBC) through miR-637/Akt1/β-catenin (cyclin D1) axis. Cell Death Dis. 2021, 12, 420. [Google Scholar] [CrossRef]
- Yao, X.; Tu, Y.; Xu, Y.; Guo, Y.; Yao, F.; Zhang, X. Endoplasmic reticulum stress-induced exosomal miR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J. Cell. Mol. Med. 2020, 24, 9560–9573. [Google Scholar] [CrossRef]
- Zhou, Y.; Yamamoto, Y.; Takeshita, F.; Yamamoto, T.; Xiao, Z.; Ochiya, T. Delivery of miR-424-5p via Extracellular Vesicles Promotes the Apoptosis of MDA-MB-231 TNBC Cells in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 844. [Google Scholar] [CrossRef]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.M.; Recht, L.; Strober, S. The Promise of Targeting Macrophages in Cancer Therapy. Clin. Cancer Res. 2017, 23, 3241–3250. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Song, X.; Li, Y.; Chen, B.; Zhao, W.; Wang, L.; Zhang, H.; Liu, Y.; Han, D.; Zhang, N.; et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer 2020, 19, 85. [Google Scholar] [CrossRef]
- Jang, J.-Y.; Lee, J.-K.; Jeon, Y.-K.; Kim, C.-W. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer 2013, 13, 421. [Google Scholar] [CrossRef] [Green Version]
- Moradi-Chaleshtori, M.; Shojaei, S.; Mohammadi-Yeganeh, S.; Hashemi, S.M. Transfer of miRNA in tumor-derived exosomes suppresses breast tumor cell invasion and migration by inducing M1 polarization in macrophages. Life Sci. 2021, 282, 119800. [Google Scholar] [CrossRef]
- Moradi-Chaleshtori, M.; Bandehpour, M.; Heidari, N.; Mohammadi-Yeganeh, S.; Hashemi, S.M. Exosome-mediated miR-33 transfer induces M1 polarization in mouse macrophages and exerts antitumor effect in 4T1 breast cancer cell line. Int. Immunopharmacol. 2021, 90, 107198. [Google Scholar] [CrossRef]
- Moradi-Chaleshtori, M.; Bandehpour, M.; Soudi, S.; Mohammadi-Yeganeh, S.; Hashemi, S.M. In vitro and in vivo evaluation of anti-tumoral effect of M1 phenotype induction in macrophages by miR-130 and miR-33 containing exosomes. Cancer Immunol. Immunother. 2021, 70, 1323–1339. [Google Scholar] [CrossRef]
- Gajria, D.; Chandarlapaty, S. HER2-amplified breast cancer: Mechanisms of trastuzumab resistance and novel targeted therapies. Expert Rev. Anticancer Ther. 2011, 11, 263–275. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Deng, K.; Huang, J.; Zeng, R.; Zuo, J. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front. Pharmacol. 2020, 11, 592912. [Google Scholar] [CrossRef]
- Malhotra, V.; Perry, M.C. Classical chemotherapy: Mechanisms, toxicities and the therapeutic window. Cancer Biol. Ther. 2003, 2 (Suppl. 1), S2–S4. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Jin, L.-J.; Zhang, X.-Y. Exosomal miRNA-205 promotes breast cancer chemoresistance and tumorigenesis through E2F1. Aging 2021, 13, 18498–18514. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, S.; Shi, Z.; Cao, L.; Liu, J.; Pan, T.; Zhou, D.; Zhang, J. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 2021, 40, 120. [Google Scholar] [CrossRef]
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 2020, 235, 6896–6904. [Google Scholar] [CrossRef]
- Hu, K.; Liu, X.; Li, Y.; Li, Q.; Xu, Y.; Zeng, W.; Zhong, G.; Yu, C. Exosomes Mediated Transfer of Circ_UBE2D2 Enhances the Resistance of Breast Cancer to Tamoxifen by Binding to MiR-200a-3p. Med. Sci. Monit. 2020, 26, e922253. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Gu, Y.; Lu, P.; Li, J.; Cao, H.; Li, X.; Qian, X.; Yu, C.; Yang, Y.; Yang, X.; et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol. Cancer 2020, 19, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Z.; Chen, M.; Xing, P.; Yan, X.; Xie, B. Increased Expression of Exosomal AGAP2-AS1 (AGAP2 Antisense RNA 1) In Breast Cancer Cells Inhibits Trastuzumab-Induced Cell Cytotoxicity. Med. Sci. Monit. 2019, 25, 2211–2220. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Y.; Ye, M.; Wu, J.; Ma, L.; Chen, H. Cisplatin-resistant MDA-MB-231 Cell-derived Exosomes Increase the Resistance of Recipient Cells in an Exosomal miR-423-5p-dependent Manner. Curr. Drug Metab. 2019, 20, 804–814. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, K.; Tang, Y.; Li, Z.; Zou, T.; Liu, D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J. Biosci. 2019, 44, 37. [Google Scholar] [CrossRef]
- Santos, J.C.; da Silva Lima, N.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Rong, Y.; Teng, Y.; Zhuang, X.; Samykutty, A.; Mu, J.; Zhang, L.; Cao, P.; Yan, J.; Miller, D.; et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 2017, 36, 639–651. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.-D.; Wu, Y.; Zhang, X.-H.; Lv, M.-M.; Chen, W.-X.; Chen, X.; Yang, S.-J.; Shen, H.; Zhong, S.-L.; Tang, J.-H.; et al. Exosomes from adriamycin-resistant breast cancer cells transmit drug resistance partly by delivering miR-222. Tumor Biol. 2016, 37, 3227–3235. [Google Scholar] [CrossRef]
- Xu, C.G.; Yang, M.F.; Ren, Y.Q.; Wu, C.H.; Wang, L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4362–4368. [Google Scholar]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- Chen, W.-X.; Cai, Y.-Q.; Lv, M.-M.; Chen, L.; Zhong, S.; Ma, T.-F.; Zhao, J.-H.; Tang, J.-H. Exosomes from docetaxel-resistant breast cancer cells alter chemosensitivity by delivering microRNAs. Tumour Biol. 2014, 35, 9649–9659. [Google Scholar] [CrossRef]
- Bliss, S.A.; Sinha, G.; Sandiford, O.A.; Williams, L.M.; Engelberth, D.J.; Guiro, K.; Isenalumhe, L.L.; Greco, S.J.; Ayer, S.; Bryan, M.; et al. Mesenchymal Stem Cell-Derived Exosomes Stimulate Cycling Quiescence and Early Breast Cancer Dormancy in Bone Marrow. Cancer Res. 2016, 76, 5832–5844. [Google Scholar] [CrossRef] [Green Version]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.-U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Yu, G.; Sun, Z.; Wang, T.; Tian, X.; Duan, X.; Zhang, C. Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother. Pharmacol. 2020, 86, 761–772. [Google Scholar] [CrossRef]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef]
- Andreeva, O.E.; Sorokin, D.V.; Mikhaevich, E.I.; Bure, I.V.; Shchegolev, Y.Y.; Nemtsova, M.V.; Gudkova, M.V.; Scherbakov, A.M.; Krasil’Nikov, M.A. Towards Unravelling the Role of ERα-Targeting miRNAs in the Exosome-Mediated Transferring of the Hormone Resistance. Molecules 2021, 26, 6661. [Google Scholar] [CrossRef]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O’Driscoll, L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef] [Green Version]
- Bao, S.; Hu, T.; Liu, J.; Su, J.; Sun, J.; Ming, Y.; Li, J.; Wu, N.; Chen, H.; Zhou, M. Genomic instability-derived plasma extracellular vesicle-microRNA signature as a minimally invasive predictor of risk and unfavorable prognosis in breast cancer. J. Nanobiotechnol. 2021, 19, 22. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Xu, F.; Wang, Y.; Leng, D. Detection significance of miR-3662, miR-146a, and miR-1290 in serum exosomes of breast cancer patients. J. Cancer Res. Ther. 2021, 17, 749–755. [Google Scholar] [CrossRef]
- Hirschfeld, M.; Rücker, G.; Weiß, D.; Berner, K.; Ritter, A.; Jäger, M.; Erbes, T. Urinary Exosomal MicroRNAs as Potential Non-invasive Biomarkers in Breast Cancer Detection. Mol. Diagn. Ther. 2020, 24, 215–232. [Google Scholar] [CrossRef]
- Li, D.; Wang, J.; Ma, L.J.; Yang, H.B.; Jing, J.F.; Jia, M.M.; Zhang, X.-J.; Guo, F.; Gao, J.-N. Identification of serum exosomal miR-148a as a novel prognostic biomarker for breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7303–7309. [Google Scholar]
- Lv, S.; Wang, Y.; Xu, W.; Dong, X. Serum Exosomal miR-17-5p as a Promising Biomarker Diagnostic Biomarker for Breast Cancer. Clin. Lab. 2020, 66, 9. [Google Scholar] [CrossRef]
- Wang, J.; Ma, G.; Han, X.; Liang, M.; Wang, X.; Xia, T.; Wang, S. The low expression of miR-1976 in plasma samples indicating its biological functions in the progression of breast cancer. Clin. Transl. Oncol. 2020, 22, 2111–2120. [Google Scholar] [CrossRef]
- Ando, W.; Kikuchi, K.; Uematsu, T.; Yokomori, H.; Takaki, T.; Sogabe, M.; Kohgo, Y.; Otori, K.; Ishikawa, S.; Okazaki, I. Novel breast cancer screening: Combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci. Rep. 2019, 9, 13595. [Google Scholar] [CrossRef]
- Li, M.; Zou, X.; Xia, T.; Wang, T.; Liu, P.; Zhou, X.; Wang, S.; Zhu, W. A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 2019, 8, 7006–7017. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Martínez, A.; De Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef]
- Ni, Q.; Stevic, I.; Pan, C.; Müller, V.; Oliviera-Ferrer, L.; Pantel, K.; Schwarzenbach, H. Different signatures of miR-16, miR-30b and miR-93 in exosomes from breast cancer and DCIS patients. Sci. Rep. 2018, 8, 12974. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.L.; Liu, L.C.; Hung, Y.; Chen, C.J.; Lin, Y.Z.; Wu, W.R.; Wang, S.C. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast 2019, 46, 64–69. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.-Q.; Chen, W.-Z.; Jiang, J.-X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.-B.; Xia, W.-J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Yang, S.; Wang, D.-D.; Zhou, S.-Y.; Zhang, Q.; Wang, J.-Y.; Zhong, S.-L.; Zhang, H.-D.; Wang, X.-Y.; Xia, X.; Chen, W.; et al. Identification of circRNA–miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics 2020, 12, 101–125. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E.; et al. miRTargetLink 2.0-interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.-P.; Keller, A. miRPathDB 2.0: A novel release of the miRNA Pathway Dictionary Database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- Xing, L.; Tang, X.; Wu, K.; Huang, X.; Yi, Y.; Huan, J. LncRNA HAND2-AS1 suppressed the growth of triple negative breast cancer via reducing secretion of MSCs derived exosomal miR-106a-5p. Aging 2020, 13, 424–436. [Google Scholar] [CrossRef]
- Wang, X.; Ji, C.; Hu, J.; Deng, X.; Zheng, W.; Yu, Y.; Hua, K.; Zhou, X.; Fang, L. Hsa_circ_0005273 facilitates breast cancer tumorigenesis by regulating YAP1-hippo signaling pathway. J. Exp. Clin. Cancer Res. 2021, 40, 29. [Google Scholar] [CrossRef]
- Servetto, A.; Kollipara, R.; Formisano, L.; Lin, C.-C.; Lee, K.-M.; Sudhan, D.R.; Gonzalez-Ericsson, P.I.; Chatterjee, S.; Guerrero-Zotano, A.; Mendiratta, S.; et al. Nuclear FGFR1 Regulates Gene Transcription and Promotes Antiestrogen Resistance in ER(+) Breast Cancer. Clin. Cancer Res. 2021, 27, 4379–4396. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, A.; Bao, P.-P.; Lin, L.; Wang, Y.; Wu, H.; Shu, X.-O.; Liu, A.; Cai, Q. MicroRNA-374b inhibits breast cancer progression through regulating CCND1 and TGFA genes. Carcinogenesis 2021, 42, 528–536. [Google Scholar] [CrossRef]
- Jia, H.; Wu, D.; Zhang, Z.; Li, S. Regulatory effect of the MAFG-AS1/miR-150-5p/MYB axis on the proliferation and migration of breast cancer cells. Int. J. Oncol. 2021, 58, 33–44. [Google Scholar] [CrossRef]
- Fallah, Y.; Demas, D.M.; Jin, L.; He, W.; Shajahan-Haq, A.N. Targeting WEE1 Inhibits Growth of Breast Cancer Cells That Are Resistant to Endocrine Therapy and CDK4/6 Inhibitors. Front. Oncol. 2021, 11, 681530. [Google Scholar] [CrossRef]
- DiGiacomo, J.W.; Godet, I.; Trautmann-Rodriguez, M.; Gilkes, D.M. Extracellular Matrix-Bound FGF2 Mediates Estrogen Receptor Signaling and Therapeutic Response in Breast Cancer. Mol. Cancer Res. 2021, 19, 136–149. [Google Scholar] [CrossRef]
- Akhter, N.; A Dar, S.; Haque, S.; Wahid, M.; Jawed, A.; Akhtar, M.S.; Alharbi, R.A.; Sindi, A.; Alruwetei, A.; Choudhry, H.M.Z.; et al. Crosstalk of Cyclin-dependent kinase inhibitor 1A (CDKN1A) gene polymorphism with p53 and CCND1 polymorphism in breast cancer. Eur. Rev. Med Pharmacol. Sci. 2021, 25, 4258–4273. [Google Scholar]
- Zhang, Q.; Li, T.; Wang, Z.; Kuang, X.; Shao, N.; Lin, Y. lncRNA NR2F1-AS1 promotes breast cancer angiogenesis through activating IGF-1/IGF-1R/ERK pathway. J. Cell. Mol. Med. 2020, 24, 8236–8247. [Google Scholar] [CrossRef]
- Jin, X.; Ge, L.P.; Li, D.Q.; Shao, Z.M.; Di, G.H.; Xu, X.E.; Jiang, Y.Z. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Mol. Cancer 2020, 19, 87. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. 2019, 38, 173. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Pang, J.-S.; Sun, Q.; Huang, Y.; Hou, J.-Y.; Chen, G.; Zeng, J.-J.; Feng, Z.-B. The clinical significance of CHEK1 in breast cancer: A high-throughput data analysis and immunohistochemical study. Int. J. Clin. Exp. Pathol. 2019, 12, 1–20. [Google Scholar]
- Turner, N.C.; Liu, Y.; Zhu, Z.; Loi, S.; Colleoni, M.; Loibl, S.; DeMichele, A.; Harbeck, N.; André, F.; Bayar, M.A.; et al. Cyclin E1 Expression and Palbociclib Efficacy in Previously Treated Hormone Receptor-Positive Metastatic Breast Cancer. J. Clin. Oncol. 2019, 37, 1169–1178. [Google Scholar] [CrossRef]
- Hong, T.; Ding, J.; Li, W. miR-7 Reverses Breast Cancer Resistance to Chemotherapy by Targeting MRP1 and BCL2. Onco Targets Ther. 2019, 12, 11097–11105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Geng, D.; Li, S.; Chen, Z.; Sun, M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018, 7, 842–855. [Google Scholar] [CrossRef] [PubMed]
- Huszno, J.; Grzybowska, E. TP53 mutations and SNPs as prognostic and predictive factors in patients with breast cancer. Oncol. Lett. 2018, 16, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tormo, E.; Adam-Artigues, A.; Ballester, S.; Pineda, B.; Zazo, S.; González-Alonso, P.; Albanell, J.; Rovira, A.; Rojo, F.; Lluch, A.; et al. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci. Rep. 2017, 7, 41309. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhfyan, A.; Alhoshani, A.; Korashy, H.M. Aryl hydrocarbon receptor/cytochrome P450 1A1 pathway mediates breast cancer stem cells expansion through PTEN inhibition and β-Catenin and Akt activation. Mol. Cancer 2017, 16, 1–18. [Google Scholar]
- Veenstra, C.; Pérez-Tenorio, G.; Stelling, A.; Karlsson, E.; Mirwani, S.M.; Nordensköljd, B.; Fornander, T.; Stål, O. Met and its ligand HGF are associated with clinical outcome in breast cancer. Oncotarget 2016, 7, 37145–37159. [Google Scholar] [CrossRef]
- Oi, R.; Koizumi, H.; Maeda, I.; Noguchi, A.; Tatsunami, S.; Iwatani, T.; Kawamoto, H.; Tsugawa, K.; Takagi, M. Clinicopathological Significance of TARBP2, APP, and ZNF395 in Breast Cancer. Breast Cancer 2016, 10, 211–221. [Google Scholar] [CrossRef]
- Justenhoven, C.; Pierl, C.B.; Haas, S.; Fischer, H.-P.; Hamann, U.; Baisch, C.; Harth, V.; Spickenheuer, A.; Rabstein, S.; Vollmert, C.; et al. Polymorphic loci of E2F2, CCND1 and CCND3 are associated with HER2 status of breast tumors. Int. J. Cancer 2009, 124, 2077–2081. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists. Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research—Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [Green Version]
- Baassiri, A.; Nassar, F.; Mukherji, D.; Shamseddine, A.; Nasr, R.; Temraz, S. Exosomal non coding RNA in LIQUID biopsies as a promising biomarker for colorectal cancer. Int. J. Mol. Sci. 2020, 21, 1398. [Google Scholar] [CrossRef] [Green Version]
- Nie, H.; Liao, Z.; Wang, Y.; Zhou, J.; He, X.; Ou, C. Exosomal long non-coding RNAs: Emerging players in cancer metastasis and potential diagnostic biomarkers for personalized oncology. Genes Dis. 2021, 8, 769–780. [Google Scholar] [CrossRef]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating methods for isolation and quantification of exosomes: A review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef]
- Hosseini, K.; Ranjbar, M.; Tazehkand, A.P.; Asgharian, P.; Montazersaheb, S.; Tarhriz, V.; Ghasemnejad, T. Evaluation of exosomal non-coding RNAs in cancer using high-throughput sequencing. J. Transl. Med. 2022, 20, 1–15. [Google Scholar] [CrossRef]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [Green Version]
- Nassar, F.; Nasr, R.; Talhouk, R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol. Ther. 2017, 172, 34–49. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [Green Version]

| Noncoding RNA | Source | Biomarker Type | Status | Clinical Evidence | Mechanism | Reference |
|---|---|---|---|---|---|---|
| miR-421, miR128-1, miR128-2 | plasma | Diagnostic/Prognostic | Upregulated | Yes | N/A | [89] |
| miR-3662, miR-146a, miR-1290 | serum | Predictive/Diagnostic | Upregulated | Yes | N/A | [90] |
| miR-424, miR-423, miR-660, let7-i | urine | Diagnostic | Up, down, down, downregulated | Yes | N/A | [91] |
| miR-148a | serum | Diagnostic/Prognostic | Downregulated | Yes | N/A | [92] |
| miR-17-5P | serum | Diagnostic | Downregulated | Yes | N/A | [93] |
| miR-1246, miR-155 | serum | Predictive/Prognostic | Upregulated | Yes | N/A | [85] |
| miR-1910-3p | serum | Diagnostic | Upregulated | Yes | downregulated myotubularin-related protein 3, activated the NF-κB and wnt/β-catenin signaling pathway, and promoted breast cancer progression | [39] |
| miR-1976 | plasma, tissues | Diagnostic | Downregulated | Yes | N/A | [94] |
| miR-21 (with MMP1) | urine | Diagnostic | Downregulated, Upregulated | Yes | [95] | |
| Let-7b-5p, miR-122-5p, miR-146b-5p, miR-210-3p, miR-215-5p | plasma | Diagnostic | N/A | No | N/A | [96] |
| miR-21, miR-222, miR-155 | serum | Diagnostic/Predictive | Upregulated | Yes | N/A | [97] |
| miR-16, miR-30b, miR-93 | serum, plasma | Diagnostic | Up, down, upregulated | Yes | N/A | [98] |
| miR-106a-3p, miR-106a-5p, miR-20b-5p, miR-92a-2-5p | plasma, serum | Diagnostic | Upregulated | Yes | N/A | [99] |
| miR-1246 | serum | Diagnostic | Upregulated | No | Suppresses the expression of cyclin-G2 (CCNG2) | [32] |
| miR-1246, miR-21 | plasma | Diagnostic | Upregulated | Yes | N/A | [100] |
| SNHG14 | serum | Diagnostic | Upregulated | Yes | N/A | [77] |
| HOTAIR | serum | Diagnostic/Prognostic | Upregulated | Yes | N/A | [75] |
| HOTAIR | plasma | Diagnostic/Prognostic | Upregulated | Yes | Positively correlated with ERBB2/HER2 expression | [101] |
| AFAP1-AS1 | serum | Diagnostic/Prognostic | Upregulated | Yes | Promotes ERBB2 translation via AUF1 binding | [72] |
| SNHG16 | peripheral blood | Prognostic | Upregulated | Yes | Promotes CD73 expression on γδ1 T cells via the TGF-β1/SMAD5 pathway, enabled via miR-16-5p sponging | [102] |
| H19 | serum | Diagnostic/Prognostic | Upregulated | Yes | N/A | [70] |
| SUMO1P3 | serum | Diagnostic/Prognostic | Upregulated | Yes | N/A | [46] |
| circFOXK2 | tissues | Diagnostic | Upregulated | No | Acts with IGF2BP3 and miR370 | [35] |
| circPSMA1 | serum | Prognostic | Upregulated | Yes | circPSMA1 sponges miR-637, activating Akt1-β-catenin (Cyclin D1) signaling | [55] |
| hsa-circRNA-0005795, hsa-circRNA-0088088 | serum | Diagnostic | Downregulated, Upregulated | Yes | N/A | [103] |
| Biological Process | ||
|---|---|---|
| GO Term | Gene Count | FDR |
| negative regulation of transcription from RNA polymerase II promoter | 10 | 4.70 × 10−5 |
| regulation of cell cycle | 7 | 4.70 × 10−5 |
| regulation of cyclin-dependent protein serine/threonine kinase activity | 5 | 4.70 × 10−5 |
| positive regulation of gene expression | 8 | 4.70 × 10−5 |
| negative regulation of G1/S transition of mitotic cell cycle | 5 | 4.70 × 10−5 |
| positive regulation of MAPK cascade | 6 | 4.70 × 10−5 |
| negative regulation of apoptotic process | 8 | 4.70 × 10−5 |
| cytokine-mediated signaling pathway | 7 | 4.70 × 10−5 |
| G1/S transition of mitotic cell cycle | 5 | 6.50 × 10−5 |
| positive regulation of protein kinase B signaling | 6 | 8.00 × 10−5 |
| cell division | 7 | 8.80 × 10−5 |
| positive regulation of protein phosphorylation | 6 | 8.80 × 10−5 |
| positive regulation of phosphatidylinositol 3-kinase signaling | 5 | 1.10 × 10−4 |
| cellular response to DNA damage stimulus | 6 | 2.40 × 10−4 |
| negative regulation of cell proliferation | 6 | 3.00 × 10−3 |
| positive regulation of transcription from RNA polymerase II promoter | 8 | 4.00 × 10−3 |
| protein phosphorylation | 6 | 4.30 × 10−3 |
| positive regulation of cell proliferation | 6 | 5.20 × 10−3 |
| response to drug | 5 | 5.40 × 10−3 |
| negative regulation of gene expression | 5 | 7.50 × 10−3 |
| nervous system development | 5 | 1.40 × 10−2 |
| positive regulation of transcription, DNA-templated | 5 | 6.30 × 10−2 |
| Molecular Function | ||
| GO Name | Gene Count | FDR |
| protein kinase binding | 7 | 2.90 × 10−4 |
| identical protein binding | 9 | 4.40 × 10−3 |
| protein binding | 20 | 1.20 × 10−2 |
| protein serine/threonine/tyrosine kinase activity | 5 | 1.50 × 10−2 |
| Cellular Component | ||
| GO Name | Gene Count | FDR |
| cyclin-dependent protein kinase holoenzyme complex | 6 | 2.30 × 10−8 |
| nucleus | 18 | 3.20 × 10−6 |
| nucleoplasm | 14 | 1.20 × 10−4 |
| cytoplasm | 15 | 6.10 × 10−4 |
| centrosome | 5 | 2.30 × 10−2 |
| macromolecular complex | 5 | 3.90 × 10−2 |
| membrane | 8 | 5.90 × 10−2 |
| extracellular region | 6 | 3.20 × 10−1 |
| KEGG Pathway | Gene Count | FDR |
|---|---|---|
| PI3K-Akt signaling pathway | 15 | 4.40 × 10−14 |
| p53 signaling pathway | 10 | 6.60 × 10−13 |
| Pathways in cancer | 14 | 1.10 × 10−10 |
| Cell cycle | 9 | 2.00 × 10−9 |
| Cellular senescence | 9 | 8.10 × 10−9 |
| MicroRNAs in cancer | 10 | 7.50 × 10−8 |
| Breast cancer | 8 | 1.50 × 10−7 |
| Proteoglycans in cancer | 8 | 1.10 × 10−6 |
| EGFR tyrosine kinase inhibitor resistance | 6 | 2.80 × 10−6 |
| Focal adhesion | 7 | 1.40 × 10−5 |
| Endocrine resistance | 5 | 1.80 × 10−4 |
| MAPK signaling pathway | 6 | 1.00 × 10−3 |
| Rap1 signaling pathway | 5 | 2.50 × 10−3 |
| Chemical carcinogenesis-receptor activation | 5 | 2.50 × 10−3 |
| Ras signaling pathway | 5 | 3.30 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashekyan, O.; Abdallah, S.; Shoukari, A.A.; Chamandi, G.; Choubassy, H.; Itani, A.R.S.; Alwan, N.; Nasr, R. Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis. Int. J. Mol. Sci. 2022, 23, 8351. https://doi.org/10.3390/ijms23158351
Ashekyan O, Abdallah S, Shoukari AA, Chamandi G, Choubassy H, Itani ARS, Alwan N, Nasr R. Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis. International Journal of Molecular Sciences. 2022; 23(15):8351. https://doi.org/10.3390/ijms23158351
Chicago/Turabian StyleAshekyan, Ohanes, Samira Abdallah, Ayman Al Shoukari, Ghada Chamandi, Hayat Choubassy, Abdul Rahman S. Itani, Nisreen Alwan, and Rihab Nasr. 2022. "Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis" International Journal of Molecular Sciences 23, no. 15: 8351. https://doi.org/10.3390/ijms23158351
APA StyleAshekyan, O., Abdallah, S., Shoukari, A. A., Chamandi, G., Choubassy, H., Itani, A. R. S., Alwan, N., & Nasr, R. (2022). Spotlight on Exosomal Non-Coding RNAs in Breast Cancer: An In Silico Analysis to Identify Potential lncRNA/circRNA-miRNA-Target Axis. International Journal of Molecular Sciences, 23(15), 8351. https://doi.org/10.3390/ijms23158351







