Response of Anthocyanin Accumulation in Pepper (Capsicum annuum) Fruit to Light Days
Abstract
:1. Introduction
2. Results
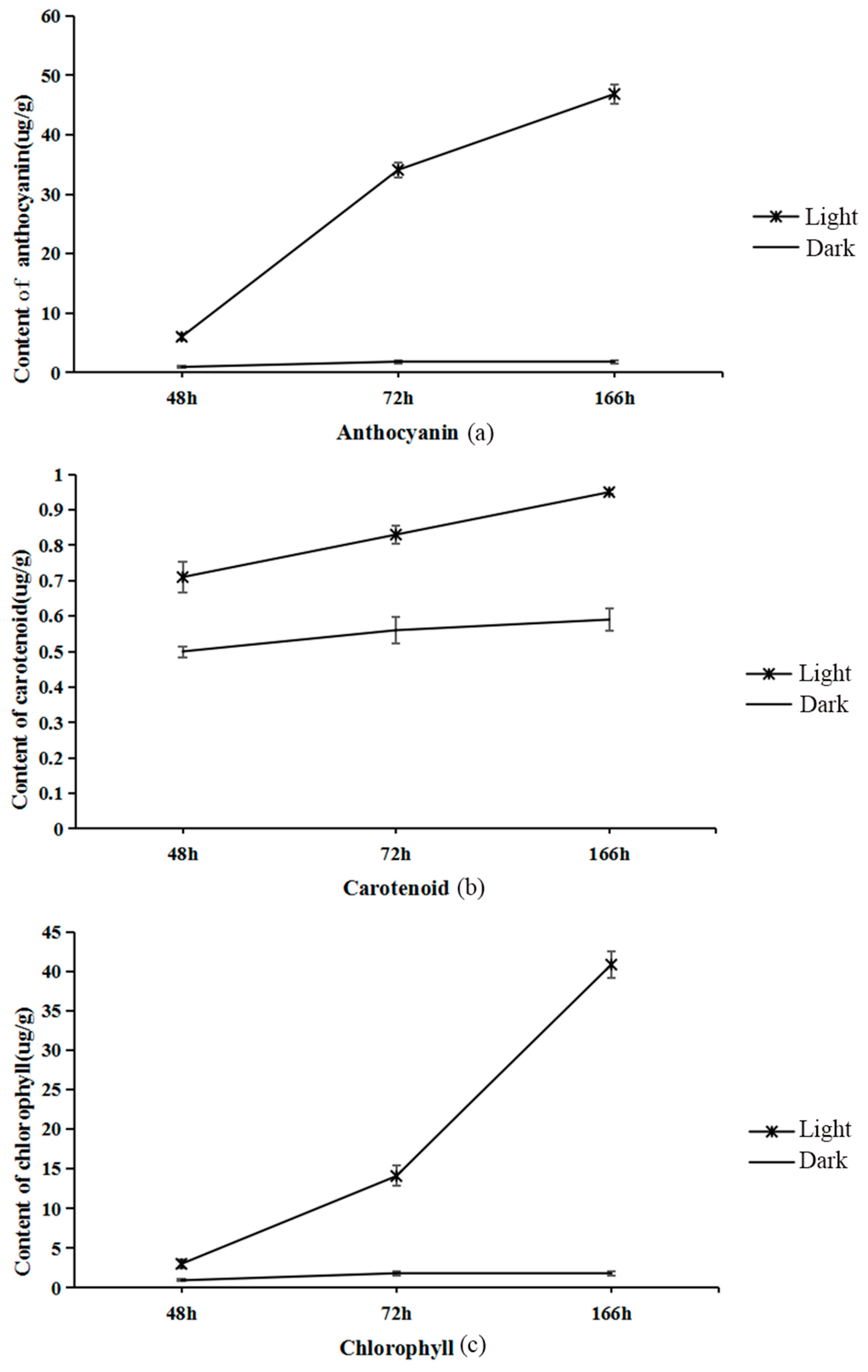
2.1. Total Anthocyanin, Chlorophyll, and Carotenoid Concentrations
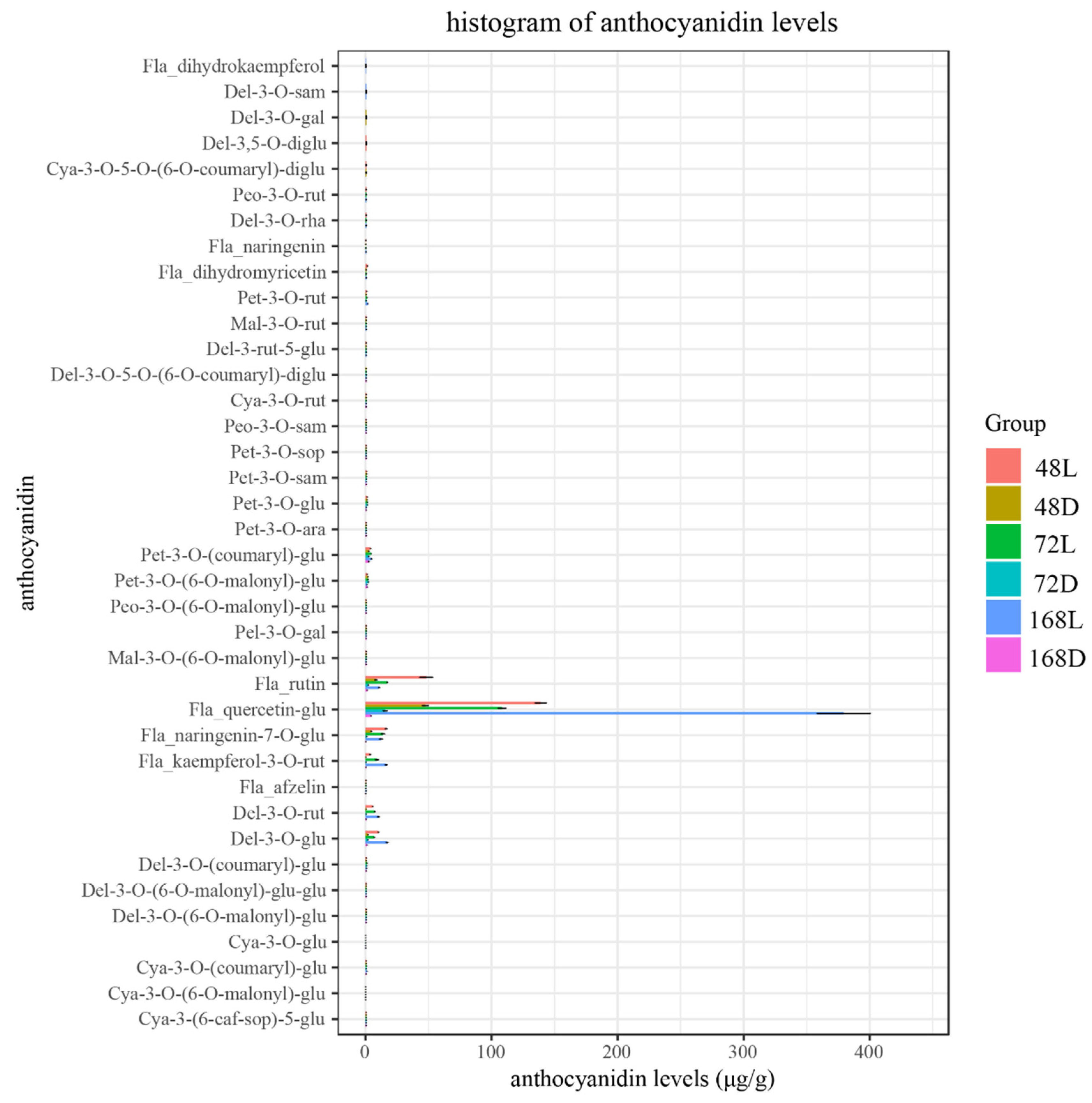
2.2. Analysis of Flavonoids in Pepper Peel Samples by Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
2.3. Analysis of Transcriptome Data Quality of Pepper Peel Samples
2.4. Comparative Statistical Analysis of Transcriptome Data of Pepper Peel Samples
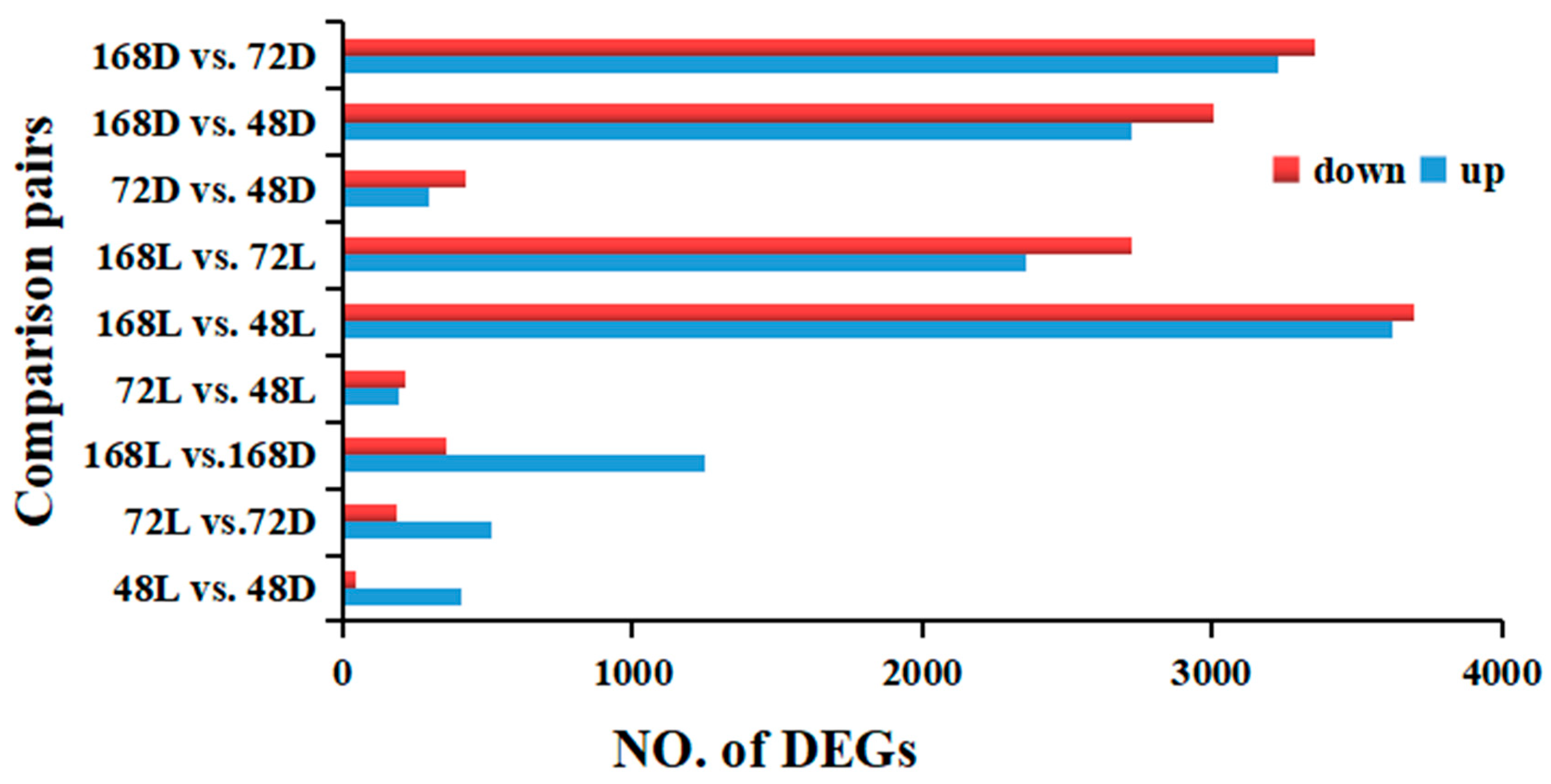
2.5. Differentially Expressed Genes (DEGs) in Pepper Peel Samples
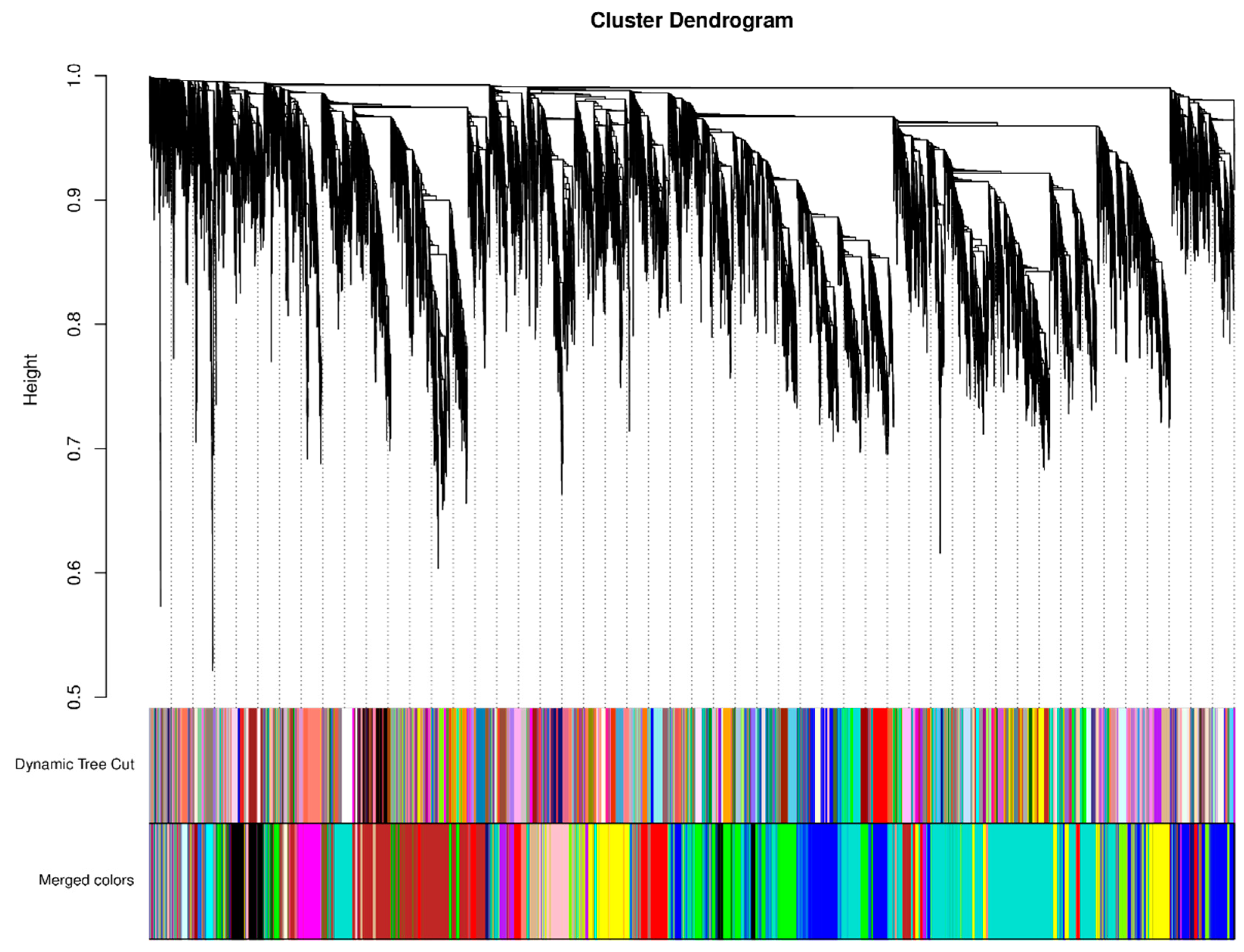
2.6. Weighted Gene Co-Expression Network Analysis (WGCNA) Module Hierarchical Clustering Tree of DEGs in Pepper Peel Samples
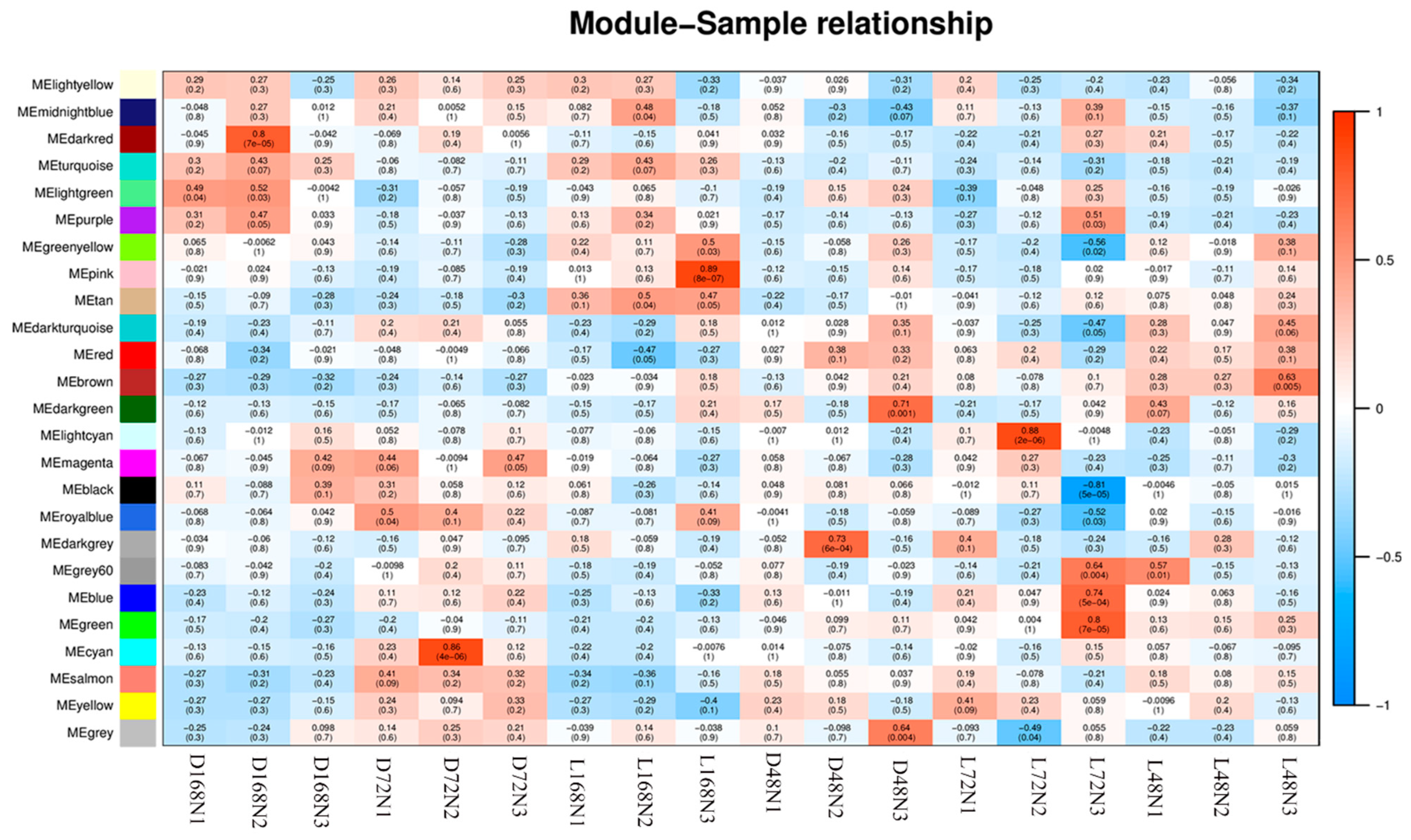
2.7. Correlation Heatmap of Pepper Peel Samples and Modules and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of Sample-Specific Modules in Different Development Stages
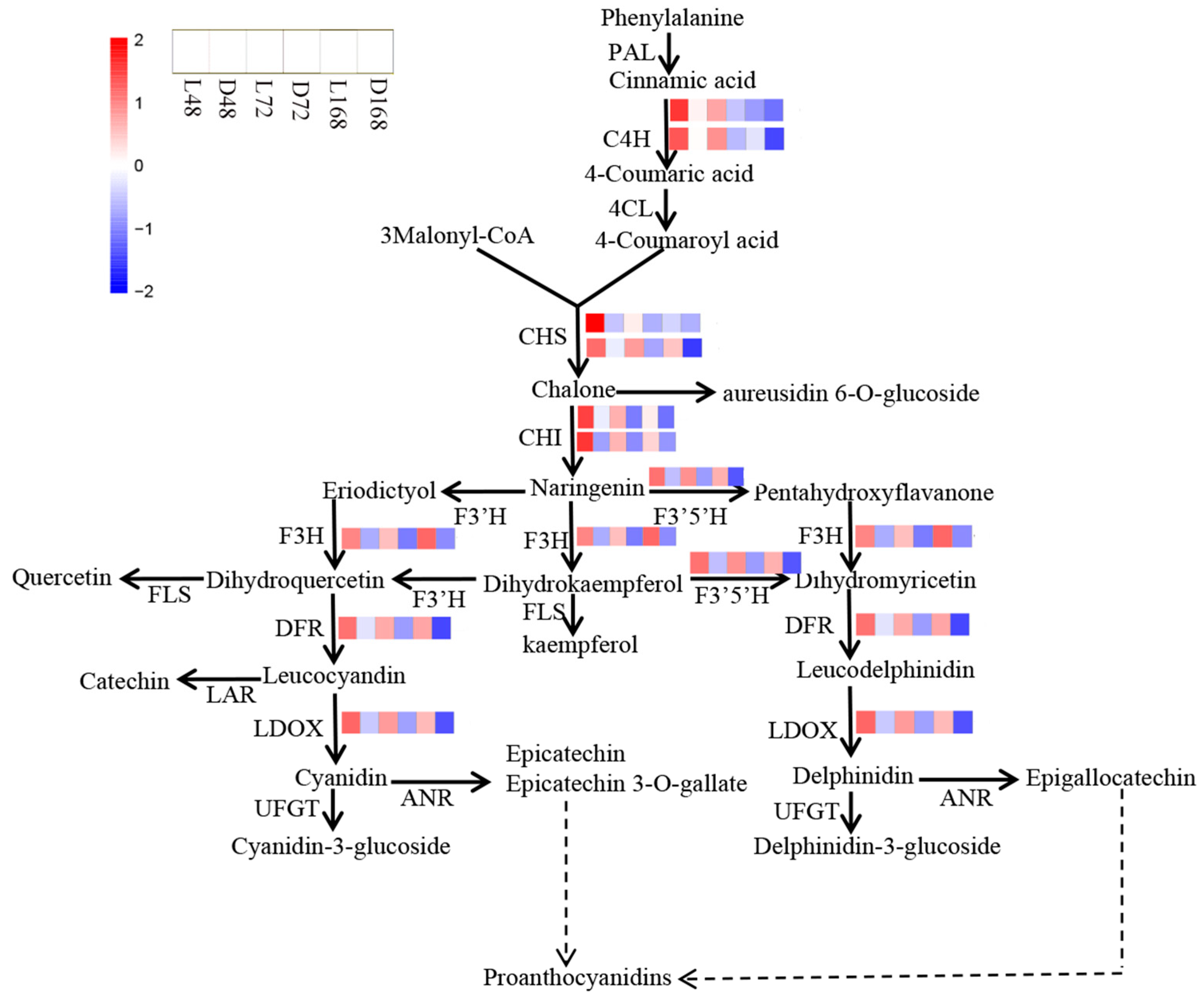
2.8. Expression of Structural Genes in the Flavonoid Synthesis Pathway in Different Periods
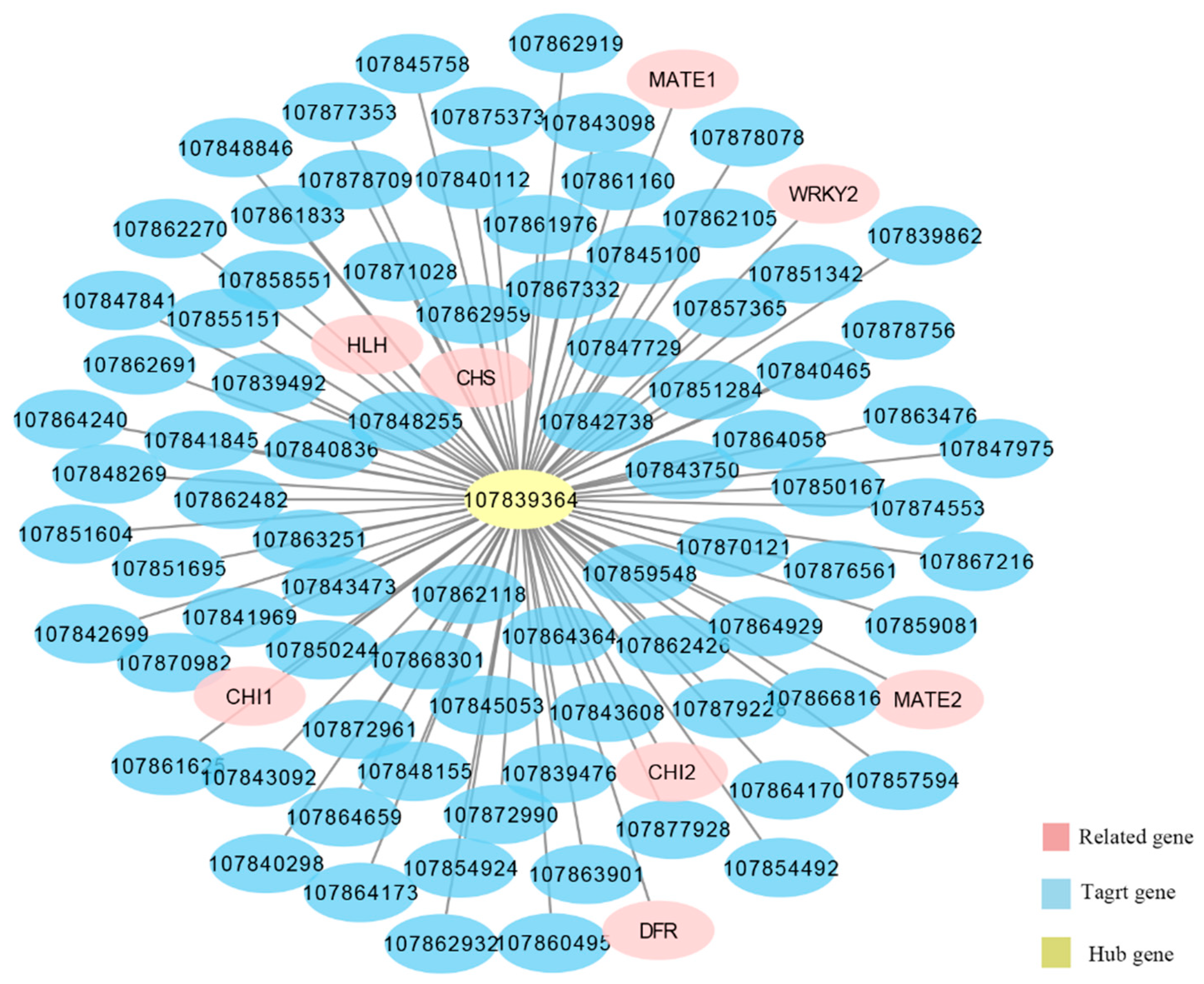
2.9. Mining of Anthocyanin-Related Light Response Genes
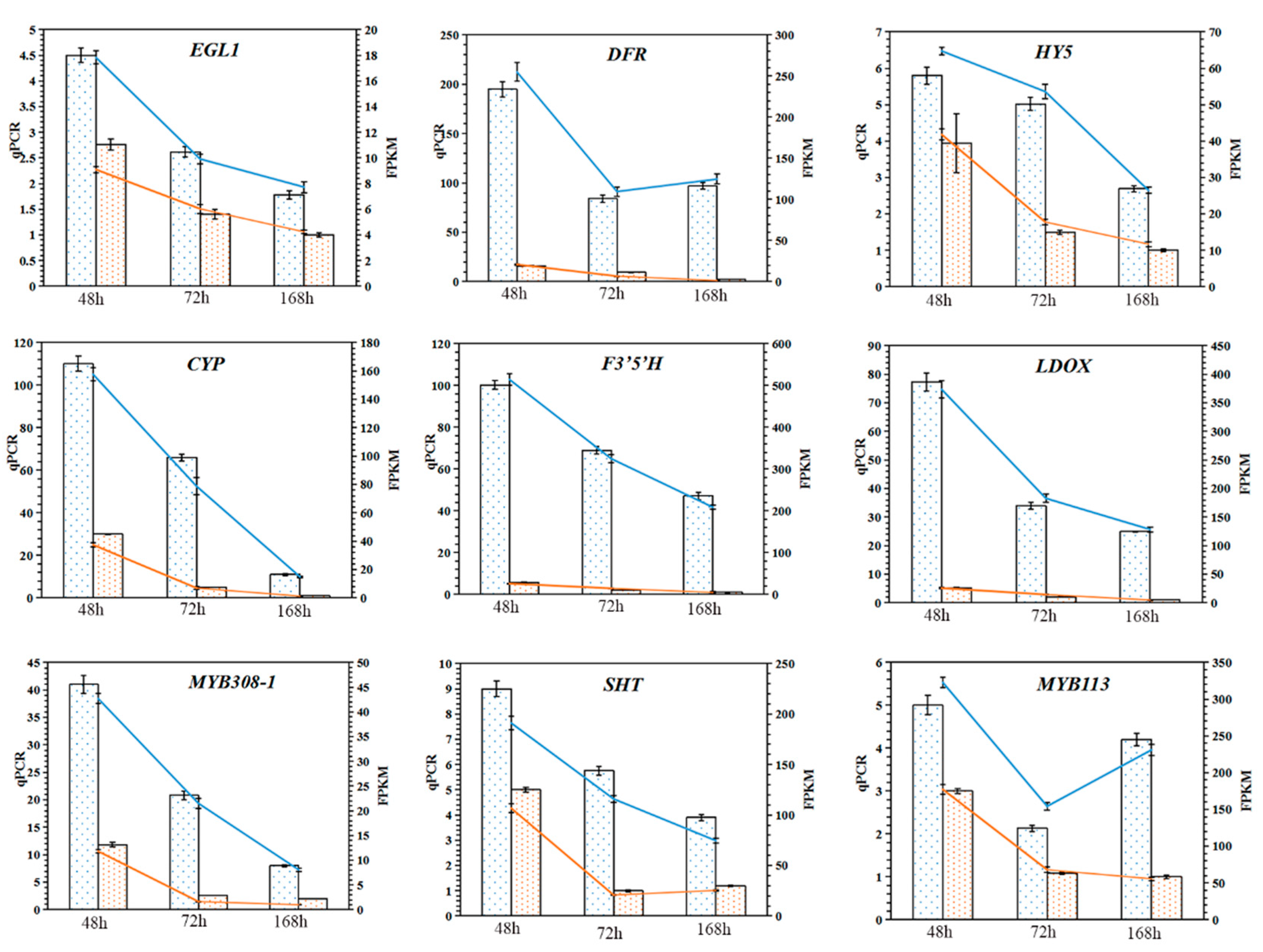
2.10. Validation of Genes Related to Anthocyanin Synthesis
3. Discussion
3.1. WGCNA Revealed Key Genes Related to Anthocyanin Synthesis
3.2. Structural Genes Related to Anthocyanin Synthesis
3.3. Regulatory Genes Related to Anthocyanin Synthesis
4. Materials and Methods
4.1. Material Preparation
4.2. Measurement of Total Anthocyanin, Chlorophyll, and Carotenoid Concentrations
4.3. Determination of Flavonoids by LC-ESI-MS/MS
4.4. RNA-Seq
4.5. Differential Expression Analysis
4.6. RNA Extraction and Quantitative Reverse Transcription PCR
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, L.; Cao, S.; Chen, W.; Yang, Z. Blue light induced anthocyanin accumulation and expression of associated genes in Chinese bayberry fruit. Sci. Hortic. 2014, 179, 98–102. [Google Scholar] [CrossRef]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Thrimawithana, A.H.; Cooney, J.M.; Jensen, D.J.; Espley, R.V.; Allan, A.C. The proanthocyanin-related transcription factors MYBC1 and WRKY44 regulate branch points in the kiwifruit anthocyanin pathway. Sci. Rep. 2020, 10, 14161. [Google Scholar] [CrossRef]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 83. [Google Scholar] [CrossRef]
- Conn, S.; Curtin, C.; Bézier, A.; Franco, C.; Zhang, W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Conejero, G.; Torregrosa, L.; Cheynier, V.; Terrier, N.; Ageorges, A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 2011, 67, 960–970. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debeaujon, I.; Peeters, A.J.; Leon-Kloosterziel, K.M.; Koornneef, M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 2001, 13, 853–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kou, M.; Liu, Y.J.; Li, Z.Y.; Zhang, Y.G.; Tang, W.; Yan, H.; Wang, X.; Chen, X.G.; Su, Z.X.; Arisha, M.H.; et al. A novel glutathione S-transferase gene from sweetpotato, IbGSTF4, is involved in anthocyanin sequestration. Plant Physiol. Biochem. 2019, 135, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, N.; Kar, D.; Deepak Mahajan, B.; Nanda, S.; Rahiman, R.; Panchakshari, N.; Bhagavatula, L.; Datta, S. The multitasking abilities of MATE transporters in plants. J. Exp. Bot. 2019, 70, 4643–4656. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [Green Version]
- Ichino, T.; Fuji, K.; Ueda, H.; Takahashi, H.; Koumoto, Y.; Takagi, J.; Tamura, K.; Sasaki, R.; Aoki, K.; Shimada, T.; et al. GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 2014, 80, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Ng, M.S.; Cheng, S.S.; Lo, A.W.; Xiao, Z.; Shin, T.S.; Chung, G.; Lam, H.M. Understanding the composition, biosynthesis, accumulation and transport of flavonoids in crops for the promotion of crops as healthy sources of flavonoids for human consumption. Nutrients 2020, 12, 1717. [Google Scholar] [CrossRef]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids--biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Emiliani, J.; Grotewold, E.; Falcone Ferreyra, M.L.; Casati, P. Flavonols protect Arabidopsis plants against UV-B deleterious effects. Mol. Plant 2013, 6, 1376–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Mu, L.; Yan, G.L.; Liang, N.N.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Rezende, F.M.; Clausen, M.H.; Rossi, M.; Furlan, C.M. The Regulation of floral colour change in Pleroma raddianum (DC.) Gardner. Molecules 2020, 25, 4664. [Google Scholar] [CrossRef] [PubMed]
- Aza-González, C.; Núñez-Palenius, H.G.; Ochoa-Alejo, N. Molecular biology of capsaicinoid biosynthesis in chili pepper (Capsicum spp.). Plant Cell Rep. 2011, 30, 695–706. [Google Scholar] [CrossRef]
- Tang, B.; Li, L.; Hu, Z.; Chen, Y.; Tan, T.; Jia, Y.; Xie, Q.; Chen, G. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple pepper. J. Agric. Food Chem. 2020, 68, 12152–12163. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Wei, K.; Cheng, H.; Wang, L.Y.; Zhang, C.C. Accumulation of catechins and expression of catechin synthetic genes in Camellia sinensis at different developmental stages. Bot. Stud. 2016, 57, 31. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Ren, Y.; Zhang, D.; Chen, X.; Huang, J.; Xu, Y.; Aucapina, C.B.; Zhang, Y.; Miao, Y. Transcriptome-based WGCNA analysis reveals regulated metabolite fluxes between floral color and scent in Narcissus tazetta Flower. Int. J. Mol. Sci. 2021, 22, 8249. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, Å.; Qian, M.; Li, J.; Li, Y.; Shen, J.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus x domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; et al. Unraveling the regulatory mechanism of color diversity in Camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Afifi, M.; Obenland, D.; El-Kereamy, A. The Complexity of modulating anthocyanin biosynthesis pathway by deficit irrigation in table grapes. Front. Plant Sci. 2021, 12, 713277. [Google Scholar] [CrossRef]
- Appelhagen, I.; Wulff-Vester, A.K.; Wendell, M.; Hvoslef-Eide, A.K.; Russell, J.; Oertel, A.; Martens, S.; Mock, H.P.; Martin, C.; Matros, A. Colour bio-factories: Towards scale-up production of anthocyanins in plant cell cultures. Metab. Eng. 2018, 48, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Shin, D.H.; Choi, S.B.; Yoo, S.D.; Choi, G.; Park, Y.I. Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells 2012, 34, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Wang, X.; Ma, B.; Wu, Z.; Zheng, L.; Qi, Z.; Wang, Y. A leucoanthocyanidin dioxygenase gene (RtLDOX2) from the feral forage plant Reaumuria trigyna promotes the accumulation of flavonoids and improves tolerance to abiotic stresses. J. Plant Res. 2021, 134, 1121–1138. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, P.; Gupta, A.P.; Bedi, Y.S.; Vishwakarma, R.A.; Gandhi, S.G. Mannitol stress directs flavonoid metabolism toward synthesis of flavones via differential regulation of two cytochrome P450 monooxygenases in Coleus forskohlii. Front. Plant Sci. 2016, 7, 985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Mumtaz, M.A.; Zhang, Y.; Yang, Z.; Hao, Y.; Shu, H.; Zhu, J.; Bao, W.; Cheng, S.; Zhu, G.; et al. Response of anthocyanin biosynthesis to light by strand-specific transcriptome and miRNA analysis in Capsicum annuum. BMC Plant Biol. 2022, 22, 79. [Google Scholar] [CrossRef]
- Du, Z.; Su, Q.; Wu, Z.; Huang, Z.; Bao, J.; Li, J.; Tu, H.; Zeng, C.; Fu, J.; He, H. Genome-wide characterization of MATE gene family and expression profiles in response to abiotic stresses in rice (Oryza sativa). BMC Ecol. Evol. 2021, 21, 141. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, K.; Shitan, N.; Yazaki, K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 2014, 31, 417–430. [Google Scholar] [CrossRef] [Green Version]
- Marinova, K.; Pourcel, L.; Weder, B.; Schwarz, M.; Barron, D.; Routaboul, J.M.; Debeaujon, I.; Klein, M. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 2007, 19, 2023–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, A.; Brown, M.; Hatlestad, G.; Akhavan, N.; Smith, T.; Hembd, A.; Moore, J.; Montes, D.; Mosley, T.; Resendez, J.; et al. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 2016, 419, 54–63. [Google Scholar] [CrossRef]
- Schwinn, K.E.; Ngo, H.; Kenel, F.; Brummell, D.A.; Albert, N.W.; McCallum, J.A.; Pither-Joyce, M.; Crowhurst, R.N.; Eady, C.; Davies, K.M. The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 2016, 7, 1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Zhu, Z.; Cao, P.; Chen, H.; Chen, C.; Zhou, X.; Mao, Y.; Lei, J.; Jiang, Y.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Hassani, D.; Fu, X.; Shen, Q.; Khalid, M.; Rose, J.K.C.; Tang, K. Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci. 2020, 25, 466–476. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Z.; Ge, H.; Liu, Y.; Chen, H. Weighted gene co-expression network analysis identifies genes related to anthocyanin biosynthesis and functional verification of hub gene SmWRKY44. Plant Sci. 2021, 309, 110935. [Google Scholar] [CrossRef]
- Zhou, Z.; Gao, H.; Ming, J.; Ding, Z.; Lin, X.; Zhan, R. Combined transcriptome and metabolome analysis of pitaya fruit unveiled the mechanisms underlying peel and pulp color formation. BMC Genom. 2020, 21, 734. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tan, L.; Zou, Y.; Tan, X.; Huang, J.; Chen, W.; Tang, Q. The effects of Ultraviolet A/B treatments on anthocyanin accumulation and gene expression in dark-purple tea cultivar ‘Ziyan’ (Camellia sinensis). Molecules 2020, 25, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.; Lam, H.M. A putative lambda class glutathione S-transferase enhances plant survival under salinity stress. Plant Cell Physiol. 2014, 55, 570–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, S.; Akita, Y.; Ishizaka, H.; Narumi, I.; Tanaka, A. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in cyclamen. J. Plant Physiol. 2012, 169, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Li, C.; Zhao, H.; Zhao, J.; Chen, H.; Bu, T.; Anhu, W.; Wu, Q. Deep sequencing of the transcriptome reveals distinct flavonoid metabolism features of black tartary buckwheat (Fagopyrum tataricum Garetn.). Prog. Biophys. Mol. Biol. 2017, 124, 49–60. [Google Scholar] [CrossRef] [PubMed]

| Sample | Clean_Reads | Clean_Bases | Q30 | GC_pct |
|---|---|---|---|---|
| L48N1 | 46,636,752 | 7.0 G | 94.36 | 42.57 |
| L48N2 | 46,049,962 | 6.91 G | 94.52 | 42.46 |
| L48N3 | 46,999,312 | 7.05 G | 94.41 | 42.43 |
| D48N1 | 54,821,486 | 8.22 G | 94.01 | 42.33 |
| D48N2 | 42,996,712 | 6.45 G | 94.39 | 42.5 |
| D48N3 | 44,649,882 | 6.7 G | 94.28 | 42.39 |
| L72N1 | 48,664,980 | 7.3 G | 94.08 | 42.44 |
| L72N2 | 45,198,608 | 6.78 G | 94.37 | 42.39 |
| L72N3 | 46,525,636 | 6.98 G | 94.27 | 42.97 |
| D72N1 | 52,924,474 | 7.94 G | 94.28 | 42.19 |
| D72N2 | 44,877,252 | 6.73 G | 94.18 | 42.42 |
| D72N3 | 46,212,326 | 6.93 G | 94.36 | 42.43 |
| L168N1 | 46,145,536 | 6.92 G | 94.11 | 42.06 |
| L168N2 | 44,256,704 | 6.64 G | 94.43 | 41.88 |
| L168N3 | 45,263,740 | 6.79 G | 94.19 | 41.98 |
| D168N1 | 46,224,450 | 6.93 G | 94.48 | 42.15 |
| D168N2 | 46,802,178 | 7.02 G | 94.56 | 42.1 |
| D168N3 | 44,491,876 | 6.67 G | 94.11 | 41.74 |
| Sample | Total_Reads | Total_Map(%) | Unique_Map(%) | Multi_Map(%) | Positive_Map(%) | Negative_Map(%) |
|---|---|---|---|---|---|---|
| L48N1 | 46,636,752 | 95.58% | 92.33% | 3.25% | 46.13% | 46.20% |
| L48N2 | 46,049,962 | 83.78% | 80.94% | 2.84% | 40.45% | 40.50% |
| L48N3 | 46,999,312 | 95.64% | 92.23% | 3.40% | 46.08% | 46.15% |
| D48N1 | 54,821,486 | 89.29% | 86.47% | 2.82% | 43.21% | 43.26% |
| D48N2 | 42,996,712 | 84.37% | 81.66% | 2.71% | 40.80% | 40.86% |
| D48N3 | 44,649,882 | 95.31% | 92.10% | 3.20% | 46.02% | 46.08% |
| L72N1 | 48,664,980 | 94.67% | 91.60% | 3.07% | 45.77% | 45.83% |
| L72N2 | 45,198,608 | 75.08% | 72.65% | 2.43% | 36.31% | 36.34% |
| L72N3 | 46,525,636 | 96.09% | 92.97% | 3.11% | 46.44% | 46.53% |
| D72N1 | 52,924,474 | 95.01% | 92.08% | 2.93% | 46.01% | 46.06% |
| D72N2 | 44,877,252 | 95.53% | 92.64% | 2.89% | 46.29% | 46.35% |
| D72N3 | 46,212,326 | 95.59% | 92.61% | 2.98% | 46.28% | 46.33% |
| L168N1 | 46,145,536 | 87.66% | 84.78% | 2.88% | 42.36% | 42.41% |
| L168N2 | 44,256,704 | 80.60% | 77.95% | 2.65% | 38.96% | 38.99% |
| L168N3 | 45,263,740 | 95.02% | 92.00% | 3.02% | 45.99% | 46.01% |
| D168N1 | 46,224,450 | 85.77% | 83.19% | 2.57% | 41.58% | 41.62% |
| D168N2 | 46,802,178 | 77.71% | 75.31% | 2.40% | 37.64% | 37.67% |
| D168N3 | 44,491,876 | 78.57% | 76.26% | 2.31% | 38.12% | 38.15% |
| Module | Sample | KEGGID | Description | GeneRatio | padj |
|---|---|---|---|---|---|
| pink | L168N2 | sly04075 | Plant hormone signal transduction | 16/88 | 2.64 × 10−4 |
| blue | L72N3 | sly03010 | Ribosome | 72/526 | 5.36 × 10−12 |
| sly03050 | Proteasome | 18/526 | 1.39 × 10−4 | ||
| sly00510 | N-Glycan biosynthesis | 14/526 | 1.23 × 10−2 | ||
| sly03060 | Protein export | 13/526 | 2.02 × 10−2 | ||
| sly04141 | Protein processing in endoplasmic reticulum | 37/526 | 3.93 × 10−2 | ||
| sly00190 | Oxidative phosphorylation | 25/526 | 4.02 × 10−2 | ||
| sly04145 | Phagosome | 18/526 | 4.02 × 10−2 | ||
| brown | L48N3 | sly00860 | Porphyrin and chlorophyll metabolism | 19/487 | 1.63 × 10−5 |
| sly00196 | Photosynthesis antenna proteins | 11/487 | 4.48 × 10−5 | ||
| sly01200 | Carbon metabolism | 52/487 | 1.10 × 10−4 | ||
| sly00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 14/487 | 1.28 × 10−3 | ||
| sly00970 | Aminoacyl-tRNA biosynthesis | 15/487 | 1.87 × 10−3 | ||
| sly00710 | Carbon fixation in photosynthetic organisms | 19/487 | 3.31 × 10−3 | ||
| sly00195 | Photosynthesis | 18/487 | 1.03 × 10−2 | ||
| sly00941 | Flavonoid biosynthesis | 12/487 | 1.44 × 10−2 | ||
| sly00750 | Vitamin B6 metabolism | 6/487 | 1.80 × 10−2 | ||
| sly04712 | Circadian rhythm—plant | 11/487 | 2.57 × 10−2 | ||
| sly00030 | Pentose phosphate pathway | 13/487 | 2.63 × 10−2 | ||
| sly00071 | Fatty acid degradation | 13/487 | 2.90 × 10−2 |
| Gene_ID | Module | Gene_Description |
|---|---|---|
| 107875625 | Blue | agmatine coumaroyltransferase-2-like%2C transcript variant X2 |
| 107850816 | Blue | omega-hydroxypalmitate O-feruloyl transferase |
| 107851884 | Brown | acetyl-CoA-benzylalcohol acetyltransferase-like |
| 107848097 | Brown | agmatine coumaroyltransferase-2-like |
| 107843659 | Brown | anthocyanidin 3-O-glucosyltransferase-like |
| 107864266 | Brown | chalcone synthase 1B |
| 107871256 | Brown | chalcone synthase 2 |
| 107852750 | brown | chalcone--flavonone isomerase%2C transcript variant X1 |
| 107844023 | brown | cytochrome P450 98A2-like |
| 107860031 | brown | dihydroflavonol-4-reductase |
| 107862334 | brown | flavonoid 3′-monooxygenase |
| 107866341 | brown | leucoanthocyanidin dioxygenase |
| 107871144 | brown | probable chalcone--flavonone isomerase 3%2C transcript variant X2 |
| 107840863 | brown | shikimate O-hydroxycinnamoyltransferase-like |
| 107860278 | green | caffeoyl-CoA O-methyltransferase%2C transcript variant X2 |
| 107853336 | green | probable caffeoyl-CoA O-methyltransferase At4g26220%2C transcript variant X1 |
| 107840262 | magenta | caffeoyl-CoA O-methyltransferase 6 |
| 107840069 | turquoise | acetyl-CoA-benzylalcohol acetyltransferase-like |
| 107839366 | turquoise | cytochrome P450 98A2-like |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Mumtaz, M.A.; Zhang, Y.; Shu, H.; Hao, Y.; Lu, X.; Cheng, S.; Zhu, G.; Wang, Z. Response of Anthocyanin Accumulation in Pepper (Capsicum annuum) Fruit to Light Days. Int. J. Mol. Sci. 2022, 23, 8357. https://doi.org/10.3390/ijms23158357
Zhou Y, Mumtaz MA, Zhang Y, Shu H, Hao Y, Lu X, Cheng S, Zhu G, Wang Z. Response of Anthocyanin Accumulation in Pepper (Capsicum annuum) Fruit to Light Days. International Journal of Molecular Sciences. 2022; 23(15):8357. https://doi.org/10.3390/ijms23158357
Chicago/Turabian StyleZhou, Yan, Muhammad Ali Mumtaz, Yonghao Zhang, Huangying Shu, Yuanyuan Hao, Xu Lu, Shanhan Cheng, Guopeng Zhu, and Zhiwei Wang. 2022. "Response of Anthocyanin Accumulation in Pepper (Capsicum annuum) Fruit to Light Days" International Journal of Molecular Sciences 23, no. 15: 8357. https://doi.org/10.3390/ijms23158357
APA StyleZhou, Y., Mumtaz, M. A., Zhang, Y., Shu, H., Hao, Y., Lu, X., Cheng, S., Zhu, G., & Wang, Z. (2022). Response of Anthocyanin Accumulation in Pepper (Capsicum annuum) Fruit to Light Days. International Journal of Molecular Sciences, 23(15), 8357. https://doi.org/10.3390/ijms23158357







