A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast
Abstract
:1. Introduction
2. Results
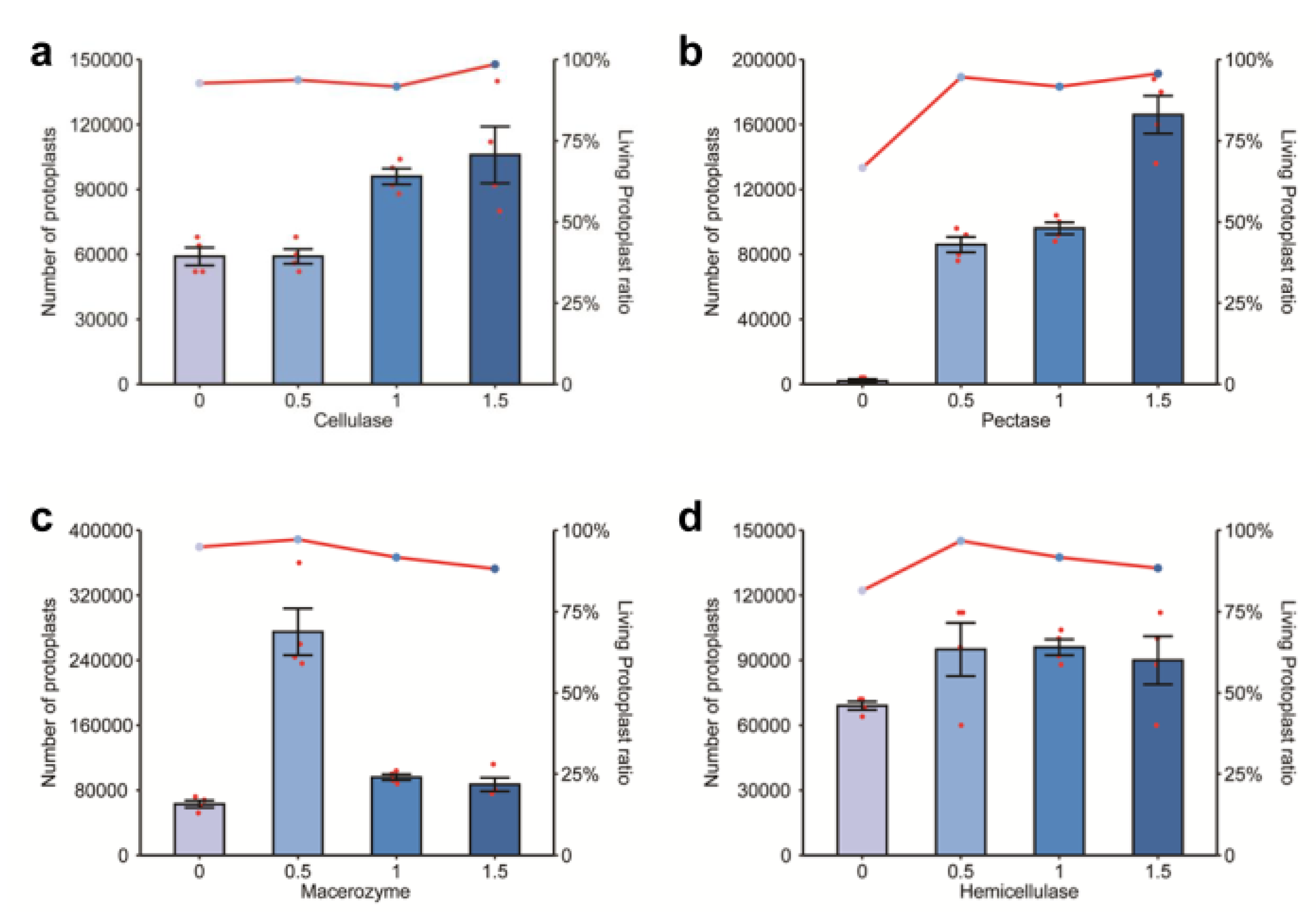
2.1. Effects of Different Enzyme Concentrations on Protoplast Extraction
2.2. Effects of Combined Varied Enzyme Concentrations on Protoplasts
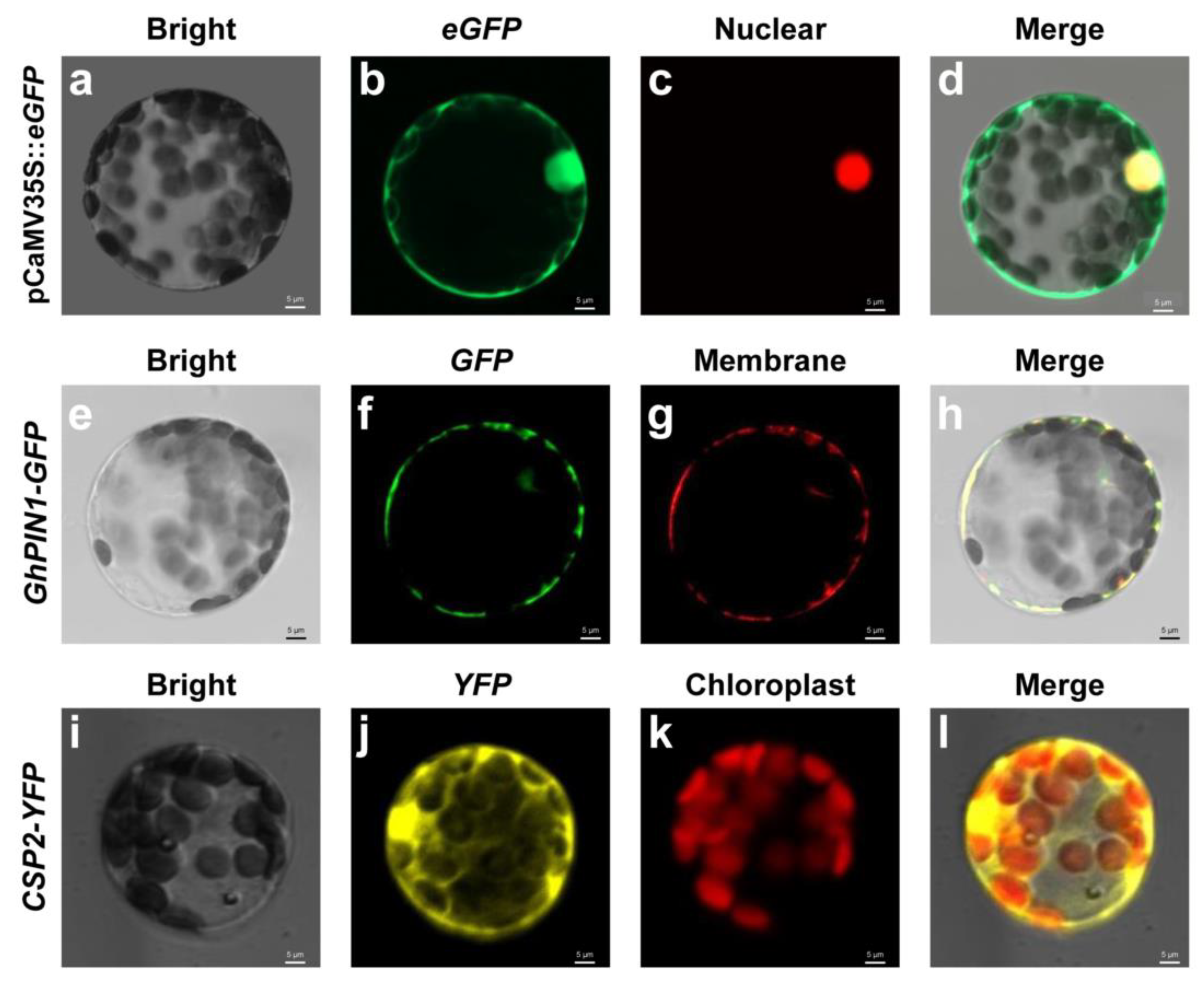
2.3. Protoplast Transformations for Subcellular Localization
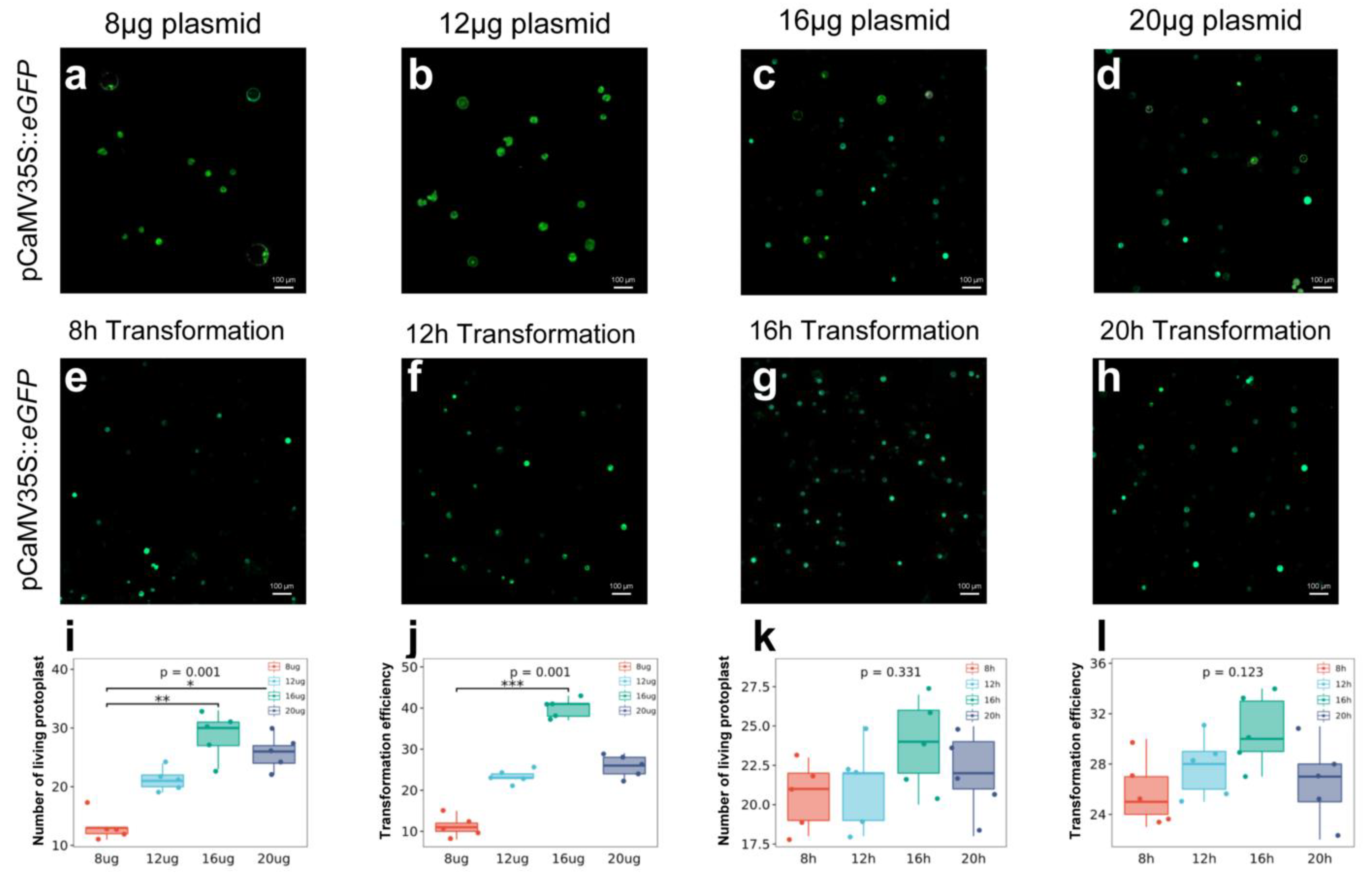
2.4. Relationship between Plasmid Concentration and Transformation Efficiency
2.5. Relationship between Transformation Time and Efficiency
3. Discussion
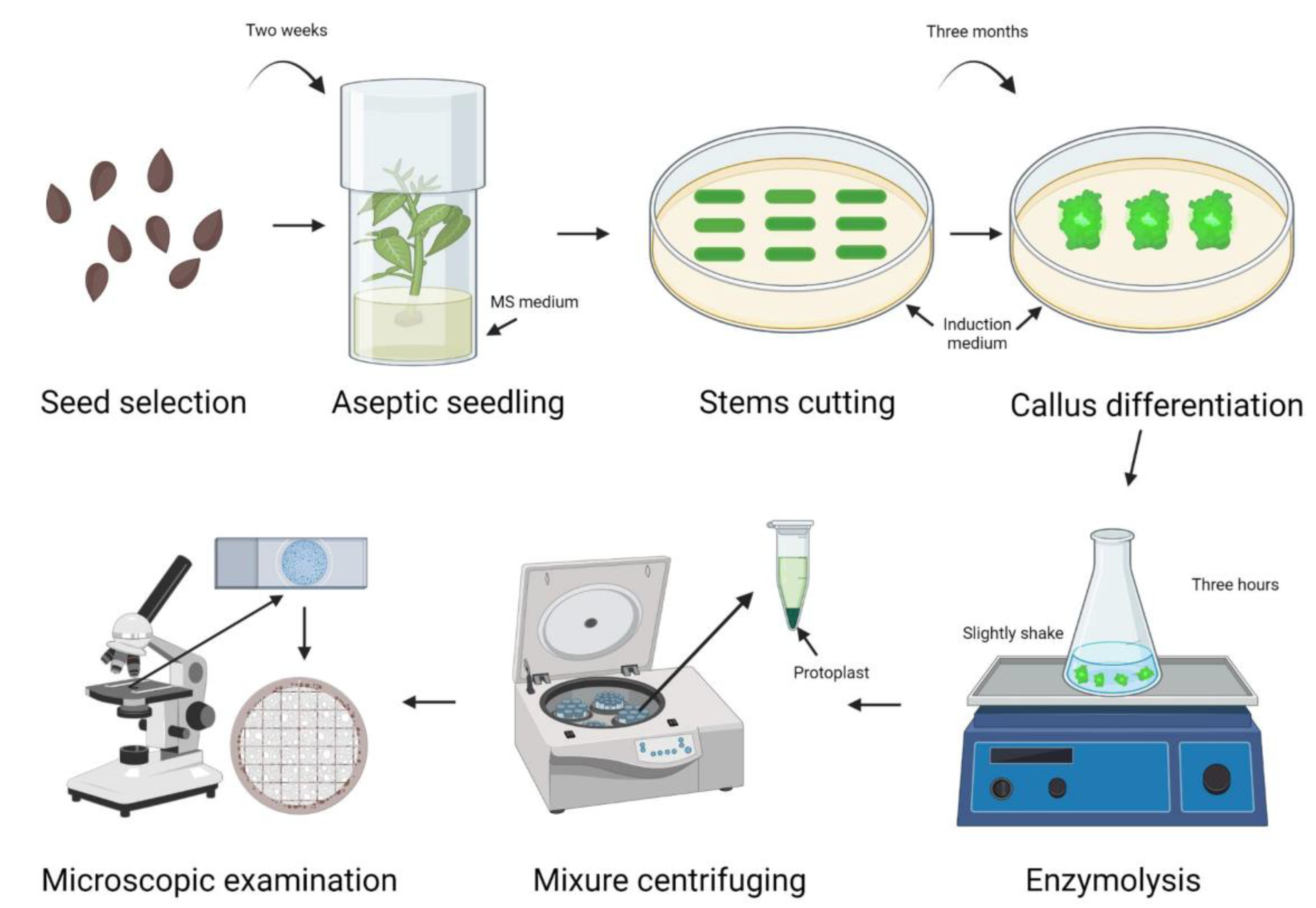
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protoplast Isolation
4.3. Vector Construction and Plasmid Isolation
4.4. Protoplast Transfection
4.5. Microscopy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAP | 6-benzylaminopurine |
| NAA | 1-naphthaleneacetic acid |
| IAA | indole-3-acetic acid |
| MS | Murashige and Skoog |
| MES | Murashige and Skoog Medium with MES |
| PEG | polyethylene glycol |
References
- Sahoo, R.K.; Tuteja, N. Development of Agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food 2012, 3, 123–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.X.; Wu, Y.; Zou, H.D.; Su, S.Z.; Li, S.P.; Shan, X.H.; Liu, H.K.; Yuan, Y.P. Transformation of GBSS I Gene Driven with Barley Endosperm-specific Promoter into Maize Inbred Line Mediated by Agrobacterium. Maize Genom. Genet. 2012, 3, 1–5. [Google Scholar] [CrossRef]
- Kalbande, B.B.; Patil, A.S. Plant tissue culture independent Agrobacterium tumefaciens mediated In-planta transformation strategy for upland cotton (Gossypium hirsutum). J. Genet. Eng. Biotechnol. 2016, 14, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sainsbury, F.; Lomonossoff, G.P. Transient expressions of synthetic biology in plants. Curr. Opin. Plant Biol. 2014, 19, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Andrieu, A.; Breitler, J.C.; Siré, C.; Meynard, D.; Gantet, P.; Guiderdoni, E. An in planta, Agrobacterium-mediated transient gene expression method for inducing gene silencing in rice (Oryza sativa L.) leaves. Rice 2012, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Bhaskar, P.B.; Venkateshwaran, M.; Wu, L.; Ané, J.M.; Jiang, J. Agrobacterium-mediated transient gene expression and silencing: A rapid tool for functional gene assay in potato. PLoS ONE 2009, 4, e5812. [Google Scholar] [CrossRef] [Green Version]
- Panwar, V.; McCallum, B.; Bakkeren, G. Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 2013, 73, 521–532. [Google Scholar] [CrossRef]
- Ditt, R.F.; Kerr, K.F.; de Figueiredo, P.; Delrow, J.; Comai, L.; Nester, E.W. The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 2006, 19, 665–681. [Google Scholar] [CrossRef] [Green Version]
- Kirienko, D.R.; Luo, A.; Sylvester, A.W. Reliable transient transformation of intact maize leaf cells for functional genomics and experimental study. Plant Physiol. 2012, 159, 1309–1318. [Google Scholar] [CrossRef] [Green Version]
- Ueki, S.; Magori, S.; Lacroix, B.; Citovsky, V. Transient gene expression in epidermal cells of plant leaves by biolistic DNA delivery. Methods Mol Biol. 2013, 940, 17–26. [Google Scholar]
- Zhao, F.L.; Li, Y.J.; Hu, Y.; Gao, Y.R.; Zang, X.W.; Ding, Q.; Wang YJWen, Y.Q. A highly efficient grapevine mesophyll protoplast system for transient gene expression and the study of disease resistance proteins. Plant Cell Tissue Organ Cult. 2015, 125, 43–57. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Burris, K.P.; Dlugosz, E.M.; Collins, A.G.; Stewart CNJr Lenaghan, S.C. Development of a rapid, low-cost protoplast transfection system for switchgrass (Panicum virgatum L.). Plant Cell Rep. 2016, 35, 693–704. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Han, N.; Wu, J.; Yang, Y.; Wang, J.; Zhu, M.; Bian, H. A transient gene expression system using barley protoplasts to evaluate microRNAs for post-transcriptional regulation of their target genes. Plant Cell Tissue Organ Cult. 2014, 119, 211–219. [Google Scholar] [CrossRef]
- Jia, X.; Zhang, X.; Qu, J.; Han, R. Optimization conditions of wheat mesophyll protoplast isolation. Agric. Sci. 2016, 7, 850–858. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Cheng, Q.; Xie, Z.; Xu, B.; Huang, B.; Zhao, B. An efficient protocol for perennial ryegrass mesophyll protoplast isolation and transformation, and its application on interaction study between LpNOL and LpNYC1. Plant Methods 2017, 13, 46. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.H.; Shen, S.C.; Lee, L.Y.; Lee, S.H.; Chan, M.T.; Lin, C.S. Tape-Arabidopsis Sandwich—A simpler Arabidopsis protoplast isolation method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Fang, L.; Wang, Q.; Hu, Y.; Jia, Y.; Chen, J.; Liu, B.; Zhang, Z.; Guan, X.; Chen, S.; Zhou, B.; et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017, 49, 1089–1098. [Google Scholar] [CrossRef]
- Shan, X.; Li, Y.; Zhou, L.; Tong, L.; Wei, C.; Qiu, L.; Gao, X.; Wang, L. Efficient isolation of protoplasts from Freesia callus and its application in transient expression assays. Plant Cell Tissue Organ Cult. 2019, 138, 529–541. [Google Scholar] [CrossRef]
- Cocking, E.C. A method for the isolation of plant protoplasts and vacuoles. Nature 1960, 187, 962–963. [Google Scholar] [CrossRef]
- Zhang, H.M.; Wang, J.L. Plant protoplast isolation, culture and application. Hebei J. Orchard Res. 2002, 17, 376–382. [Google Scholar]
- Li, R.J.; Zhang, Z.X.; Shi, Y.H. A preliminary report on the separation of cotton mesphyll protoplast. Hereditas 1980, 2, 36. [Google Scholar]
- Sun, Q.; Zhang, L.; Huang, C.; Nie, Y.C.; Guo, X.P. Plant regeneration via somatic embryogenesis from protoplasts of six explants in Coker 201 (Gossypium hirsutum). Plant Cell Tissue Organ Cult. 2005, 82, 309–315. [Google Scholar] [CrossRef]
- Gao, X.Q.; Wheeler, T.; Li, Z.H.; Kenerley, C.M.; He, P.; Shan, L.B. Silencing GhNDR1 and GhMKK2 compromised cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Ding, L.; Zhang, Z.; Guo, W. Isolation of mesophyll protoplast and establishment of gene transient expression system in cotton. Acta Agron. Sin. 2014, 40, 231–239. [Google Scholar] [CrossRef]
- Wang, J.E.; Sun, Y.Q.; Yan, S.F.; Dao, D.; Shen, X.J.; Zhu, J. Study on the plant regeneration from protoplasts of upland cotton (Gossypium hirsutum L.) via somatic embryogenesis. Cotton Sci. 2008, 20, 403–407. [Google Scholar]
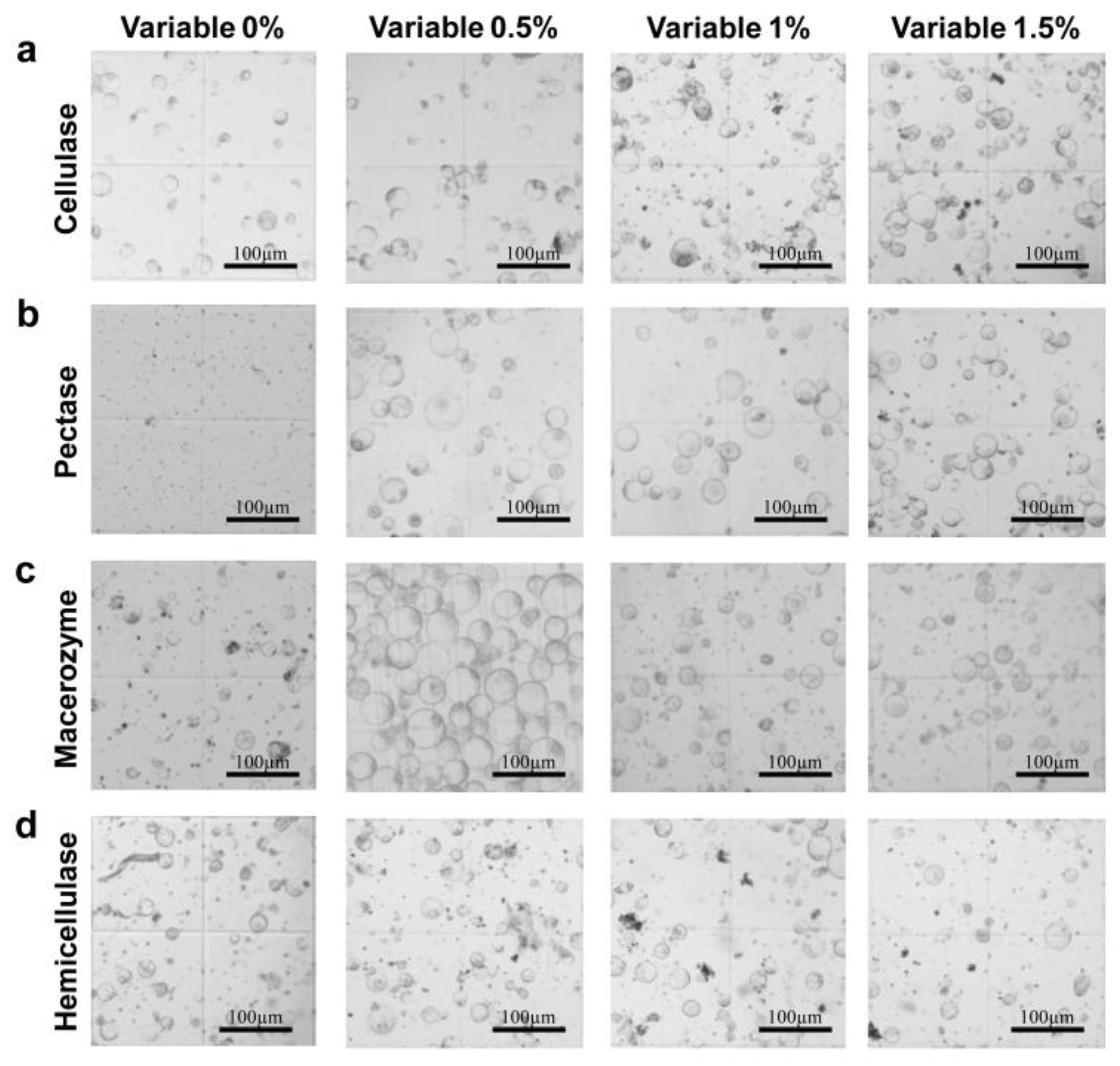

| Group | Cellulase | Pectase | Macerozym | Hemicellulase |
|---|---|---|---|---|
| Group 1 | 0 | 1 | 1 | 1 |
| Group 2 | 0.5 | 1 | 1 | 1 |
| Group 3 | 1 | 1 | 1 | 1 |
| Group 4 | 1.5 | 1 | 1 | 1 |
| Group 5 | 1 | 0 | 1 | 1 |
| Group 6 | 1 | 0.5 | 1 | 1 |
| Group 7 | 1 | 1 | 1 | 1 |
| Group 8 | 1 | 1.5 | 1 | 1 |
| Group 9 | 1 | 1 | 0 | 1 |
| Group 10 | 1 | 1 | 0.5 | 1 |
| Group 11 | 1 | 1 | 1 | 1 |
| Group 12 | 1 | 1 | 1.5 | 1 |
| Group 13 | 1 | 1 | 1 | 0 |
| Group 14 | 1 | 1 | 1 | 0.5 |
| Group 15 | 1 | 1 | 1 | 1 |
| Group 16 | 1 | 1 | 1 | 1.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Pu, Y.; Abid, M.A.; Kang, L.; Ye, Y.; Zhang, M.; Liang, C.; Wei, Y.; Zhang, R.; Meng, Z. A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast. Int. J. Mol. Sci. 2022, 23, 8368. https://doi.org/10.3390/ijms23158368
Wang P, Pu Y, Abid MA, Kang L, Ye Y, Zhang M, Liang C, Wei Y, Zhang R, Meng Z. A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast. International Journal of Molecular Sciences. 2022; 23(15):8368. https://doi.org/10.3390/ijms23158368
Chicago/Turabian StyleWang, Peilin, Yuanchun Pu, Muhammad Ali Abid, Linglin Kang, Yulu Ye, Man Zhang, Chengzhen Liang, Yunxiao Wei, Rui Zhang, and Zhigang Meng. 2022. "A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast" International Journal of Molecular Sciences 23, no. 15: 8368. https://doi.org/10.3390/ijms23158368
APA StyleWang, P., Pu, Y., Abid, M. A., Kang, L., Ye, Y., Zhang, M., Liang, C., Wei, Y., Zhang, R., & Meng, Z. (2022). A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast. International Journal of Molecular Sciences, 23(15), 8368. https://doi.org/10.3390/ijms23158368







