Genome-Wide Identification and Characterization of Members of the ACS Gene Family in Cucurbita maxima and Their Transcriptional Responses to the Specific Treatments
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification of ACSs in Pumpkin
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of CmaACS Gene Family
2.3. Gene Structure and Conserved Motif Composition of CmaACS Genes Family
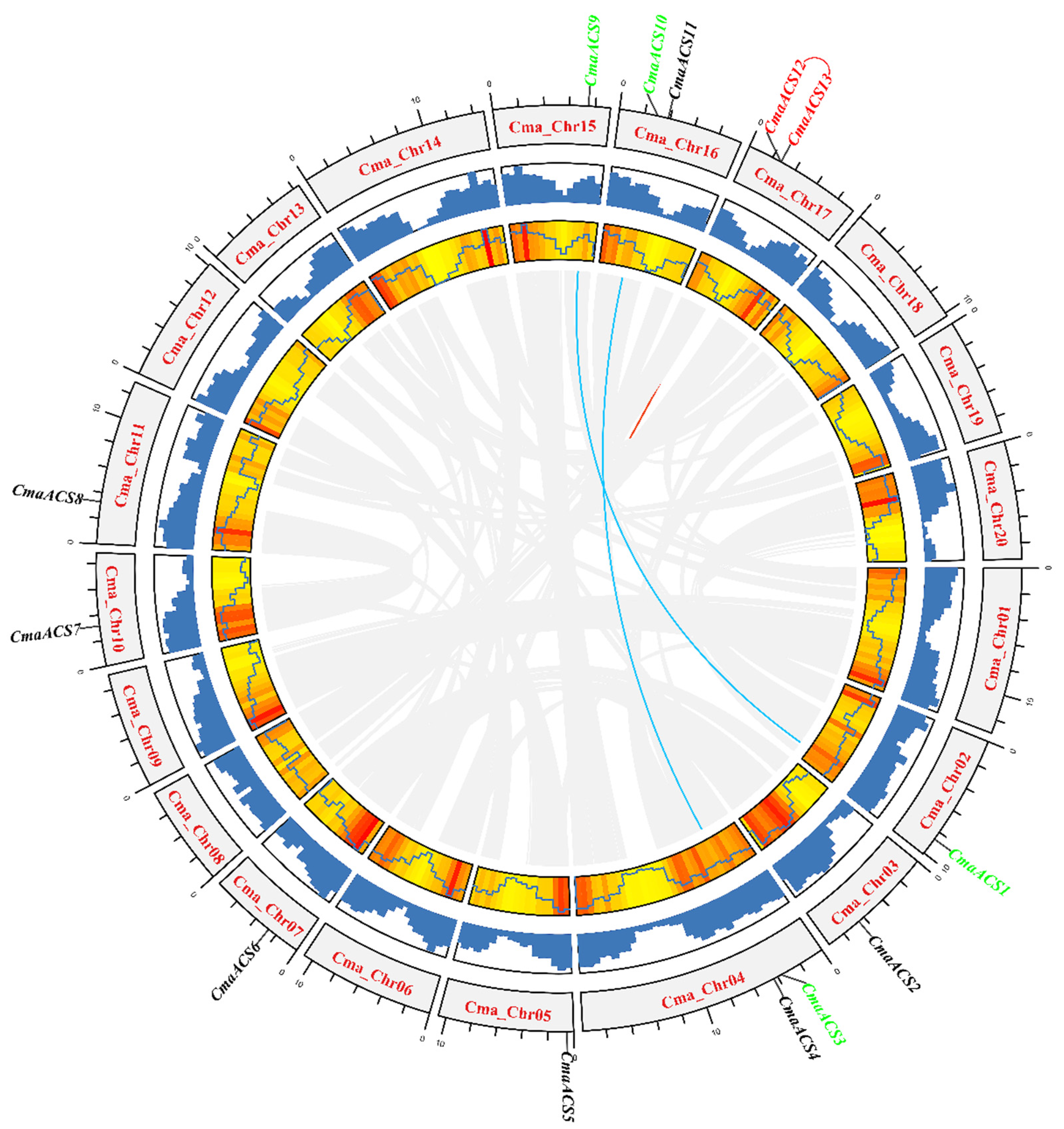
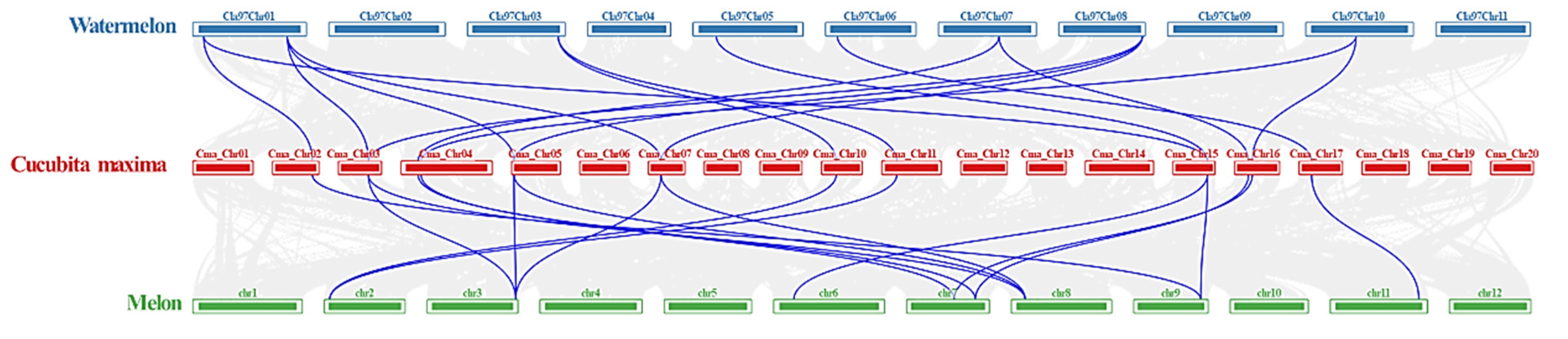
2.4. Chromosomal Distribution and Synteny Analysis of CmaACS Genes
2.5. Cis-Elements in the Promoters of CmaACSs
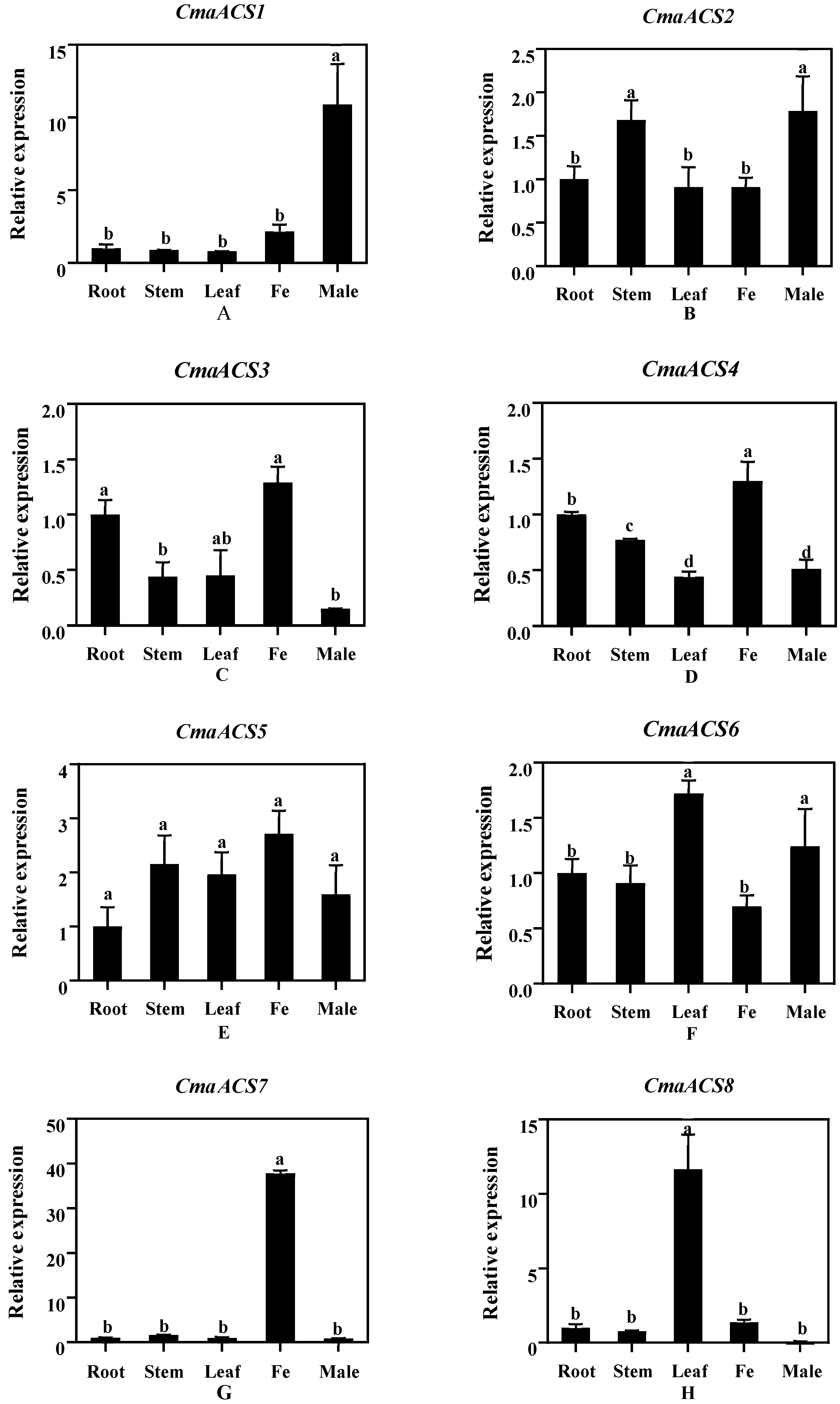
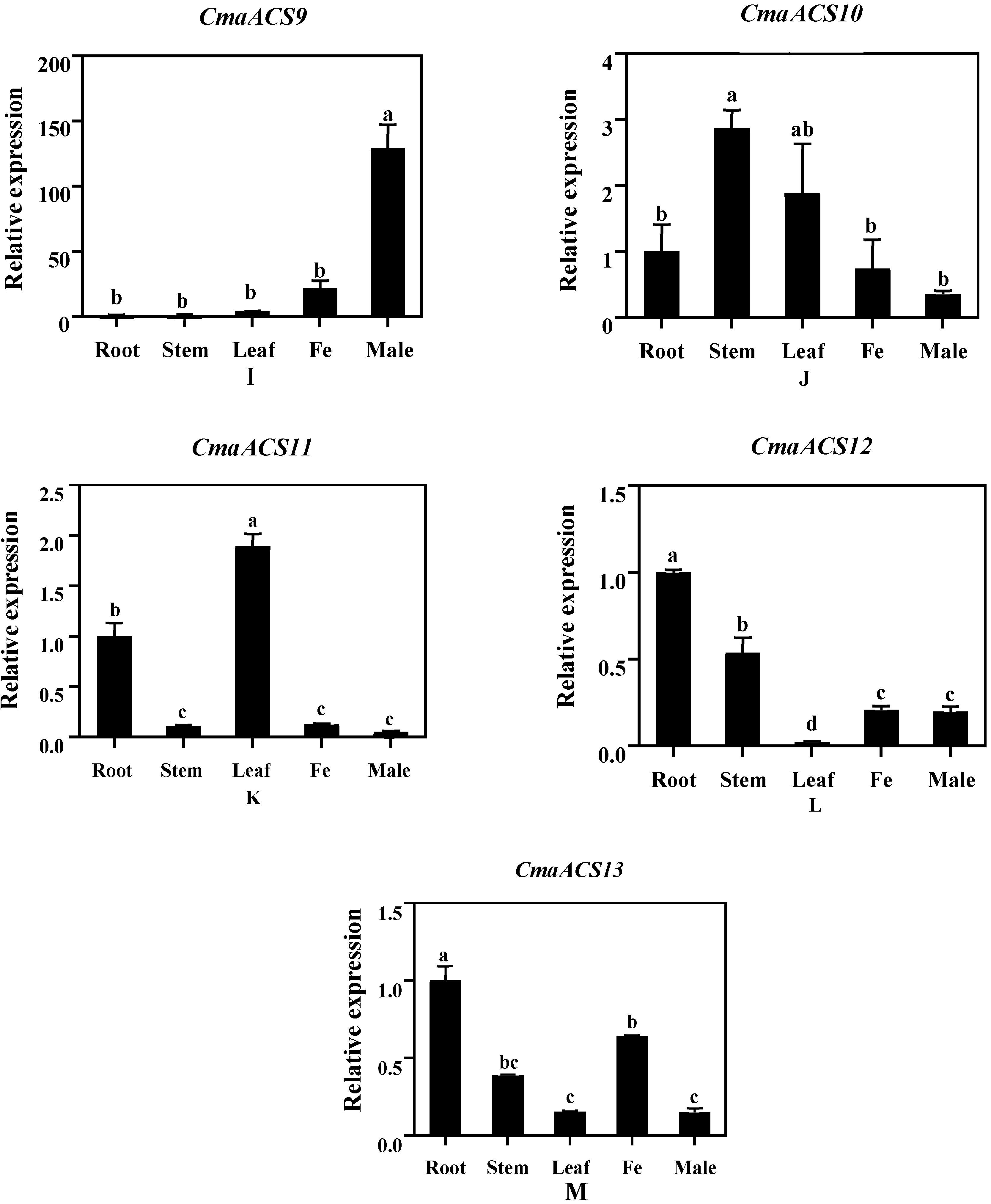
2.6. Expression Analysis of CmaACSs in Pumpkin Tissue
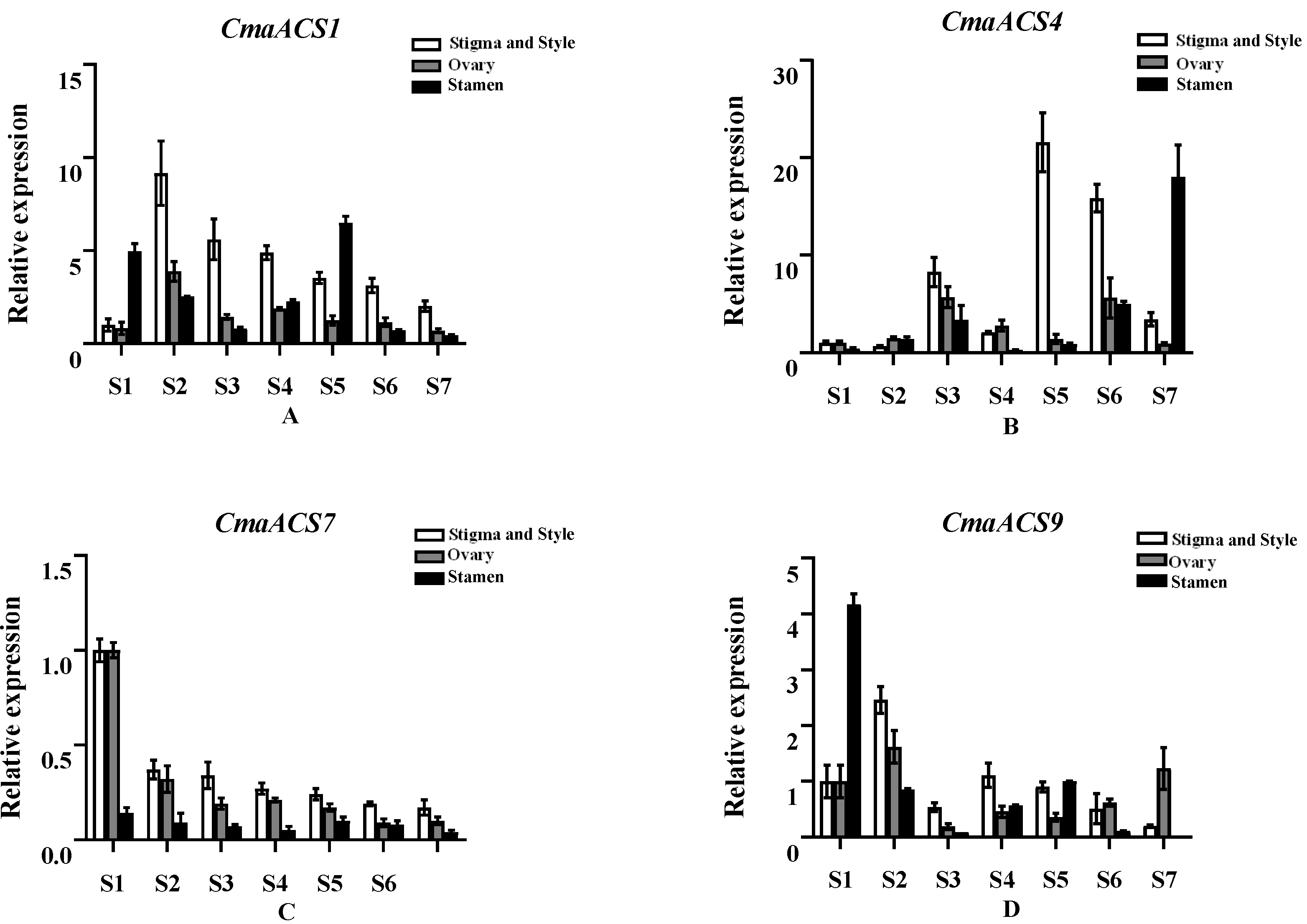
2.7. Expression Profiles of the CmaACSs of C. maxima at Different Flower Developmental Stages
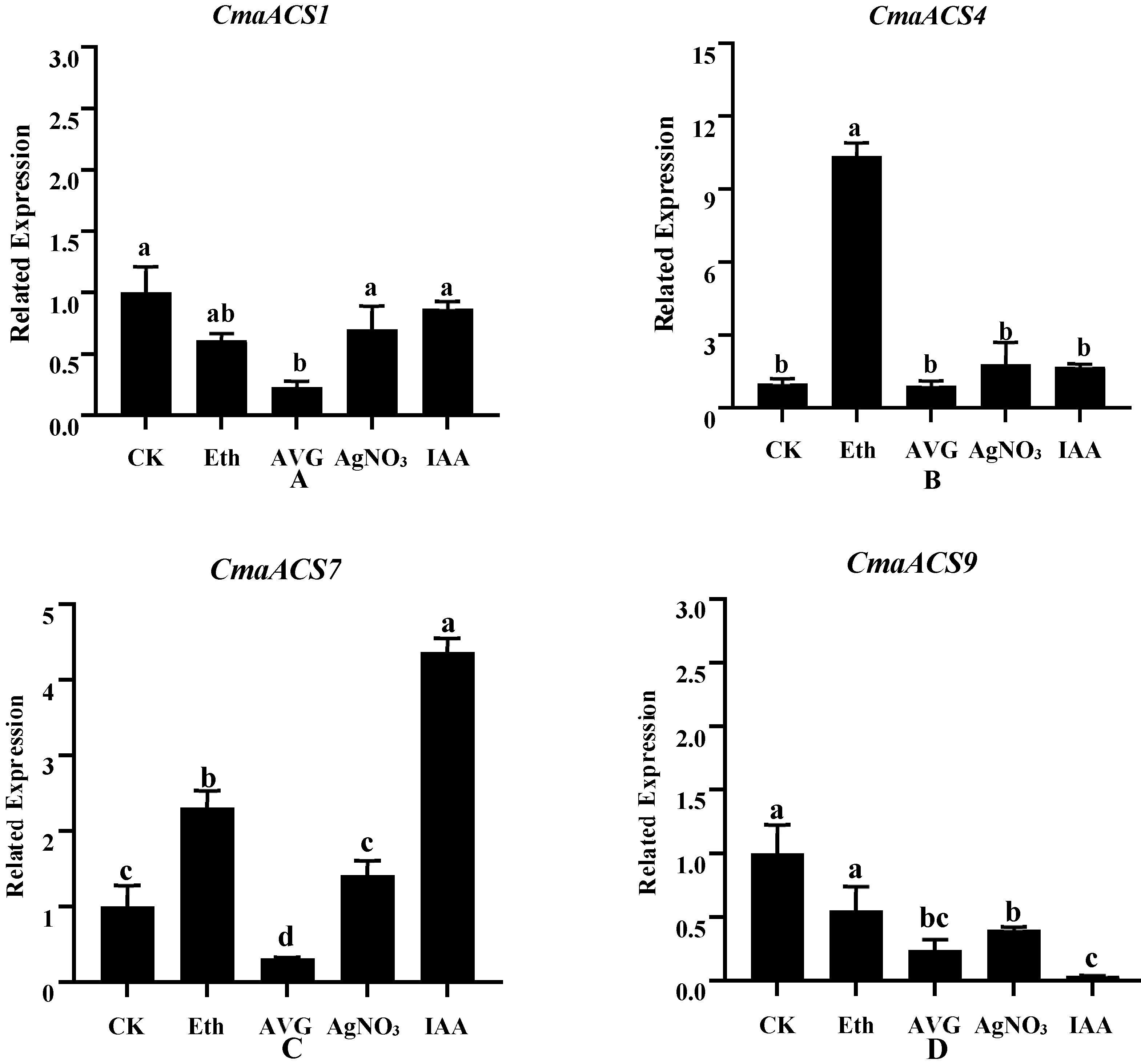
2.8. Expression Profiles of CmaACSs in Pumpkin under Phytohormones and Ethylene Inhibitors Treatment
3. Discussion
4. Materials and Methods
4.1. Prediction and Identification of CmaACSs in C. maxima
4.2. Sequence Analysis and Structural Characterization
4.3. Phylogenetic Analysis
4.4. Promoter Sequence Analysis
4.5. Analysis of the Collinearity and Selection Pressure of the ACS Gene Family
4.6. Expression Pattern of CmaACSs in C. maxima
4.7. RNA Extraction and Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, S.J.; Chang, C.L.; Wang, P.H.; Tsai, M.C.; Hsu, P.H.; Chang, I.F. A type III ACC synthase, ACS7, is involved in root gravitropism in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 4343–4360. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Cui, X.; Sun, Y.; Dong, C.-H. Ethylene signaling and regulation in plant growth and stress responses. Plant Cell Rep. 2013, 32, 1099–1109. [Google Scholar] [CrossRef]
- Yin, T.; Quinn, J.A. Tests of a mechanistic model of one hormone regulating both sexes in Cucumis sativus (Cucurbitaceae). Am. J. Bot. 1995, 82, 1537–1546. [Google Scholar] [CrossRef]
- Malepszy, S.; Niemirowicz-Szczytt, K. Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Sci. 1991, 80, 39–47. [Google Scholar] [CrossRef]
- Kater, M.M.; Franken, J.; Carney, K.J.; Colombo, L.; Angenent, G.C. Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 2001, 13, 481–493. [Google Scholar] [CrossRef] [Green Version]
- García, A.; Aguado, E.; Garrido, D.; Martínez, C.; Jamilena, M. Two androecious mutations reveal the crucial role of ethylene receptors in the initiation of female flower development in Cucurbita pepo. Plant J. 2020, 103, 1548–1560. [Google Scholar] [CrossRef]
- Byers, R.E.; Baker, L.R.; Sell, H.M.; Herner, R.C.; Dilley, D.R. Ethylene: A Natural Regulator of Sex Expression of Cucumis melo L. Proc. Natl. Acad. Sci. USA 1972, 69, 717–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudich, J.; Halevy, A.; Kedar, N. Increase in femaleness of three cucurbits by treatment with ethrel, an ethylene releasing compound. Planta 1969, 86, 69–76. [Google Scholar] [CrossRef]
- Rudich, J. Biochemical aspects of hormonal regulation of sex expression in Cucurbits. Biol. Util. Cucurbitaceae 1990, 269–280. [Google Scholar] [CrossRef]
- Martínez, C.; Manzano, S.; Megías, Z.; Garrido, D.; Picó, B.; Jamilena, M. Involvement of ethylene biosynthesis and signalling in fruit set and early fruit development in zucchini squash (Cucurbita pepo L.). BMC Plant Biol. 2013, 13, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Den Nijs, A.; Visser, D. Induction of male flowering in gynoecious cucumbers (Cucumis sativus L.) by silver ions. Euphytica 1980, 29, 273–280. [Google Scholar] [CrossRef]
- Manzano, S.; Martínez, C.; Megías, Z.; Gómez, P.; Garrido, D.; Jamilena, M. The role of ethylene and brassinosteroids in the control of sex expression and flower development in Cucurbita pepo. Plant Growth Regul. 2011, 65, 213–221. [Google Scholar] [CrossRef]
- Manzano, S.; Martínez, C.; García, J.M.; Megías, Z.; Jamilena, M. Involvement of ethylene in sex expression and female flower development in watermelon (Citrullus lanatus). Plant Physiol. Biochem. 2014, 85, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jianting, S.; Gaojie, J.; Zhang, H.; Guoyi, G.; Shaogui, G.; Yi, R.; Jianguang, F.; Shouwei, T.; Yong, X. Modulation of sex expression in four forms of watermelon by gibberellin, ethephone and silver nitrate. Hortic. Plant J. 2017, 3, 91–100. [Google Scholar] [CrossRef]
- Khan, Z.I.; Ahmad, K.; Ashraf, M.; Parveen, R.; Bibi, Z.; Mustafa, I.; Noorka, I.R.; Tahir, H.M.; Akram, N.A.; Ullah, M.F.; et al. Risk assessment of heavy metal and metalloid toxicity through a contaminated vegetable (Cucurbita maxima) from wastewater irrigated area: A case study for a site-specific risk assessment in Jhang, Pakistan. Hum. Ecol. Risk Assess 2016, 22, 86–98. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14 (Suppl. 1), S131–S151. [Google Scholar] [CrossRef] [Green Version]
- Zarembinski, T.I.; Theologis, A. Ethylene biosynthesis and action: A case of conservation. In Signals and Signal Transduction Pathways in Plants; Springer: Berlin/Heidelberg, Germany, 1994; pp. 343–361. [Google Scholar]
- Adams, D.; Yang, S. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.R.; Roy, S.; Sengupta, D.N. C-Terminal phosphorylation is essential for regulation of ethylene synthesizing ACC synthase enzyme. Plant Signal. Behav. 2013, 8, 491–511. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.-P.; Lin, T.-Y.; Wang, N.-N.; Shih, M.-C. Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Mol. Biol. 2005, 58, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; Shih, M.-C.; Li, N. The GUS reporter-aided analysis of the promoter activities of Arabidopsis ACC synthase genes AtACS4, AtACS5, and AtACS7 induced by hormones and stresses. J. Exp. Bot. 2005, 56, 909–920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagami, T.; Tsuchisaka, A.; Yamada, K.; Haddon, W.F.; Harden, L.A.; Theologis, A. Biochemical Diversity among the 1-Amino-cyclopropane-1-Carboxylate Synthase Isozymes Encoded by the Arabidopsis Gene Family. J. Biol. Chem. 2003, 278, 49102–49112. [Google Scholar] [CrossRef] [Green Version]
- Tsuchisaka, A.; Theologis, A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2275–2280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzano, S.; Aguado, E.; Martinez, C.; Megias, Z.; Garcia, A.; Jamilena, M. The ethylene biosynthesis gene CitACS4 regulates monoecy/andromonoecy in watermelon (Citrullus lanatus). PLoS ONE 2016, 11, e0154362. [Google Scholar] [CrossRef] [Green Version]
- Ji, G.; Zhang, J.; Zhang, H.; Sun, H.; Gong, G.; Shi, J.; Tian, S.; Guo, S.; Ren, Y.; Shen, H. Mutation in the gene encoding 1-aminocyclopropane-1-carboxylate synthase 4 (CitACS4) led to andromonoecy in watermelon. J. Integr. Plant Biol. 2016, 58, 762–765. [Google Scholar] [CrossRef] [Green Version]
- Martínez, C.; Manzano, S.; Megías, Z.; Barrera, A.; Boualem, A.; Garrido, D.; Bendahmane, A.; Jamilena, M. Molecular and functional characterization of CpACS27A gene reveals its involvement in monoecy instability and other associated traits in squash (Cucurbita pepo L.). Planta 2014, 239, 1201–1215. [Google Scholar] [CrossRef]
- Boualem, A.; Troadec, C.; Kovalski, I.; Sari, M.-A.; Perl-Treves, R.; Bendahmane, A. A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS ONE 2009, 4, e6144. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Fergany, M.; Fernandez, R.; Troadec, C.; Martin, A.; Morin, H.; Sari, M.-A.; Collin, F.; Flowers, J.M.; Pitrat, M. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 2008, 321, 836–838. [Google Scholar] [CrossRef]
- Boualem, A.; Troadec, C.; Camps, C.; Lemhemdi, A.; Morin, H.; Sari, M.A.; Fraenkel-Zagouri, R.; Kovalski, I.; Dogimont, C.; Perl-Treves, R. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 2015, 350, 688–691. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.C.; Choi, D.; Han, J.H.; Jung, Y.; Lee, S. RNA expression, protein activity, and interactions in the ACC synthase gene family in cucumber (Cucumis sativus L.). Hortic. Environ. Biotechnol. 2018, 59, 81–91. [Google Scholar] [CrossRef]
- Wang, Z.; Yadav, V.; Yan, X.; Cheng, D.; Wei, C.; Zhang, X. Systematic genome-wide analysis of the ethylene-responsive ACS gene family: Contributions to sex form differentiation and development in melon and watermelon. Gene 2021, 805, 145910. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Li, W.; Zhang, C.; Wei, Y.; Wang, Y.; Hu, H. Phylogenetic Analysis of ACS Gene Family in Musa acuminata. Mol. Plant Breed. 2017, 15, 425–432. [Google Scholar]
- Zhai, J. The Cloning and Expression Analysis of the ACS Family Gene of Vitis Tinifera; Liaoning Normal University, Liaoning, China, 2012.
- Robert; Finn; Penelope; Coggill; Ruth; Eberhardt; Sean, The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [CrossRef]
- Chae, H.S.; Kieber, J.J. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 2005, 10, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Booker, M.A.; DeLong, A. Producing the ethylene signal: Regulation and diversification of ethylene biosynthetic enzymes. Plant Physiol. 2015, 169, 42–50. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1237. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Tao, Q.; Niu, H.; Zhang, Z.; Li, D.; Gong, Z.; Weng, Y.; Li, Z. A novel allele of monoecious (m) locus is responsible for elongated fruit shape and perfect flowers in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2015, 128, 2483–2493. [Google Scholar] [CrossRef]
- Salman-Minkov, A.; Levi, A.; Wolf, S.; Trebitsh, T. ACC synthase genes are polymorphic in watermelon (Citrullus spp.) and differentially expressed in flowers and in response to auxin and gibberellin. Plant Cell Physiol. 2008, 49, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Sheng, Y.; Niu, H.; Li, Z. Gene interactions regulating sex determination in cucurbits. Front. Plant Sci. 2019, 10, 1231. [Google Scholar] [CrossRef]
- Dong, H.; Zhen, Z.; Peng, J.; Chang, L.; Gong, Q.; Wang, N.N. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 2011, 62, 4875–4887. [Google Scholar] [CrossRef] [Green Version]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Li, Z. Cloning and Characterization of Unisexual Flower-Controlling M/m Gene in Cucumber. Ph.D. Thesis, Shanghai Jiao Tong University, Shanghai, China, 2010. [Google Scholar]
- Li, Z.; Wang, S.; Tao, Q.; Pan, J.; Si, L.; Gong, Z.; Cai, R. A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 2012, 63, 4475–4484. [Google Scholar] [CrossRef] [PubMed]
- Tatsuki, M.; Nakajima, N.; Fujii, H.; Shimada, T.; Nakano, M.; Hayashi, K.-I.; Hayama, H.; Yoshioka, H.; Nakamura, Y. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J. Exp. Bot. 2013, 64, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, C.; Zou, B.; Wang, C.; Xu, W.; Cui, C.; Qu, S. Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression. Int. Int. J. Mol. Sci. 2019, 20, 3185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, K.; Peterson, C.; Tolla, G. Induction of perfect flowers on gynoecious muskmelon by silver nitrate and aminoethoxyvinylglycine. HortScience 1980, 15, 654–655. [Google Scholar]
- Baker, J.E.; Lieberman, M.; Anderson, J.D. Inhibition of Ethylene Production in Fruit Slices by a Rhizobitoxine Analog and Free Radical Scavengers. Plant Physiol. 1978, 61, 886–888. [Google Scholar] [CrossRef] [Green Version]
- Bangerth, F. The effect of a substituted amino acid on ethylene biosynthesis, respiration, ripening and preharvest drop of apple fruits. J. Am. Soc. Hortic. Sci. 1978, 103, 401–404. [Google Scholar]
- Schaller, G.E.; Binder, B.M. Inhibitors of ethylene biosynthesis and signaling. In Ethylene Signaling; Springer: Berlin/Heidelberg, Germany, 2017; pp. 223–235. [Google Scholar]
- Beyer Jr, E.M. Effect of silver ion, carbon dioxide, and oxygen on ethylene action and metabolism. Plant Physiol. 1979, 63, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivica, L.; Peer, B. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar]
- Sara, E.G.; Jaina, M.; Alex, B.; Eddy, S.R.; Aurélien, L.; Potter, S.C.; Matloob, Q.; Richardson, L.J.; Salazar, G.A.; Alfredo, S. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, K.B. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 2003, 19, 1585–1586. [Google Scholar] [CrossRef] [Green Version]
- Balakrishnan, S.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Wu, C.; Liu, H.; Hu, Z.; Sun, G. Cloning and Molecular Characterization of a SERK Gene Transcriptionally Induced During Somatic Embryogenesis in Ananas comosus cv. Shenwan. Plant Mol. Biol. Report. 2012, 30, 195–203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2 (-delta delta c (t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Gene | Gene ID | Chromosomal Localization | CDS Length (bp) | Protein Length (aa) | Mw (kDa) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| CmaACS1 | CmaCh02G015170.1 | Chr02: 8650601~8653154 | 1650 | 549 | 60.14 | 5.65 | Chloroplast |
| CmaACS2 | CmaCh03G008840.1 | Chr03: 6510606~6512326 | 1437 | 478 | 53.80 | 7.98 | Nucleus |
| CmaACS3 | CmaCh04G007260.1 | Chr04: 3694549~3696868 | 1455 | 484 | 54.31 | 6.61 | Cytoplasm |
| CmaACS4 | CmaCh04G008430.1 | Chr04: 4336601~4338975 | 1410 | 469 | 53.05 | 8.96 | Nucleus |
| CmaACS5 | CmaCh05G001180.1 | Chr05: 508990~511072 | 1329 | 442 | 49.88 | 9.42 | Cytoplasm |
| CmaACS6 | CmaCh07G006250.1 | Chr07: 2713056~2714884 | 1428 | 475 | 53.48 | 8.43 | Nucleus |
| CmaACS7 | CmaCh10G007020.1 | Chr10: 3155561~3157085 | 1338 | 445 | 49.92 | 5.86 | Cytoplasm |
| CmaACS8 | CmaCh11G006760.1 | Chr11: 3270641~3272363 | 1326 | 441 | 49.81 | 5.61 | Chloroplast |
| CmaACS9 | CmaCh15G011790.1 | Chr15: 7462460~7465739 | 1650 | 549 | 60.27 | 5.76 | Chloroplast |
| CmaACS10 | CmaCh16G005820.1 | Chr16: 3012078~3013989 | 1455 | 484 | 54.22 | 7.60 | Cytoplasm |
| CmaACS11 | CmaCh16G007300.1 | Chr16: 3845420~3847307 | 1491 | 496 | 56.18 | 8.57 | Nucleus |
| CmaACS12 | CmaCh17G004550.1 | Chr17: 2713968~2716904 | 1395 | 464 | 52.53 | 6.90 | Chloroplast |
| CmaACS13 | CmaCh17G004560.1 | Chr17: 2729331~2731675 | 1482 | 493 | 55.90 | 6.77 | Chloroplast |
| Treatments | Number of Female Flowers Per Plant | First Female Flower Position | Number of Bisexual Flowers Per Plant |
|---|---|---|---|
| Ethephon | 14.8 ± 0.6a | 5.7 ± 0.5c | 0.5 ± 0.2b |
| IAA | 14.3 ± 1.2a | 6.8 ± 1.4c | 1.3 ± 0.4b |
| AVG | 9.8 ± 0.8b | 11.3 ± 0.9b | 5.4 ± 0.9a |
| AgNO3 | 6.5 ± 0.2c | 12.9 ± 1.0a | 6.4 ± 0.9a |
| CK | 10.2 ± 0.8b | 10.1 ± 1.1b | 5.2 ± 1.1a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Li, W.; Chen, F.; Cheng, Y.; Huang, X.; Zou, B.; Wang, Y.; Xu, W.; Qu, S. Genome-Wide Identification and Characterization of Members of the ACS Gene Family in Cucurbita maxima and Their Transcriptional Responses to the Specific Treatments. Int. J. Mol. Sci. 2022, 23, 8476. https://doi.org/10.3390/ijms23158476
Wang C, Li W, Chen F, Cheng Y, Huang X, Zou B, Wang Y, Xu W, Qu S. Genome-Wide Identification and Characterization of Members of the ACS Gene Family in Cucurbita maxima and Their Transcriptional Responses to the Specific Treatments. International Journal of Molecular Sciences. 2022; 23(15):8476. https://doi.org/10.3390/ijms23158476
Chicago/Turabian StyleWang, Chaojie, Wenling Li, Fangyuan Chen, Yaqian Cheng, Xin Huang, Bingxue Zou, Yunli Wang, Wenlong Xu, and Shuping Qu. 2022. "Genome-Wide Identification and Characterization of Members of the ACS Gene Family in Cucurbita maxima and Their Transcriptional Responses to the Specific Treatments" International Journal of Molecular Sciences 23, no. 15: 8476. https://doi.org/10.3390/ijms23158476
APA StyleWang, C., Li, W., Chen, F., Cheng, Y., Huang, X., Zou, B., Wang, Y., Xu, W., & Qu, S. (2022). Genome-Wide Identification and Characterization of Members of the ACS Gene Family in Cucurbita maxima and Their Transcriptional Responses to the Specific Treatments. International Journal of Molecular Sciences, 23(15), 8476. https://doi.org/10.3390/ijms23158476






