Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points
Abstract
:1. Introduction
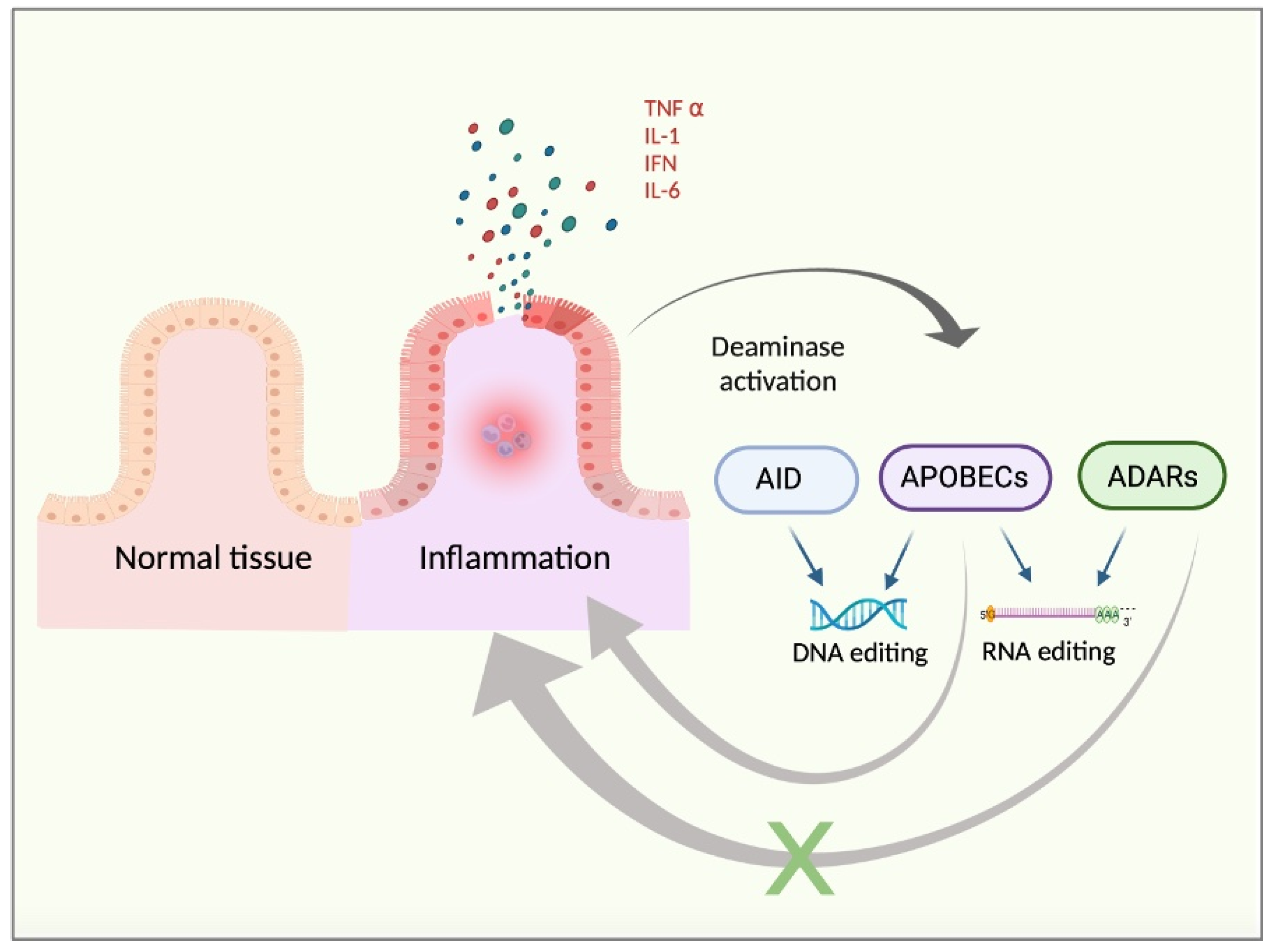
2. Deaminases and Cancer Inflammation
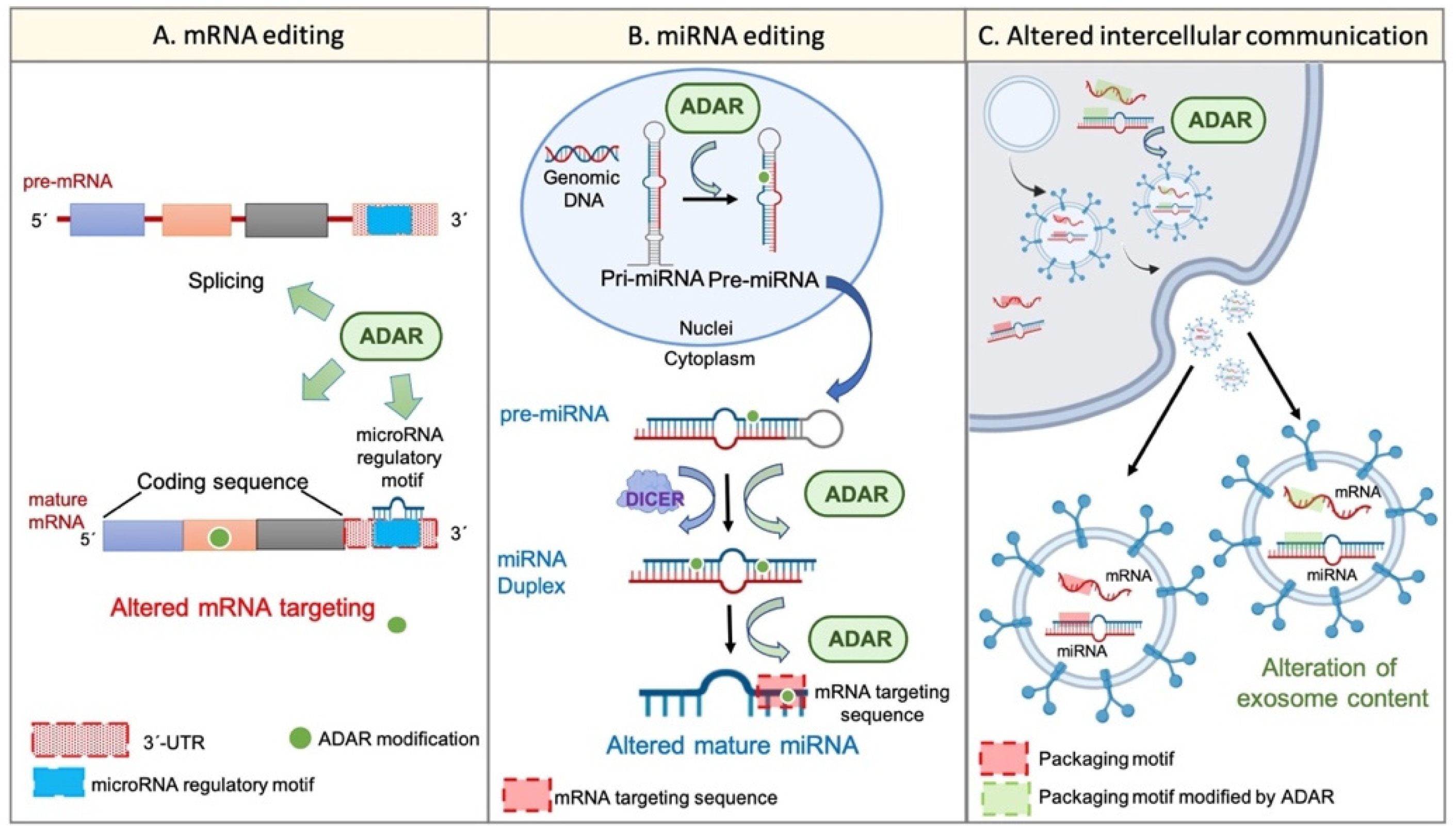
3. RNA Editing, Cellular Transcriptome, and Cancer Stem Cells
4. Deaminases and Our Understanding of Tumour Progression
5. Diagnostic and Therapeutic Considerations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADARs | adenosine deaminases acting on RNA |
| AID | activation-induced cytidine deaminase |
| APOBEC | apolipoprotein B mRNA editing catalytic polypeptide-like |
| CSCs | cancer stem cells |
| EMT | epithelial-to-mesenchymal transition |
| iPSCs | induced pluripotent stem cells |
| CAFs | cancer-associated fibroblasts |
| TAMs | tumour-associated macrophages |
| TME | tumour microenvironment |
References
- Rios, L.A.d.S.; Cloete, B.; Mowla, S. Activation-Induced Cytidine Deaminase: In Sickness and in Health. J. Cancer Res. Clin. Oncol. 2020, 146, 2721–2730. [Google Scholar] [CrossRef]
- Wong, L.; Vizeacoumar, F.S.; Vizeacoumar, F.J.; Chelico, L. APOBEC1 Cytosine Deaminase Activity on Single-Stranded DNA Is Suppressed by Replication Protein A. Nucleic Acids Res. 2021, 49, 322–339. [Google Scholar] [CrossRef]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic Landscape and Regulation of RNA Editing in Mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Shiromoto, Y.; Sakurai, M.; Minakuchi, M.; Ariyoshi, K.; Nishikura, K. ADAR1 RNA Editing Enzyme Regulates R-Loop Formation and Genome Stability at Telomeres in Cancer Cells. Nat. Commun. 2021, 12, 1654. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Godsmark, G.; DE Souza Rios, L.A.; Mowla, S. Activation-Induced Cytidine Deaminase Promotes Proliferation and Enhances Chemoresistance and Migration in B-Cell Lymphoma. Anticancer Res. 2021, 41, 237–247. [Google Scholar] [CrossRef]
- Ito, F.; Fu, Y.; Kao, S.-C.A.; Yang, H.; Chen, X.S. Family-Wide Comparative Analysis of Cytidine and Methylcytidine Deamination by Eleven Human APOBEC Proteins. J. Mol. Biol. 2017, 429, 1787–1799. [Google Scholar] [CrossRef]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.T.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Neufert, C.; Heichler, C.; Brabletz, T.; Scheibe, K.; Boonsanay, V.; Greten, F.R.; Neurath, M.F. Inducible Mouse Models of Colon Cancer for the Analysis of Sporadic and Inflammation-Driven Tumor Progression and Lymph Node Metastasis. Nat. Protoc. 2021, 16, 61–85. [Google Scholar] [CrossRef]
- Liu, C.-H.; Chen, Z.; Chen, K.; Liao, F.-T.; Chung, C.-E.; Liu, X.; Lin, Y.-C.; Keohavong, P.; Leikauf, G.D.; Di, Y.P. Lipopolysaccharide-Mediated Chronic Inflammation Promotes Tobacco Carcinogen-Induced Lung Cancer and Determines the Efficacy of Immunotherapy. Cancer Res. 2021, 81, 144–157. [Google Scholar] [CrossRef]
- Zong, C.; Kimura, Y.; Kinoshita, K.; Takasu, S.; Zhang, X.; Sakurai, T.; Sekido, Y.; Ichihara, S.; Endo, G.; Ichihara, G. Exposure to 1,2-Dichloropropane Upregulates the Expression of Activation-Induced Cytidine Deaminase (AID) in Human Cholangiocytes Co-Cultured With Macrophages. Toxicol. Sci. 2019, 168, 137–148. [Google Scholar] [CrossRef]
- Petljak, M.; Alexandrov, L.B.; Brammeld, J.S.; Price, S.; Wedge, D.C.; Grossmann, S.; Dawson, K.J.; Ju, Y.S.; Iorio, F.; Tubio, J.M.C.; et al. Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis. Cell 2019, 176, 1282–1294.e20. [Google Scholar] [CrossRef] [Green Version]
- Pfaller, C.K.; George, C.X.; Samuel, C.E. Adenosine Deaminases Acting on RNA (ADARs) and Viral Infections. Annu. Rev. Virol. 2021, 8, 239–264. [Google Scholar] [CrossRef]
- Paul, R.; Dorsey, J.F.; Fan, Y. Cell Plasticity, Senescence, and Quiescence in Cancer Stem Cells: Biological and Therapeutic Implications. Pharmacol. Ther. 2022, 231, 107985. [Google Scholar] [CrossRef]
- Jiang, Q.; Isquith, J.; Ladel, L.; Mark, A.; Holm, F.; Mason, C.; He, Y.; Mondala, P.; Oliver, I.; Pham, J.; et al. Inflammation-Driven Deaminase Deregulation Fuels Human Pre-Leukemia Stem Cell Evolution. Cell Rep. 2021, 34, 108670. [Google Scholar] [CrossRef]
- Varela, I.; Menendez, P.; Sanjuan-Pla, A. Intratumoral Heterogeneity and Clonal Evolution in Blood Malignancies and Solid Tumors. Oncotarget 2017, 8, 66742–66746. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.G. Understanding Cancer Stem Cell Heterogeneity and Plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef]
- Vermeulen, L.; de Sousa e Melo, F.; Richel, D.J.; Medema, J.P. The Developing Cancer Stem-Cell Model: Clinical Challenges and Opportunities. Lancet. Oncol. 2012, 13, e83–e89. [Google Scholar] [CrossRef]
- Hernández-Camarero, P.; Jiménez, G.; López-Ruiz, E.; Barungi, S.; Marchal, J.A.; Perán, M. Revisiting the Dynamic Cancer Stem Cell Model: Importance of Tumour Edges. Crit. Rev. Oncol. Hematol. 2018, 131, 35–45. [Google Scholar] [CrossRef]
- Nishikori, A.; Nishimura, Y.; Shibata, R.; Ohshima, K.-I.; Gion, Y.; Ikeda, T.; Nishimura, M.F.; Yoshino, T.; Sato, Y. Upregulated Expression of Activation-Induced Cytidine Deaminase in Ocular Adnexal Marginal Zone Lymphoma with IgG4-Positive Cells. Int. J. Mol. Sci. 2021, 22, 4083. [Google Scholar] [CrossRef]
- Mohri, T.; Nagata, K.; Kuwamoto, S.; Matsushita, M.; Sugihara, H.; Kato, M.; Horie, Y.; Murakami, I.; Hayashi, K. Aberrant Expression of AID and AID Activators of NF-ΚB and PAX5 Is Irrelevant to EBV-Associated Gastric Cancers, but Is Associated with Carcinogenesis in Certain EBV-Non-Associated Gastric Cancers. Oncol. Lett. 2017, 13, 4133–4140. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Su, N.; Cui, M.; Li, H.; Zhang, Q.; Yu, N.; Wu, S.; Cao, Z. Activation-Induced Cytidine Deaminase Expression in Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 4119–4124. [Google Scholar]
- Messerschmidt, C.; Obermayer, B.; Klinghammer, K.; Ochsenreither, S.; Treue, D.; Stenzinger, A.; Glimm, H.; Fröhling, S.; Kindler, T.; Brandts, C.H.; et al. Distinct Immune Evasion in APOBEC-Enriched, HPV-Negative HNSCC. Int. J. Cancer 2020, 147, 2293–2302. [Google Scholar] [CrossRef]
- Li, S.; Bao, X.; Wang, D.; You, L.; Li, X.; Yang, H.; Bian, J.; Wang, Y.; Yang, Y. APOBEC3B and IL-6 Form a Positive Feedback Loop in Hepatocellular Carcinoma Cells. Sci. China. Life Sci. 2017, 60, 617–626. [Google Scholar] [CrossRef]
- Faure-Dupuy, S.; Riedl, T.; Rolland, M.; Hizir, Z.; Reisinger, F.; Neuhaus, K.; Schuehle, S.; Remouchamps, C.; Gillet, N.; Schönung, M.; et al. Control of APOBEC3B Induction and CccDNA Decay by NF-ΚB and MiR-138-5p. JHEP Rep. Innov. Hepatol. 2021, 3, 100354. [Google Scholar] [CrossRef]
- Mussil, B.; Suspène, R.; Aynaud, M.-M.; Gauvrit, A.; Vartanian, J.-P.; Wain-Hobson, S. Human APOBEC3A Isoforms Translocate to the Nucleus and Induce DNA Double Strand Breaks Leading to Cell Stress and Death. PLoS ONE 2013, 8, e73641. [Google Scholar] [CrossRef] [Green Version]
- Alqassim, E.Y.; Sharma, S.; Khan, A.N.M.N.H.; Emmons, T.R.; Cortes Gomez, E.; Alahmari, A.; Singel, K.L.; Mark, J.; Davidson, B.A.; Robert McGray, A.J.; et al. RNA Editing Enzyme APOBEC3A Promotes Pro-Inflammatory M1 Macrophage Polarization. Commun. Biol. 2021, 4, 102. [Google Scholar] [CrossRef]
- Hou, S.; Lee, J.M.; Myint, W.; Matsuo, H.; Kurt Yilmaz, N.; Schiffer, C.A. Structural Basis of Substrate Specificity in Human Cytidine Deaminase Family APOBEC3s. J. Biol. Chem. 2021, 297, 100909. [Google Scholar] [CrossRef]
- Zipeto, M.A.; Court, A.C.; Sadarangani, A.; Delos Santos, N.P.; Balaian, L.; Chun, H.-J.; Pineda, G.; Morris, S.R.; Mason, C.N.; Geron, I.; et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell 2016, 19, 177–191. [Google Scholar] [CrossRef]
- Jiang, L.; Park, M.J.; Cho, C.J.; Lee, K.; Jung, M.K.; Pack, C.G.; Myung, S.-J.; Chang, S. ADAR1 Suppresses Interferon Signaling in Gastric Cancer Cells by MicroRNA-302a-Mediated IRF9/STAT1 Regulation. Int. J. Mol. Sci. 2020, 21, 6195. [Google Scholar] [CrossRef]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of ADAR1 in Tumours Overcomes Resistance to Immune Checkpoint Blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.-P.; Cottrell, K.A.; Ryu, S.; Bramel, E.R.; Kladney, R.D.; Bao, E.A.; Freeman, E.C.; Sabloak, T.; Maggi, L.J.; Weber, J.D. Evaluating the Therapeutic Potential of ADAR1 Inhibition for Triple-Negative Breast Cancer. Oncogene 2021, 40, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Yujie Ding, M.M.; Shi, X.; Ji, J.; Su, Y. ADAR1p150 Regulates the Biosynthesis and Function of MiRNA-149* in Human Melanoma. Biochem. Biophys. Res. Commun. 2020, 523, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, J.; Liu, S.; Li, X.; Zhang, D.; Wang, X.; Jiang, J.; Hu, W.; Zhang, Y.; Jin, B.; et al. ADAR1 Attenuates Allogeneic Graft Rejection by Suppressing MiR-21 Biogenesis in Macrophages and Promoting M2 Polarization. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 5162–5173. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Shi, R.; Zhao, T.; Wang, Y.; Anastasov, N.; Rosemann, M.; Fang, W. Integrated Analysis of Single-Cell RNA-Seq and Bulk RNA-Seq Unravels Tumour Heterogeneity plus M2-like Tumour-Associated Macrophage Infiltration and Aggressiveness in TNBC. Cancer Immunol. Immunother. 2021, 70, 189–202. [Google Scholar] [CrossRef]
- Takeda, S.; Shigeyasu, K.; Okugawa, Y.; Yoshinaga, K.; Mori, Y.; Yano, S.; Noma, K.; Umeda, Y.; Kondo, Y.; Kishimoto, H.; et al. Activation of AZIN1 RNA Editing Is a Novel Mechanism That Promotes Invasive Potential of Cancer-Associated Fibroblasts in Colorectal Cancer. Cancer Lett. 2019, 444, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Camarero, P.; López-Ruiz, E.; Marchal, J.A.; Perán, M. Cancer: A Mirrored Room between Tumor Bulk and Tumor Microenvironment. J. Exp. Clin. Cancer Res. 2021, 40, 217. [Google Scholar] [CrossRef]
- Guallar, D.; Fuentes-Iglesias, A.; Souto, Y.; Ameneiro, C.; Freire-Agulleiro, O.; Pardavila, J.A.; Escudero, A.; Garcia-Outeiral, V.; Moreira, T.; Saenz, C.; et al. ADAR1-Dependent RNA Editing Promotes MET and IPSC Reprogramming by Alleviating ER Stress. Cell Stem Cell 2020, 27, 300–314.e11. [Google Scholar] [CrossRef]
- Crews, L.A.; Jiang, Q.; Zipeto, M.A.; Lazzari, E.; Court, A.C.; Ali, S.; Barrett, C.L.; Frazer, K.A.; Jamieson, C.H.M. An RNA Editing Fingerprint of Cancer Stem Cell Reprogramming. J. Transl. Med. 2015, 13, 52. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Fu, Y.; Huang, J.; Wu, M.; Zhang, Z.; Xu, R.; Zhang, P.; Zhao, S.; Liu, L.; Jiang, H. ADAR1 Promotes the Epithelial-to-Mesenchymal Transition and Stem-like Cell Phenotype of Oral Cancer by Facilitating Oncogenic MicroRNA Maturation. J. Exp. Clin. Cancer Res. 2019, 38, 315. [Google Scholar] [CrossRef]
- Shen, P.; Yang, T.; Chen, Q.; Yuan, H.; Wu, P.; Cai, B.; Meng, L.; Huang, X.; Liu, J.; Zhang, Y.; et al. CircNEIL3 Regulatory Loop Promotes Pancreatic Ductal Adenocarcinoma Progression via MiRNA Sponging and A-to-I RNA-Editing. Mol. Cancer 2021, 20, 51. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Moya, J.; Baker, A.R.; Slack, F.J.; Santisteban, P. ADAR1-Mediated RNA Editing Is a Novel Oncogenic Process in Thyroid Cancer and Regulates MiR-200 Activity. Oncogene 2020, 39, 3738–3753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fumagalli, D.; Gacquer, D.; Rothé, F.; Lefort, A.; Libert, F.; Brown, D.; Kheddoumi, N.; Shlien, A.; Konopka, T.; Salgado, R.; et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep. 2015, 13, 277–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemlich, Y.; Baruch, E.N.; Besser, M.J.; Shoshan, E.; Bar-Eli, M.; Anafi, L.; Barshack, I.; Schachter, J.; Ortenberg, R.; Markel, G. ADAR1-Mediated Regulation of Melanoma Invasion. Nat. Commun. 2018, 9, 2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Liu, H.F.; Qiao, L.B.; Wang, F.B.; Wang, L.; Lin, Y.; Liu, J. Global RNA Editing Identification and Characterization during Human Pluripotent-to-Cardiomyocyte Differentiation. Mol. Ther.-Nucleic Acids 2021, 26, 879–891. [Google Scholar] [CrossRef]
- Roberts, J.T.; Patterson, D.G.; King, V.M.; Amin, S.V.; Polska, C.J.; Houserova, D.; Crucello, A.; Barnhill, E.C.; Miller, M.M.; Sherman, T.D.; et al. ADAR Mediated RNA Editing Modulates MicroRNA Targeting in Human Breast Cancer. Processes 2018, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Tassinari, V.; Cesarini, V.; Silvestris, D.A.; Scafidi, A.; Cucina, L.; Gallo, A. MicroRNA Editing Detection and Function: A Combined In Silico and Experimental Approach for the Identification and Validation of Putative Oncogenic Targets. Methods Mol. Biol. 2021, 2181, 253–267. [Google Scholar] [CrossRef]
- Farace, C.; Pisano, A.; Griñan-Lison, C.; Solinas, G.; Jiménez, G.; Serra, M.; Carrillo, E.; Scognamillo, F.; Attene, F.; Montella, A.; et al. Deregulation of Cancer-Stem-Cell-Associated MiRNAs in Tissues and Sera of Colorectal Cancer Patients. Oncotarget 2020, 11, 116–130. [Google Scholar] [CrossRef] [Green Version]
- Shiromoto, Y.; Sakurai, M.; Qu, H.; Kossenkov, A.V.; Nishikura, K. Processing of Alu Small RNAs by DICER/ADAR1 Complexes and Their RNAi Targets. RNA 2020, 26, 1801–1814. [Google Scholar] [CrossRef]
- Kapoor, U.; Licht, K.; Amman, F.; Jakobi, T.; Martin, D.; Dieterich, C.; Jantsch, M.F. ADAR-Deficiency Perturbs the Global Splicing Landscape in Mouse Tissues. Genome Res. 2020, 30, 1107–1118. [Google Scholar] [CrossRef]
- Ramírez-Moya, J.; Miliotis, C.; Baker, A.R.; Gregory, R.I.; Slack, F.J.; Santisteban, P. An ADAR1-Dependent RNA Editing Event in the Cyclin-Dependent Kinase CDK13 Promotes Thyroid Cancer Hallmarks. Mol. Cancer 2021, 20, 115. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Camarero, P.; López-Ruiz, E.; Griñán-Lisón, C.; García, M.Á.; Chocarro-Wrona, C.; Marchal, J.A.; Kenyon, J.; Perán, M. Pancreatic (pro)Enzymes Treatment Suppresses BXPC-3 Pancreatic Cancer Stem Cell Subpopulation and Impairs Tumour Engrafting. Sci. Rep. 2019, 9, 11359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafazadeh, M.; Samadi, N.; Kahroba, H.; Baradaran, B.; Haiaty, S.; Nouri, M. Potential Roles and Prognostic Significance of Exosomes in Cancer Drug Resistance. Cell Biosci. 2021, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ning, Z.; Ma, L.; Liu, W.; Shao, C.; Shu, Y.; Shen, H. Exosomal MiRNAs and MiRNA Dysregulation in Cancer-Associated Fibroblasts. Mol. Cancer 2017, 16, 148. [Google Scholar] [CrossRef]
- Ramachandran, S.; Palanisamy, V. Horizontal Transfer of RNAs: Exosomes as Mediators of Intercellular Communication. Wiley Interdiscip. Rev. RNA 2012, 3, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Ströbel, T.; Erkan, E.P.; Fan, J.-B.; Breakefield, X.O.; Saydam, O. MiR-1289 and “Zipcode”-like Sequence Enrich MRNAs in Microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Tang, G.; Xie, B.; Hong, X.; Qin, H.; Wang, J.; Huang, H.; Hao, P.; Li, X. Creating RNA Specific C-to-U Editase from APOBEC3A by Separation of Its Activities on DNA and RNA Substrates. ACS Synth. Biol. 2021, 10, 1106–1115. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, T.; Huang, L.; Wang, Z.; Yan, Z.; Guan, Y.; Hu, W.; Fan, Z.; Zhu, P. Circular RNA Cia-MAF Drives Self-Renewal and Metastasis of Liver Tumor-Initiating Cells via Transcription Factor MAFF. J. Clin. Investig. 2021, 131, e148020. [Google Scholar] [CrossRef]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A Novel Hypoxic Long Noncoding RNA KB-1980E6.3 Maintains Breast Cancer Stem Cell Stemness via Interacting with IGF2BP1 to Facilitate c-Myc MRNA Stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell 2018, 33, 817–828.e7. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The Repertoire of Mutational Signatures in Human Cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Ren, W.; Liu, D.; Li, X.; Li, W.; Wang, X.; Meng, F.-L.; Yeap, L.-S.; Hou, Y.; Zhu, S.; et al. Genome-Wide Mutational Signatures Revealed Distinct Developmental Paths for Human B Cell Lymphomas. J. Exp. Med. 2021, 218, e20200573. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, M.; Patnaik, S.K.; Ishikawa, T.; Takabe, K. Different Members of the APOBEC3 Family of DNA Mutators Have Opposing Associations with the Landscape of Breast Cancer. Am. J. Cancer Res. 2021, 11, 5111–5125. [Google Scholar] [PubMed]
- Delgado, P.; Álvarez-Prado, Á.F.; Marina-Zárate, E.; Sernandez, I.V.; Mur, S.M.; de la Barrera, J.; Sanchez-Cabo, F.; Cañamero, M.; de Molina, A.; Belver, L.; et al. Interplay between UNG and AID Governs Intratumoral Heterogeneity in Mature B Cell Lymphoma. PLoS Genet. 2020, 16, e1008960. [Google Scholar] [CrossRef]
- Mamrot, J.; Balachandran, S.; Steele, E.J.; Lindley, R.A. Molecular Model Linking Th2 Polarized M2 Tumour-Associated Macrophages with Deaminase-Mediated Cancer Progression Mutation Signatures. Scand. J. Immunol. 2019, 89, e12760. [Google Scholar] [CrossRef]
- Argyris, P.P.; Wilkinson, P.E.; Jarvis, M.C.; Magliocca, K.R.; Patel, M.R.; Vogel, R.I.; Gopalakrishnan, R.; Koutlas, I.G.; Harris, R.S. Endogenous APOBEC3B Overexpression Characterizes HPV-Positive and HPV-Negative Oral Epithelial Dysplasias and Head and Neck Cancers. Mod. Pathol. 2021, 34, 280–290. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Yu, C.P.; Chang, S.; Xie, L.L.; Wang, S. Genome-wide expression changes mediated by A-to-I RNA editing correlate with hepatic oncogenesis. Transl. Cancer Res. 2021, 10, 2725–2737. [Google Scholar] [CrossRef]
- Schubert, M.; Hackl, H.; Gassner, F.J.; Greil, R.; Geisberger, R. Investigating Epigenetic Effects of Activation-Induced Deaminase in Chronic Lymphocytic Leukemia. PLoS ONE 2018, 13, e0208753. [Google Scholar] [CrossRef]
- Che, Z.; Fan, J.; Zhou, Z.; Li, Q.; Ma, Z.; Hu, Z.; Wu, Y.; Jin, Y.; Su, Y.; Liang, P.; et al. Activation-Induced Cytidine Deaminase Expression Facilitates the Malignant Phenotype and Epithelial-to-Mesenchymal Transition in Clear Cell Renal Cell Carcinoma. DNA Cell Biol. 2020, 39, 1299–1312. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; Ma, Z.; Zhou, Z.; Fan, J.; Jin, Y.; Wu, Y.; Cheng, F.; Liang, P. AID Modulates Carcinogenesis Network via DNA Demethylation in Bladder Urothelial Cell Carcinoma. Cell Death Dis. 2019, 10, 251. [Google Scholar] [CrossRef] [Green Version]
- Nandy, S.B.; Lakshmanaswamy, R. Cancer Stem Cells and Metastasis. Prog. Mol. Biol. Transl. Sci. 2017, 151, 137–176. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Lee, L.J.; Xiong, H.; Su, H.; Rao, J.; Xiao, D.; He, J.; Wu, K.; Liu, D. Characterization of RNA Editome in Primary and Metastatic Lung Adenocarcinomas. Oncotarget 2017, 8, 11517–11529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhaoting, Y.; Zhang, C.; Liu, X.; Che, N.; Feng, Y.; Xuan, Y. SETD5 Regulates Glycolysis in Breast Cancer Stem-Like Cells and Fuels Tumor Growth. Am. J. Pathol. 2022, 192, 712–721. [Google Scholar] [CrossRef]
- Alonso-Curbelo, D.; Ho, Y.; Burdziak, C.; Maag, J.L.V.; Iv, J.P.M.; Chandwani, R.; Chen, H.; Tsanov, K.M.; Barriga, F.M.; Luan, W.; et al. A Gene—Environment-Induced Epigenetic Program Initiates Tumorigenesis. Nature 2021, 590, 642–648. [Google Scholar] [CrossRef]
- Cho, C.J.; Myung, S.J.; Chang, S. ADAR1 and MicroRNA; a Hidden Crosstalk in Cancer. Int. J. Mol. Sci. 2017, 18, 799. [Google Scholar] [CrossRef] [Green Version]
- Kanabe, B.O.; Ozaslan, M.; Aziz, S.A.; Al-Attar, M.S.; Kılıç, İ.H.; Khailany, R.A. Expression patterns of LncRNA-GAS5 and its target APOBEC3C gene through miR-103 in breast cancer patients. Cell Mol. Biol. 2021, 67, 5–10. [Google Scholar] [CrossRef]
- Liu, J.J.; Ho, J.Y.; Lee, J.E.; Hur, S.Y.; Yoo, J.; Kim, K.R.; Ryu, D.; Kim, T.M.; Choi, Y.J. Genomic, Transcriptomic, and Viral Integration Profiles Associated with Recurrent/Metastatic Progression in High-Risk Human Papillomavirus Cervical Carcinomas. Cancer Med. 2020, 9, 8243–8257. [Google Scholar] [CrossRef]
- Tan, Y.; Chen, H.; Zhang, J.; Cai, L.; Jin, S.; Song, D.; Yang, T.; Guo, Z.; Wang, X. Platinum(IV) complexes as inhibitors of CD47-SIRPα axis for chemoimmunotherapy of cancer. Eur. J. Med. Chem. 2022, 229, 114047. [Google Scholar] [CrossRef]
- Lu, Y.; Fan, D.; Wang, W.; Gao, X.; Li, H.; Guo, S.; Zhao, L.; Guo, Y.; Li, B.; Zhong, Y.; et al. The protein 4.1R downregulates VEGFA in M2 macrophages to inhibit colon cancer metastasis. Exp. Cell Res. 2021, 409, 112896. [Google Scholar] [CrossRef]
- Zammataro, L.; Lopez, S.; Bellone, S.; Pettinella, F.; Bonazzoli, E.; Perrone, E.; Zhao, S.; Menderes, G.; Altwerger, G.; Han, C.; et al. Whole-Exome Sequencing of Cervical Carcinomas Identifies Activating ERBB2 and PIK3CA Mutations as Targets for Combination Therapy. Proc. Natl. Acad. Sci. USA 2019, 116, 22730–22736. [Google Scholar] [CrossRef]
- Grillo, M.J.; Jones, K.F.M.; Carpenter, M.A.; Harris, R.S.; Harki, D.A. The Current Toolbox for APOBEC Drug Discovery. Trends Pharmacol. Sci. 2022, 43, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G Base Editors for Inducing Targeted DNA Transversions in Human Cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic Characterization of A-to-I RNA Editing Hotspots in MicroRNAs across Human Cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Yao, J.; Bao, R.; Dong, Y.; Zhang, T.; Du, Y.; Wang, G.; Ni, D.; Xun, Z.; Niu, X.; et al. Cross-Talk of Four Types of RNA Modification Writers Defines Tumor Microenvironment and Pharmacogenomic Landscape in Colorectal Cancer. Mol. Cancer 2021, 20, 29. [Google Scholar] [CrossRef] [PubMed]
- Costa Cruz, P.H.; Kato, Y.; Nakahama, T.; Shibuya, T.; Kawahara, Y. A Comparative Analysis of ADAR Mutant Mice Reveals Site-Specific Regulation of RNA Editing. RNA 2020, 26, 454–469. [Google Scholar] [CrossRef]
- Leong, W.-M.; Ripen, A.M.; Mirsafian, H.; Mohamad, S.B.; Merican, A.F. Transcriptogenomics Identification and Characterization of RNA Editing Sites in Human Primary Monocytes Using High-Depth next Generation Sequencing Data. Genomics 2019, 111, 899–905. [Google Scholar] [CrossRef]
- Reautschnig, P.; Wahn, N.; Wettengel, J.; Schulz, A.E.; Latifi, N.; Vogel, P.; Kang, T.-W.; Pfeiffer, L.S.; Zarges, C.; Naumann, U.; et al. CLUSTER Guide RNAs Enable Precise and Efficient RNA Editing with Endogenous ADAR Enzymes in Vivo. Nat. Biotechnol. 2022, 40, 759–768. [Google Scholar] [CrossRef]
- Inoue, J.; Inazawa, J. Cancer-Associated MiRNAs and Their Therapeutic Potential. J. Hum. Genet. 2021, 66, 937–945. [Google Scholar] [CrossRef]
- Jiménez, G.; Hackenberg, M.; Catalina, P.; Boulaiz, H.; Griñán-Lisón, C.; García, M.Á.; Perán, M.; López-Ruiz, E.; Ramírez, A.; Morata-Tarifa, C.; et al. Mesenchymal Stem Cell’s Secretome Promotes Selective Enrichment of Cancer Stem-like Cells with Specific Cytogenetic Profile. Cancer Lett. 2018, 429, 78–88. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Camarero, P.; López-Ruiz, E.; Marchal, J.A.; Perán, M. Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points. Int. J. Mol. Sci. 2022, 23, 8720. https://doi.org/10.3390/ijms23158720
Hernández-Camarero P, López-Ruiz E, Marchal JA, Perán M. Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points. International Journal of Molecular Sciences. 2022; 23(15):8720. https://doi.org/10.3390/ijms23158720
Chicago/Turabian StyleHernández-Camarero, Pablo, Elena López-Ruiz, Juan Antonio Marchal, and Macarena Perán. 2022. "Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points" International Journal of Molecular Sciences 23, no. 15: 8720. https://doi.org/10.3390/ijms23158720





