OsWRKY114 Inhibits ABA-Induced Susceptibility to Xanthomonas oryzae pv. oryzae in Rice
Abstract
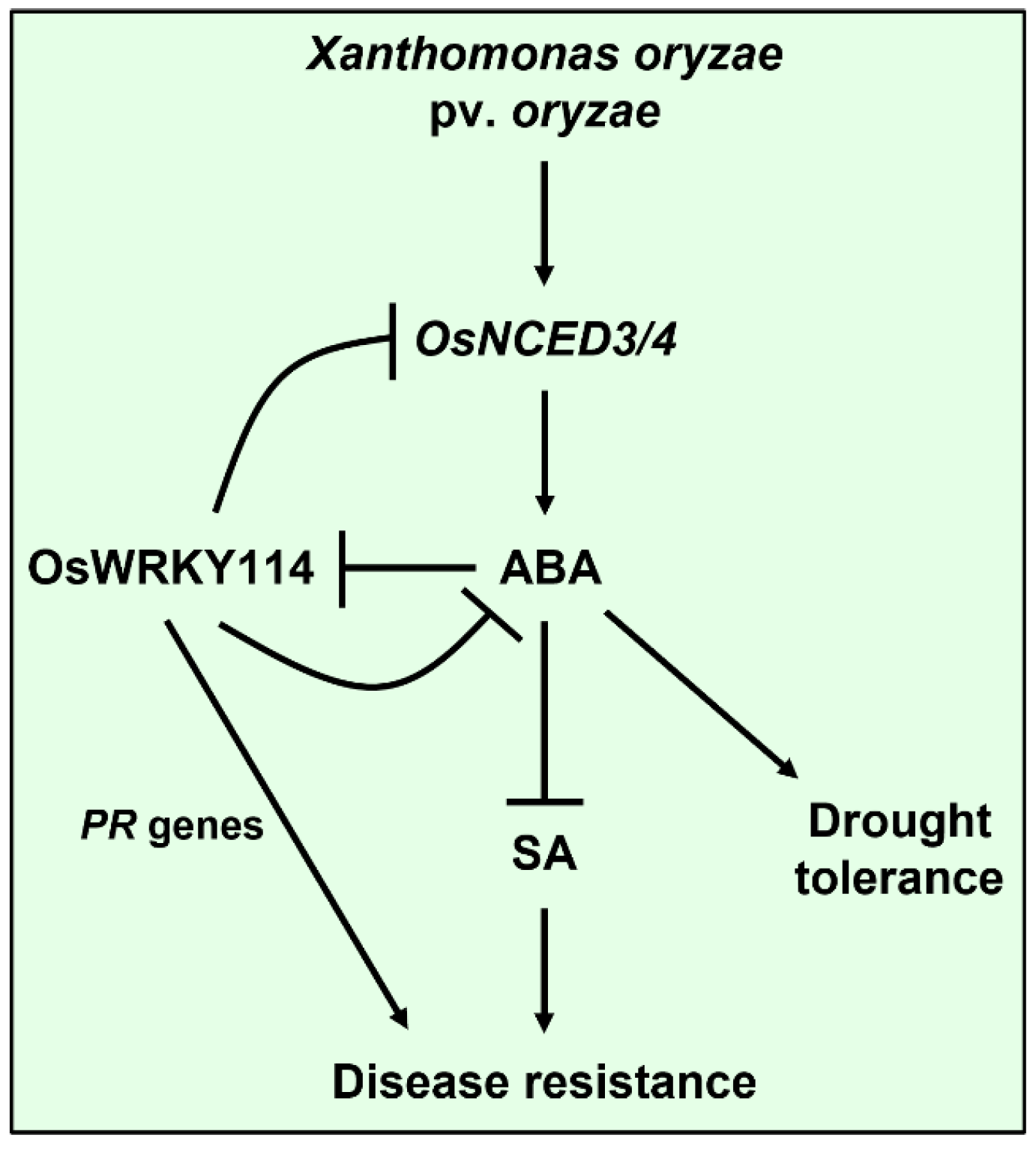
:1. Introduction
2. Results
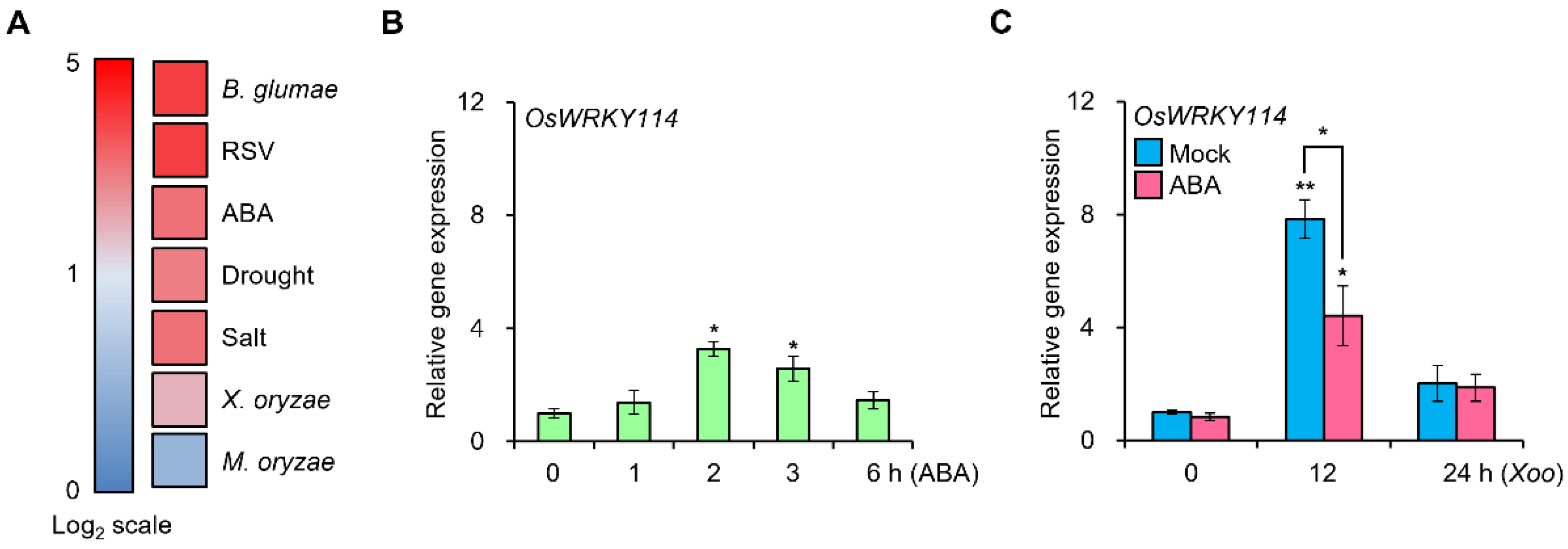
2.1. OsWRKY114 Expression Is Modulated by ABA
2.2. OsWRKY114 Alleviates ABA-Dependent Downregulation of Basal Defense Genes during Xoo Infection
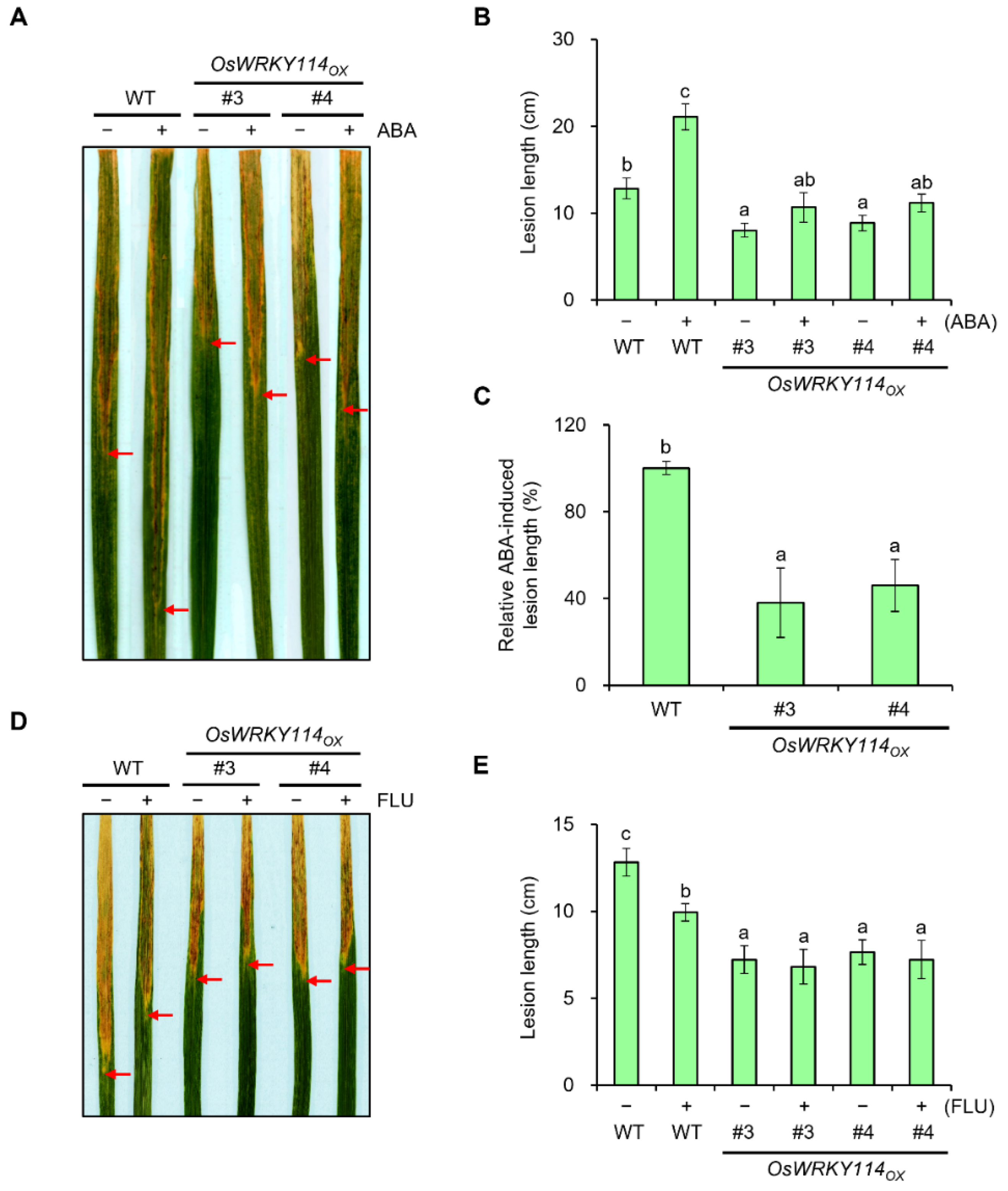
2.3. ABA-Induced Susceptibility to Xoo Is Repressed in OsWRKY114-Overexpressing Plants
2.4. The Negative Effect of ABA on SA Defense Mechanism Is Weaker in OsWRKY114-Overexpressing Plants
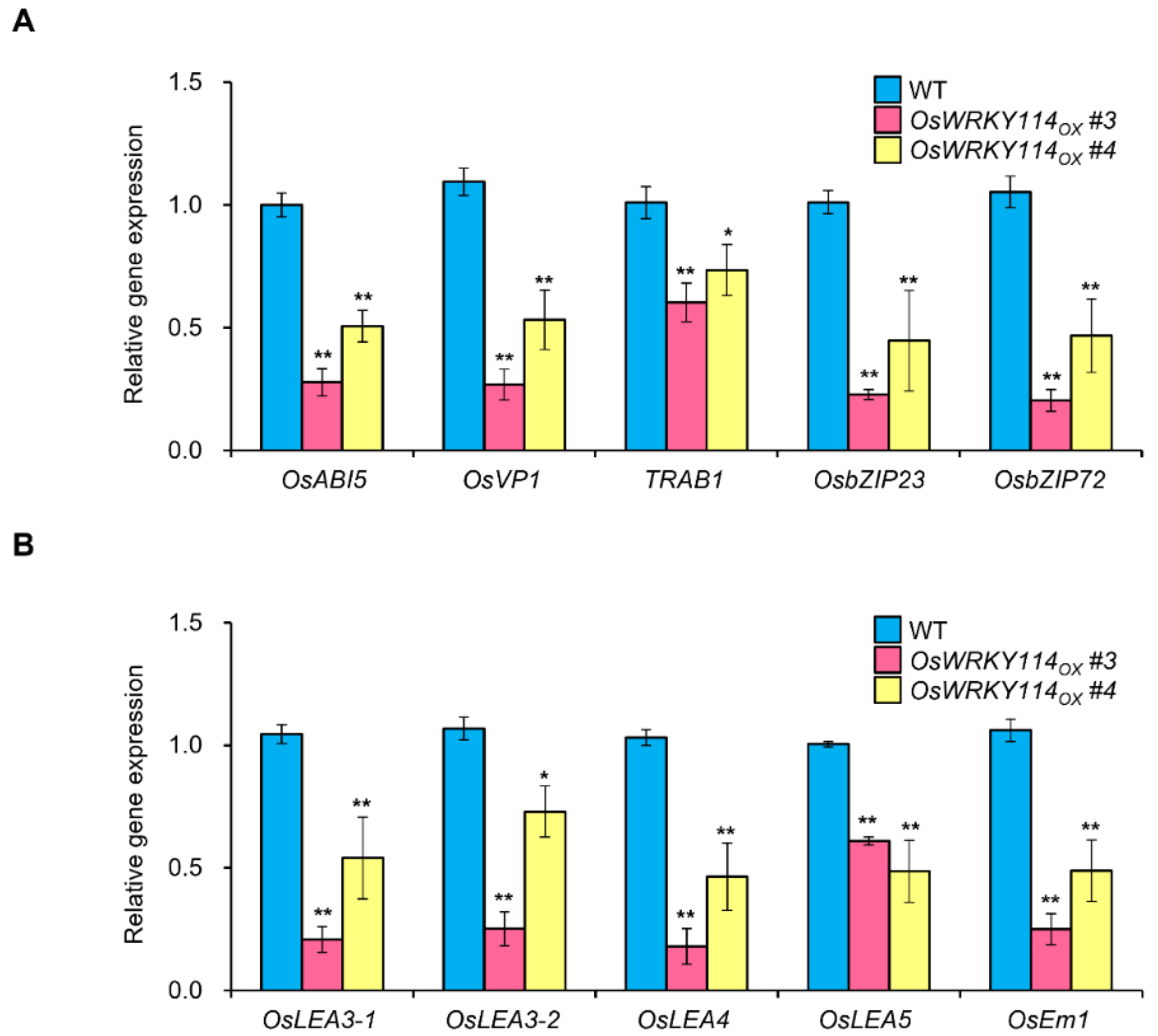
2.5. Various ABA-Response and ABA-Related Genes Are Downregulated in OsWRKY114-Overexpressing Plants
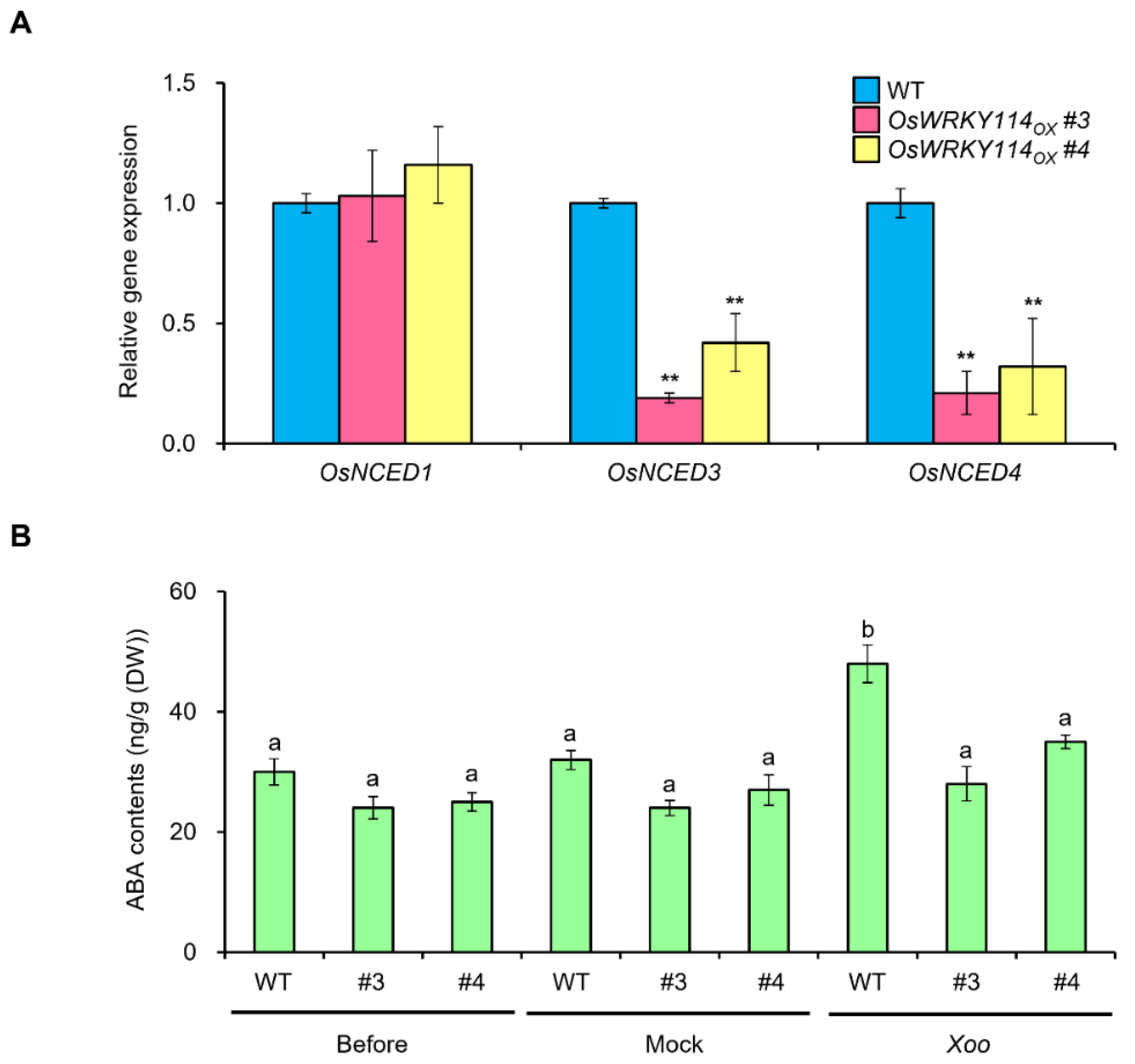
2.6. ABA Biosynthesis Is Attenuated in OsWRKY114-Overexpressing Plants during Xoo Infection
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Gene Expression Analysis
4.3. Phytohormone and Chemical Treatments
4.4. Pathogen Inoculation and Disease Assay
4.5. Transient Gene Expression Assay in Protoplasts
4.6. Analysis of ABA Contents
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weyers, J.D.B.; Paterson, N.W. Plant hormones and the control of physiological processes. New Phytol. 2001, 152, 375–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemhauser, J.L.; Hong, F.; Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 2006, 126, 467–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Env. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuteja, N. Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [Green Version]
- Flors, V.; Ton, J.; van Doorn, R.; Jakab, G.; Garcia-Agustin, P.; Mauch-Mani, B. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2008, 54, 81–92. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Kruler, V.; Winkelmuller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audenaert, K.; de Meyer, G.B.; Hofte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Asselbergh, B.; Achuo, A.E.; Hofte, M.; van Gijsegem, F. Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol. Plant Pathol. 2008, 9, 11–24. [Google Scholar] [CrossRef]
- Asselbergh, B.; De Vleesschauwer, D.; Höfte, M. Global switches and fine-tuning—ABA modulates plant pathogen defense. Mol. Plant-Microbe Interact. 2008, 21, 709–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, M.; Ishikawa, A.; Jikumaru, Y.; Seki, M.; Umezawa, T.; Asami, T.; Maruyama-Nakashita, A.; Kudo, T.; Shinozaki, K.; Yoshida, S.; et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 2008, 20, 1678–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, N. Abscisic acid-mediated suppression of systemic acquired resistance signaling. Plant Cell 2008, 20, 1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henfling, J.; Bostock, R.; Kuc, J. Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology 1980, 70, 1074–1078. [Google Scholar] [CrossRef]
- Matsumoto, K.; Suzuki, Y.; Mase, S.; Watanabe, T.; Sekizawa, Y. On the relationship between plant hormones and rice blast resistance. Jpn. J. Phytopathol. 1980, 46, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Ward, E.W.; Cahill, D.M.; Bhattacharyya, M.K. Abscisic Acid Suppression of Phenylalanine Ammonia-Lyase Activity and mRNA, and Resistance of Soybeans to Phytophthora megasperma f.sp. glycinea. Plant Physiol. 1989, 91, 23–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Koga, H.; Dohi, K.; Mori, M. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea. Physiol. Mol. Plant Pathol. 2004, 65, 3–9. [Google Scholar] [CrossRef]
- Achuo, E.; Prinsen, E.; Höfte, M. Influence of drought, salt stress and abscisic acid on the resistance of tomato to Botrytis cinerea and Oidium neolycopersici. Plant Pathol. 2006, 55, 178–186. [Google Scholar] [CrossRef]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Rodriguez Egea, P.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Hill, L.; Crooks, C.; Doerner, P.; Lamb, C. Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 2009, 150, 1750–1761. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Andrade, J.; González, B.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Vera, P. The role of ABA in plant immunity is mediated through the PYR1 receptor. Int. J. Mol. Sci. 2020, 21, 5852. [Google Scholar] [CrossRef] [PubMed]
- De Torres Zabala, M.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [CrossRef]
- Xu, J.; Audenaert, K.; Hofte, M.; de Vleesschauwer, D. Abscisic Acid Promotes Susceptibility to the Rice Leaf Blight Pathogen Xanthomonas oryzae pv oryzae by Suppressing Salicylic Acid-Mediated Defenses. PLoS ONE 2013, 8, e67413. [Google Scholar]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Sun, P.W.; Tang, X.L.; Gao, Z.H.; Zhang, Z.; Wei, J.H. Genome-wide analysis of WRKY transcription factors in Aquilaria sinensis (Lour.) Gilg. Sci. Rep. 2020, 10, 3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Falak, N.; Imran, Q.M.; Hussain, A.; Yun, B.W. Transcription Factors as the “Blitzkrieg” of Plant Defense: A Pragmatic View of Nitric Oxide’s Role in Gene Regulation. Int. J. Mol. Sci. 2021, 22, 522. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Direct 2015, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Kang, K.; Kim, S.H.; An, G.; Paek, N.C. OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway. Int. J. Mol. Sci. 2019, 20, 4437. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Liu, X.; Miao, R.; Mou, C.; Nguyen, T.; Wang, J.; Zhang, X.; et al. WRKY Transcription Factor OsWRKY29 Represses Seed Dormancy in Rice by Weakening Abscisic Acid Response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [Google Scholar] [CrossRef]
- Son, S.; An, H.K.; Seol, Y.J.; Park, S.R.; Im, J.H. Rice transcription factor WRKY114 directly regulates the expression of OsPR1a and Chitinase to enhance resistance against Xanthomonas oryzae pv. oryzae. Biochem. Biophys. Res. Commun. 2020, 533, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Son, S.; Lee, K.S.; Park, Y.J.; Suh, E.J.; Lee, S.I.; Park, S.R. OsWRKY114 Negatively Regulates Drought Tolerance by Restricting Stomatal Closure in Rice. Plants 2022, 11, 1938. [Google Scholar] [CrossRef]
- Ye, N.; Jia, L.; Zhang, J. ABA signal in rice under stress conditions. Rice 2012, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.Y.; Kim, D.H.; Hwang, I. ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 2013, 32, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Hewage, K.A.H.; Yang, J.F.; Wang, D.; Hao, G.F.; Yang, G.F.; Zhu, J.K. Chemical manipulation of abscisic acid signaling: A new approach to abiotic and biotic stress management in agriculture. Adv. Sci. 2020, 7, 2001265. [Google Scholar] [CrossRef]
- Chern, M.S.; Fitzgerald, H.A.; Yadav, R.C.; Canlas, P.E.; Dong, X.; Ronald, P.C. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 2001, 27, 101–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, H.A.; Chern, M.-S.; Navarre, R.; Ronald, P.C. Overexpression of (At) NPR1 in rice leads to a BTH-and environment-induced lesion-mimic/cell death phenotype. Mol. Plant Microbe Interact. 2004, 17, 140–151. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zhong, S.; Li, Q.; Zhu, Z.; Lou, Y.; Wang, L.; Wang, J.; Wang, M.; Li, Q.; Yang, D. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 2007, 5, 313–324. [Google Scholar] [CrossRef]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef]
- Milborrow, B.V. The pathway of biosynthesis of abscisic acid in vascular plants: A review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 2001, 52, 1145–1164. [Google Scholar] [CrossRef]
- Qin, X.; Zeevaart, J.A. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Andujar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.-S.; Park, S.R.; Lee, S.; Hwang, D.-J. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Kou, Y.; Liu, H.; Li, X.; Xiao, J.; Wang, S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 2011, 62, 4863–4874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Kalde, M.; Barth, M.; Somssich, I.E.; Lippok, B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe Interact. 2003, 16, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Li, M.Y.; Wu, P.; Xu, Z.S.; Que, F.; Wang, F.; Xiong, A.S. Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genom. 2016, 17, 788. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [Green Version]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, N.Y.; Lee, E.; Lee, S.G.; Choi, C.H.; Park, S.R.; Ahn, I.; Bae, S.C.; Hwang, C.H.; Hwang, D.J. Genome-Wide Expression Profiling of OsWRKY Superfamily Genes during Infection with Xanthomonas oryzae pv. oryzae Using Real-Time PCR. Front. Plant Sci. 2017, 8, 1628. [Google Scholar] [CrossRef] [PubMed]
- Son, S.; Moon, S.J.; Kim, H.; Lee, K.S.; Park, S.R. Identification of a novel NPR1 homolog gene, OsNH5N16, which contributes to broad-spectrum resistance in rice. Biochem. Biophys. Res. Commun. 2021, 549, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Kishwar, A.; Gujjar, R.; Ram, N.; Madhuban, G.; Aruna, T. A rapid method for estimation of abscisic acid and characterization of ABA regulated gene in response to water deficit stress from rice. Am. J. Plant Physiol. 2011, 6, 144–156. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, S.; Im, J.H.; Song, G.; Nam, S.; Park, S.R. OsWRKY114 Inhibits ABA-Induced Susceptibility to Xanthomonas oryzae pv. oryzae in Rice. Int. J. Mol. Sci. 2022, 23, 8825. https://doi.org/10.3390/ijms23158825
Son S, Im JH, Song G, Nam S, Park SR. OsWRKY114 Inhibits ABA-Induced Susceptibility to Xanthomonas oryzae pv. oryzae in Rice. International Journal of Molecular Sciences. 2022; 23(15):8825. https://doi.org/10.3390/ijms23158825
Chicago/Turabian StyleSon, Seungmin, Jong Hee Im, Giha Song, Suhyeon Nam, and Sang Ryeol Park. 2022. "OsWRKY114 Inhibits ABA-Induced Susceptibility to Xanthomonas oryzae pv. oryzae in Rice" International Journal of Molecular Sciences 23, no. 15: 8825. https://doi.org/10.3390/ijms23158825






