Identification of Key Functions Required for Production and Utilization of the Siderophore Piscibactin Encoded by the High-Pathogenicity Island irp-HPI in Vibrionaceae
Abstract
:1. Introduction

2. Results
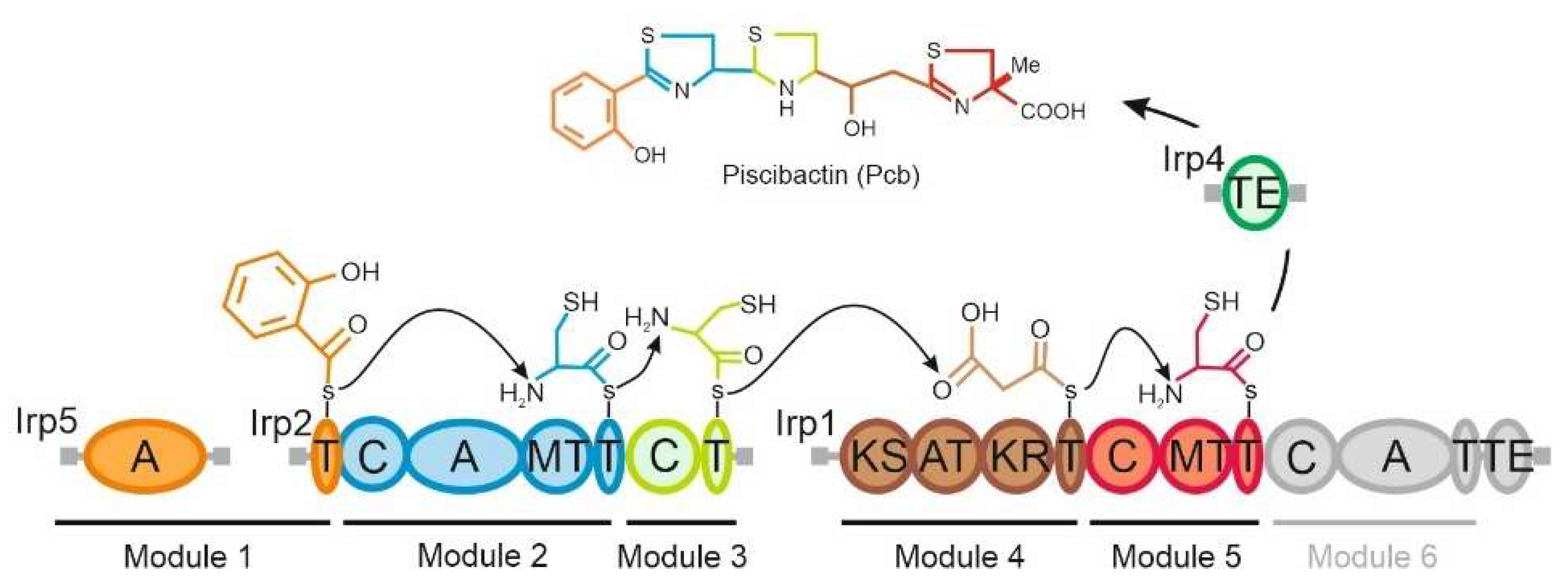
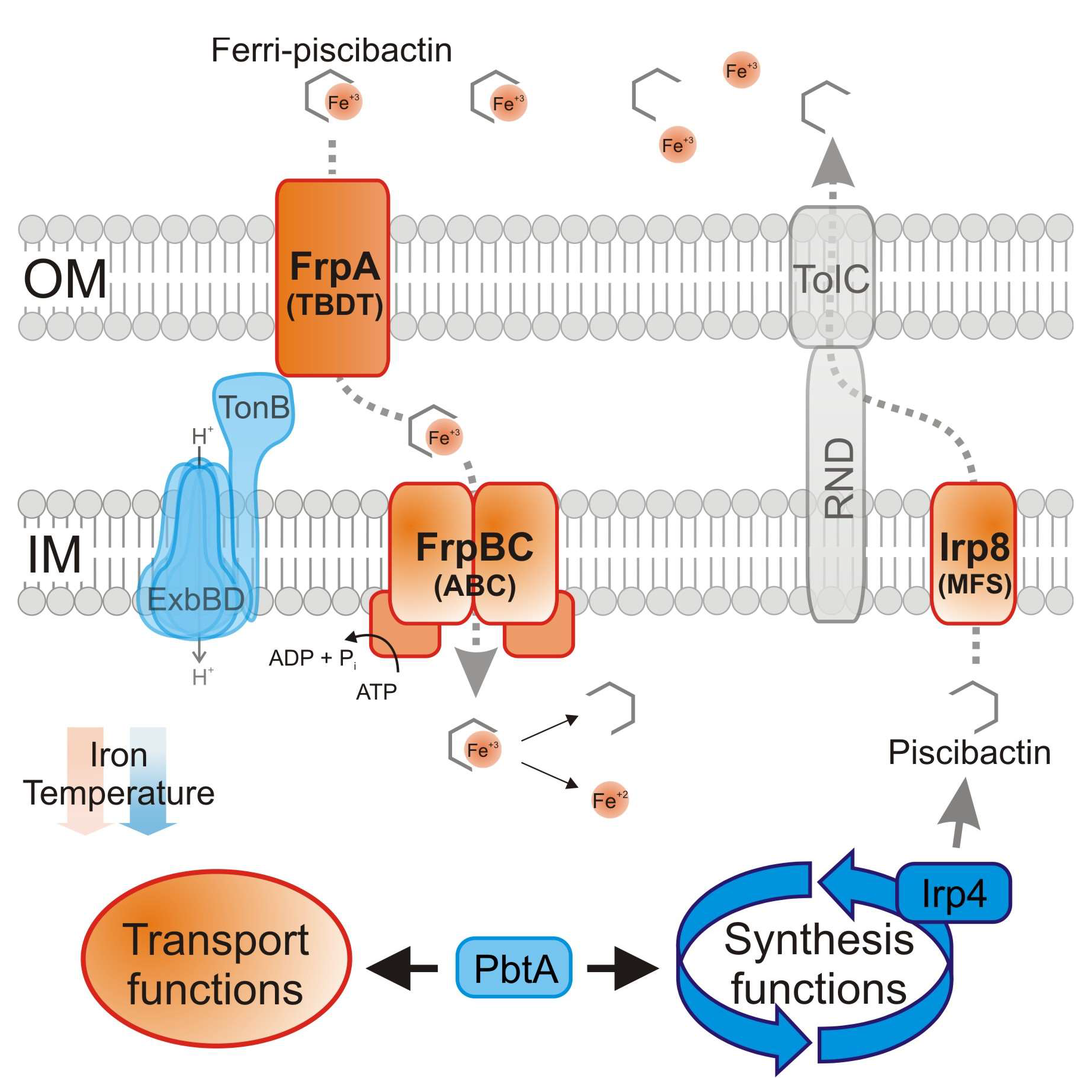
2.1. The Thioesterase Irp4 Is Required for Piscibactin Biosynthesis
2.2. Irp8 Is Required for Piscibactin Secretion
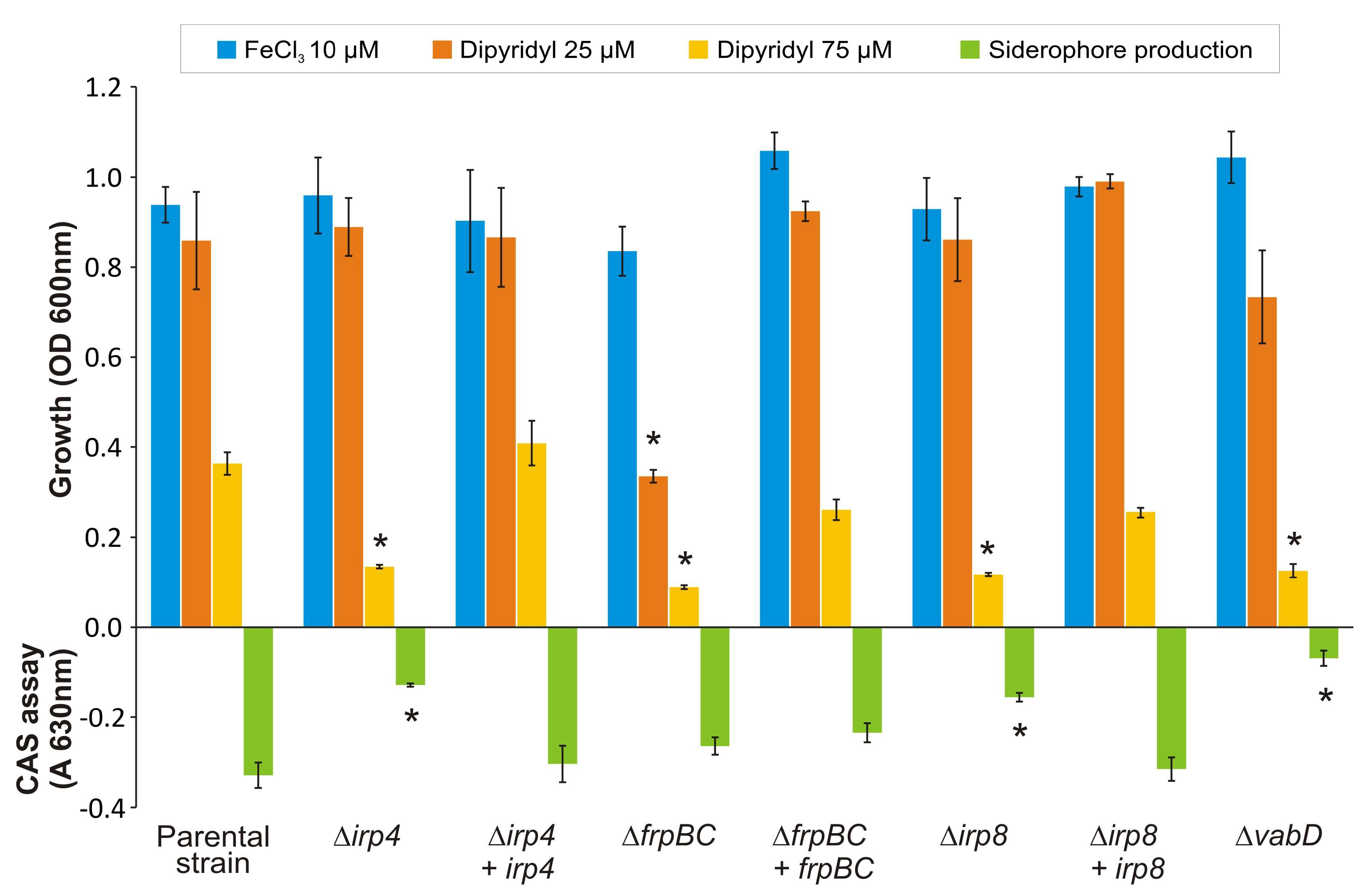
2.3. Mutants with Deleted Irp4 or Irp8 Genes Are Unable to Produce Piscibactin
2.4. Inactivation of FrpBC Disables Ferri-Piscibactin Utilization as Iron Source
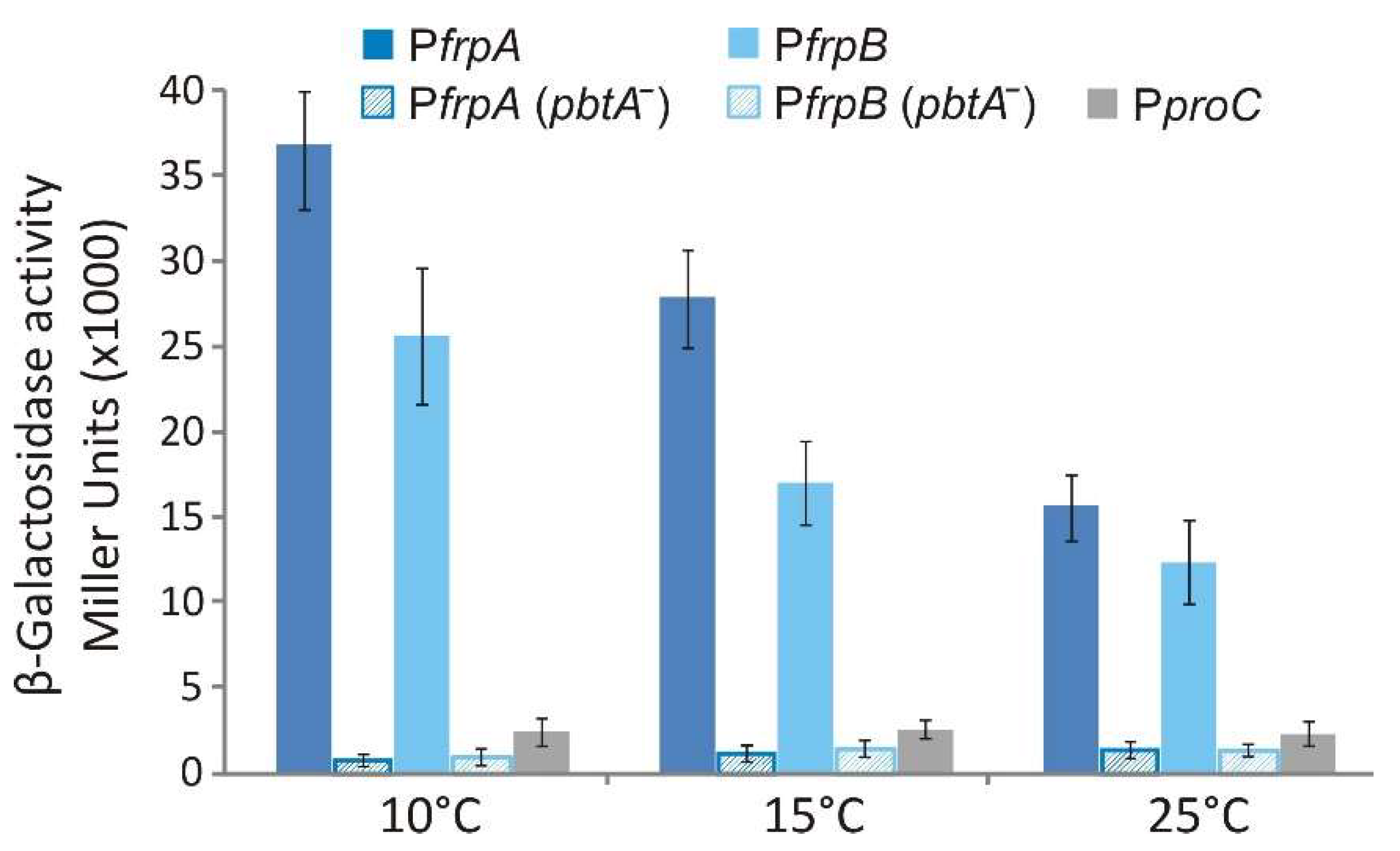
2.5. Transcriptional Regulation of FrpBC Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Media
4.2. Construction of Irp4, Irp8 and FrpBC Defective Mutants and Mutants Reversion
4.3. Growth Ability and Siderophore Production Assays
4.4. Cross-Feeding Assays
4.5. LacZ Transcriptional Fusions and β-Galactosidase Assays
4.6. Analysis of the Presence of Piscibactin by SPE-HLB/HPLC-HRMS
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lemos, M.L.; Balado, M. Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J. Appl. Microbiol. 2020, 129, 104–115. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef]
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657. [Google Scholar] [CrossRef]
- Souto, A.; Montaos, M.A.; Rivas, A.J.; Balado, M.; Osorio, C.R.; Rodríguez, J.; Lemos, M.L.; Jiménez, C. Structure and biosynthetic assembly of piscibactin, a siderophore from Photobacterium damselae subsp. piscicida, predicted from genome analysis. Eur. J. Org. Chem. 2012, 2012, 5693–5700. [Google Scholar] [CrossRef]
- Osorio, C.R.; Juiz-Rio, S.; Lemos, M.L. A siderophore biosynthesis gene cluster from the fish pathogen Photobacterium damselae subsp. piscicida is structurally and functionally related to the Yersinia high-pathogenicity island. Microbiology 2006, 152, 3327–3341. [Google Scholar] [CrossRef] [Green Version]
- Osorio, C.R.; Rivas, A.J.; Balado, M.; Fuentes-Monteverde, J.C.; Rodríguez, J.; Jiménez, C.; Lemos, M.L.; Waldor, M.K. A transmissible plasmid-borne pathogenicity island confers piscibactin biosynthesis in the fish pathogen Photobacterium damselae subsp. piscicida. Appl. Environ. Microbiol. 2015, 81, 5867–5879. [Google Scholar] [CrossRef] [Green Version]
- Balado, M.; Lages, M.A.; Fuentes-Monteverde, J.C.; Martínez-Matamoros, D.; Rodríguez, J.; Jiménez, C.; Lemos, M.L. The siderophore piscibactin is a relevant virulence factor for Vibrio anguillarum favored at low temperatures. Front. Microbiol. 2018, 9, 1766. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, P.; Balado, M.; Fuentes-Monteverde, J.C.; Toranzo, A.E.; Rodríguez, J.; Jiménez, C.; Avendaño-Herrera, R.; Lemos, M.L. The fish pathogen Vibrio ordalii under iron deprivation produces the siderophore piscibactin. Microorganisms 2019, 7, 313. [Google Scholar] [CrossRef] [Green Version]
- Galvis, F.; Ageitos, L.; Rodríguez, J.; Jiménez, C.; Barja, J.L.; Lemos, M.L.; Balado, M. Vibrio neptunius produces piscibactin and amphibactin and both siderophores contribute significantly to virulence for clams. Front. Cell. Infect. Microbiol. 2021, 11, 1049. [Google Scholar] [CrossRef]
- Thode, S.K.; Rojek, E.; Kozlowski, M.; Ahmad, R.; Haugen, P. Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives. PLoS ONE 2018, 13, e0191860. [Google Scholar] [CrossRef] [Green Version]
- Lages, M.A.; Lemos, M.L.; Balado, M. The temperature-dependent expression of the high-pathogenicity island encoding piscibactin in Vibrionaceae results from the combined effect of the AraC-Like transcriptional activator PbtA and regulatory factors from the recipient genome. Front. Microbiol. 2021, 12, 748147. [Google Scholar] [CrossRef]
- Shi, Y.M.; Hirschmann, M.; Shi, Y.N.; Ahmed, S.; Abebew, D.; Tobias, N.J.; Grün, P.; Crames, J.J.; Pöschel, L.; Kuttenlochner, W.; et al. Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat. Chem. 2022, 14, 701–712. [Google Scholar] [CrossRef]
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef]
- Drake, E.J.; Miller, B.R.; Shi, C.; Tarrasch, J.T.; Sundlov, J.A.; Leigh Allen, C.; Skiniotis, G.; Aldrich, C.C.; Gulick, A.M. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 2016, 529, 235–238. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Lages, M.A.; de la Fuente, M.C.; Ageitos, L.; Martínez-Matamoros, D.; Rodríguez, J.; Balado, M.; Jiménez, C.; Lemos, M.L. FrpA is the outer membrane piscibactin transporter in Vibrio anguillarum: Structural elements in synthetic piscibactin analogues required for transport. J. Biol. Inorg. Chem. 2022, 27, 133–142. [Google Scholar] [CrossRef]
- Geoffroy, V.A.; Fetherston, J.D.; Perry, R.D. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 2000, 68, 4452–4461. [Google Scholar] [CrossRef] [Green Version]
- Claxton, H.B.; Akey, D.L.; Silver, M.K.; Admiraal, S.J.; Smith, J.L. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 2009, 284, 5021–5029. [Google Scholar] [CrossRef] [Green Version]
- Furrer, J.L.; Sanders, D.N.; Hook-Barnard, I.G.; McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 2002, 44, 1225–1234. [Google Scholar] [CrossRef]
- Balado, M.; Osorio, C.R.; Lemos, M.L. A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 2006, 152, 3517–3528. [Google Scholar] [CrossRef] [Green Version]
- Lamb, A.L. Breaking a pathogen’s iron will: Inhibiting siderophore production as an antimicrobial strategy. Biochim. Biophys. Acta—Proteins Proteom. 2015, 1854, 1054–1070. [Google Scholar] [CrossRef] [Green Version]
- Mislin, G.L.A.; Schalk, I.J. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 2014, 6, 408–420. [Google Scholar] [CrossRef]
- Pelludat, C.; Rakin, A.; Jacobi, C.A.; Schubert, S.; Heesemann, J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: Organization and siderophore-dependent regulation. J. Bacteriol. 1998, 180, 538–546. [Google Scholar] [CrossRef] [Green Version]
- Bobrov, A.G.; Geoffroy, V.A.; Perry, R.D. Yersiniabactin production requires the thioesterase domain of HMWP2 and YbtD, a putative phosphopantetheinylate transferase. Infect. Immun. 2002, 70, 4204–4214. [Google Scholar] [CrossRef] [Green Version]
- Ohlemacher, S.I.; Xu, Y.; Kober, D.L.; Malik, M.; Nix, J.C.; Brett, T.J.; Henderson, J.P. YbtT is a low-specificity type II thioesterase that maintains production of the metallophore yersiniabactin in pathogenic enterobacteria. J. Biol. Chem. 2018, 293, 19572–19585. [Google Scholar] [CrossRef] [Green Version]
- Seeger, M.A.; van Veen, H.W. Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta 2009, 1794, 725–737. [Google Scholar] [CrossRef]
- Fluman, N.; Bibi, E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta 2009, 1794, 738–747. [Google Scholar] [CrossRef]
- Horiyama, T.; Nishino, K. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS ONE 2014, 9, e108642. [Google Scholar] [CrossRef] [Green Version]
- Bleuel, C.; Grosse, C.; Taudte, N.; Scherer, J.; Wesenberg, D.; Krauss, G.J.; Nies, D.H.; Grass, G. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 2005, 187, 6701–6707. [Google Scholar] [CrossRef] [Green Version]
- Koronakis, V.; Eswaran, J.; Hughes, C. Structure and function of TolC: The bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004, 73, 467–489. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Glupczynski, Y.; Plésiat, P.; Pechère, J.C.; Tulkens, P.M. Antibiotic efflux pumps in prokaryotic cells: Occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J. Antimicrob. Chemother. 2003, 51, 1055–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunkle, D.E.; Bina, X.R.; Bina, J.E. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 2017, 8, e00126-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, H.; Miyamoto, K.; Yasunobe, M.; Murata, M.; Myojin, T.; Tsuchiya, T.; Tanabe, T.; Funahashi, T.; Sato, T.; Azuma, T.; et al. The RND protein is involved in the vulnibactin export system in Vibrio vulnificus M2799. Microb. Pathog. 2014, 75, 59–67. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Q. Iron acquisition strategies of Vibrio anguillarum. Front. Cell. Infect. Microbiol. 2017, 7, 342. [Google Scholar] [CrossRef] [Green Version]
- Schalk, I.J. Siderophore–antibiotic conjugates: Exploiting iron uptake to deliver drugs into bacteria. Clin. Microbiol. Infect. 2018, 24, 801–802. [Google Scholar] [CrossRef] [Green Version]
- Shin, M.; Park, J.; Jin, Y.; Kim, I.J.; Payne, S.M.; Kim, K.H. Biochemical characterization of bacterial FeoBs: A perspective on nucleotide specificity. Arch. Biochem. Biophys. 2020, 685, 108350. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Guillon, L. Fate of ferrisiderophores after import across bacterial outer membranes: Different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 2013, 44, 1267–1277. [Google Scholar] [CrossRef]
- Lages, M.A.; Balado, M.; Lemos, M.L. The expression of virulence factors in Vibrio anguillarum is dually regulated by iron levels and temperature. Front. Microbiol. 2019, 10, 2335. [Google Scholar] [CrossRef] [Green Version]
- Lemos, M.L.; Salinas, P.; Toranzo, A.E.; Barja, J.L.; Crosa, J.H. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 1988, 170, 1920–1925. [Google Scholar] [CrossRef] [Green Version]
- Balado, M.; Osorio, C.R.; Lemos, M.L. Biosynthetic and regulatory elements involved in the production of the siderophore vanchrobactin in Vibrio anguillarum. Microbiology 2008, 154, 1400–1413. [Google Scholar] [CrossRef] [Green Version]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.F.; Kushner, S.R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 1991, 100, 195–199. [Google Scholar] [CrossRef]
- Mouriño, S.; Osorio, C.R.; Lemos, M.L. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 2004, 186, 6159–6167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parales, R.E.; Harwood, C.S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram-bacteria. Gene 1993, 133, 23–30. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Miller, J.H. A Short Course in Bacterial Genetics; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1992. [Google Scholar]



| Strains | Relevant Characteristics | Source |
|---|---|---|
| V. anguillarum | ||
| RV22 | Wild-type serotype O2 strain isolated from diseased turbot (Spain) | [39] |
| MB14 | RV22 with in-frame deletion of vabF gene | [20] |
| MB67 | RV22 with in-frame deletion of vabD gene | [40] |
| ML178 | MB14 with in-frame deletion of irp4 gene | This study |
| ML772 | MB14 with in-frame deletion of irp8 gene | This study |
| ML886 | MB14 with in-frame deletion of frpBC genes | This study |
| ML575 | ML178 revertant strain by the reintroduction of irp4 wild type gene | This study |
| ML955 | ML886 revertant strain by the reintroduction of frpBC wild type genes | This study |
| ML979 | ML772 irp8 genes revertant to the irp8 parental phenotype | This study |
| E. coli | ||
| DH5α | Cloning strain | Laboratory strain |
| S17-1- λpir | RP4 (Km::Tn7, Tc::Mu-1) pro-82 λpir recA1 end A1 thiE1 hsdR17 creC510 | [41] |
| Plasmids | ||
| pWKS30 | Low-copy number cloning vector | [42] |
| pNidKan | Suicide vector derived from pCVD441 | [43] |
| pHRP309 | Low-copy number lacZ reporter plasmid, mob Gmr | [44] |
| pMB276 | frpA promoter (PfrpA) fused to the promoterless lacZ gene in pHRP309 | [40] |
| pML212 | frpBC promoter (PfrpBC) fused to the promoterless lacZ gene in pHRP309 | This study |
| Oligonucleotide | Sequence (5’ -> 3’) a | Size (bp) |
|---|---|---|
| irp4 mutant construction | ||
| 1_Irp4ang_XbaI | CGCTCTAGAGTCTCATTGCAAATGCGCCA | 723 |
| 2_Irp4ang_PstI | CGCCTGCAGGGCACCATTCTGATAAAGTG | |
| 3_Irp4ang_PstI | CGCCTGCAGCTGATAGCCATATCAGGCGA | 861 |
| 4_Irp4ang_XhoI | CCGCTCGAGAGCTTGAGCATGAAAGAGCG | |
| irp8 mutant construction | ||
| 1_Irp8_F_XbaI | GGCTCTAGATCGCTTAGCTGACAACATGG | 824 |
| 2_Irp8_R_BamHI | CGCGGATCCAGTGATGCCCTGTTGTCGAA | |
| 3_Irp8_F_BamHI | CCGGGATCCCTGCTGACATTCTCCGTTAC | 819 |
| 4_Irp8_R_XhoI | GCCCTCGAGCATGGCTTGTTCAGCGTCAT | |
| frpBC mutant construction | ||
| 1_FrpBC_R_NotI | CCGGCGGCCGCTCTCAGCACGTGGAAAGCGA | 1320 |
| 2_FrpBC_F_PstI | GGCCTGCAGGCTGCGCAGTTTATCCATTC | |
| 3_FrpBC_R_PstI | CCGCTGCAGGCGCCTATCTTACTGCTTGA | 995 |
| 4_FrpBC_F_KpnI | GCCGGTACCGACCAATATCTCACCGTGAC | |
| irp4 complementation | ||
| Irp4_comp_F_NotI | CCGGCGGCCGCGTCCAATACCGAGTCAACAG | 2511 |
| Irp4_comp_R_ApaI | GCGGGGCCCAGCGGCATGTTCGGCAATTT | |
| irp8 complementation | ||
| Irp8_comp_F_NotI | CCGGCGGCCGCTCGCTTAGCTGACAACATGG | 2558 |
| Irp8_comp_R_ApaI | GCCGGGCCCCATGGCTTGTTCAGCGTCAT | |
| frpBC complementation | ||
| FrpBC_comp_F_ApaI | GCCGGGCCCGACCAATATCTCACCGTGAC | 5062 |
| 1_FrpBC_R_NotI | CCGGCGGCCGCTCTCAGCACGTGGAAAGCGA | |
| PfrpB promoter fusion construction | ||
| Transp_F_BamHI | CCGGGATCCCGATAAGGTGACGCGATTTC | 746 |
| Transp_R_XbaI | CGCTCTAGAAGCGGATGGTCAAGACTTTG | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lages, M.A.; Ageitos, L.; Rodríguez, J.; Jiménez, C.; Lemos, M.L.; Balado, M. Identification of Key Functions Required for Production and Utilization of the Siderophore Piscibactin Encoded by the High-Pathogenicity Island irp-HPI in Vibrionaceae. Int. J. Mol. Sci. 2022, 23, 8865. https://doi.org/10.3390/ijms23168865
Lages MA, Ageitos L, Rodríguez J, Jiménez C, Lemos ML, Balado M. Identification of Key Functions Required for Production and Utilization of the Siderophore Piscibactin Encoded by the High-Pathogenicity Island irp-HPI in Vibrionaceae. International Journal of Molecular Sciences. 2022; 23(16):8865. https://doi.org/10.3390/ijms23168865
Chicago/Turabian StyleLages, Marta A., Lucía Ageitos, Jaime Rodríguez, Carlos Jiménez, Manuel L. Lemos, and Miguel Balado. 2022. "Identification of Key Functions Required for Production and Utilization of the Siderophore Piscibactin Encoded by the High-Pathogenicity Island irp-HPI in Vibrionaceae" International Journal of Molecular Sciences 23, no. 16: 8865. https://doi.org/10.3390/ijms23168865
APA StyleLages, M. A., Ageitos, L., Rodríguez, J., Jiménez, C., Lemos, M. L., & Balado, M. (2022). Identification of Key Functions Required for Production and Utilization of the Siderophore Piscibactin Encoded by the High-Pathogenicity Island irp-HPI in Vibrionaceae. International Journal of Molecular Sciences, 23(16), 8865. https://doi.org/10.3390/ijms23168865










