Extraction and Biological Evaluation of Matrix-Bound Nanovesicles (MBVs) from High-Hydrostatic Pressure-Decellularized Tissues
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
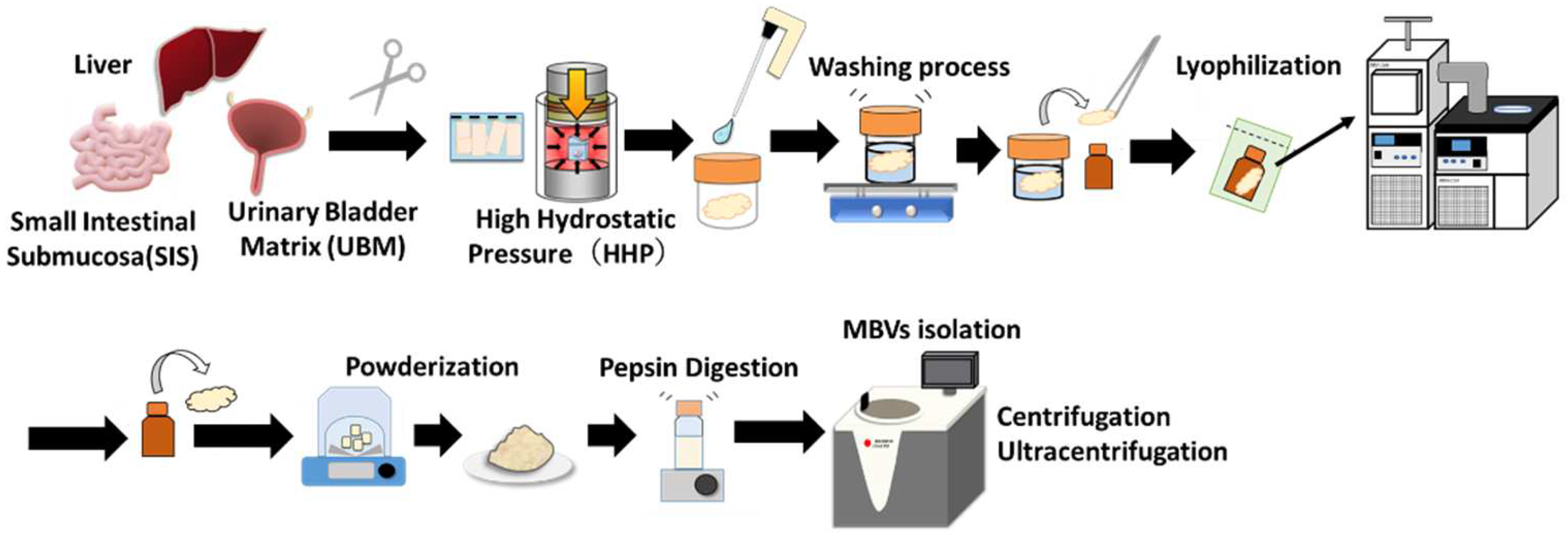
4.2. Preparation and Lyophilization of Decellularized Tissues
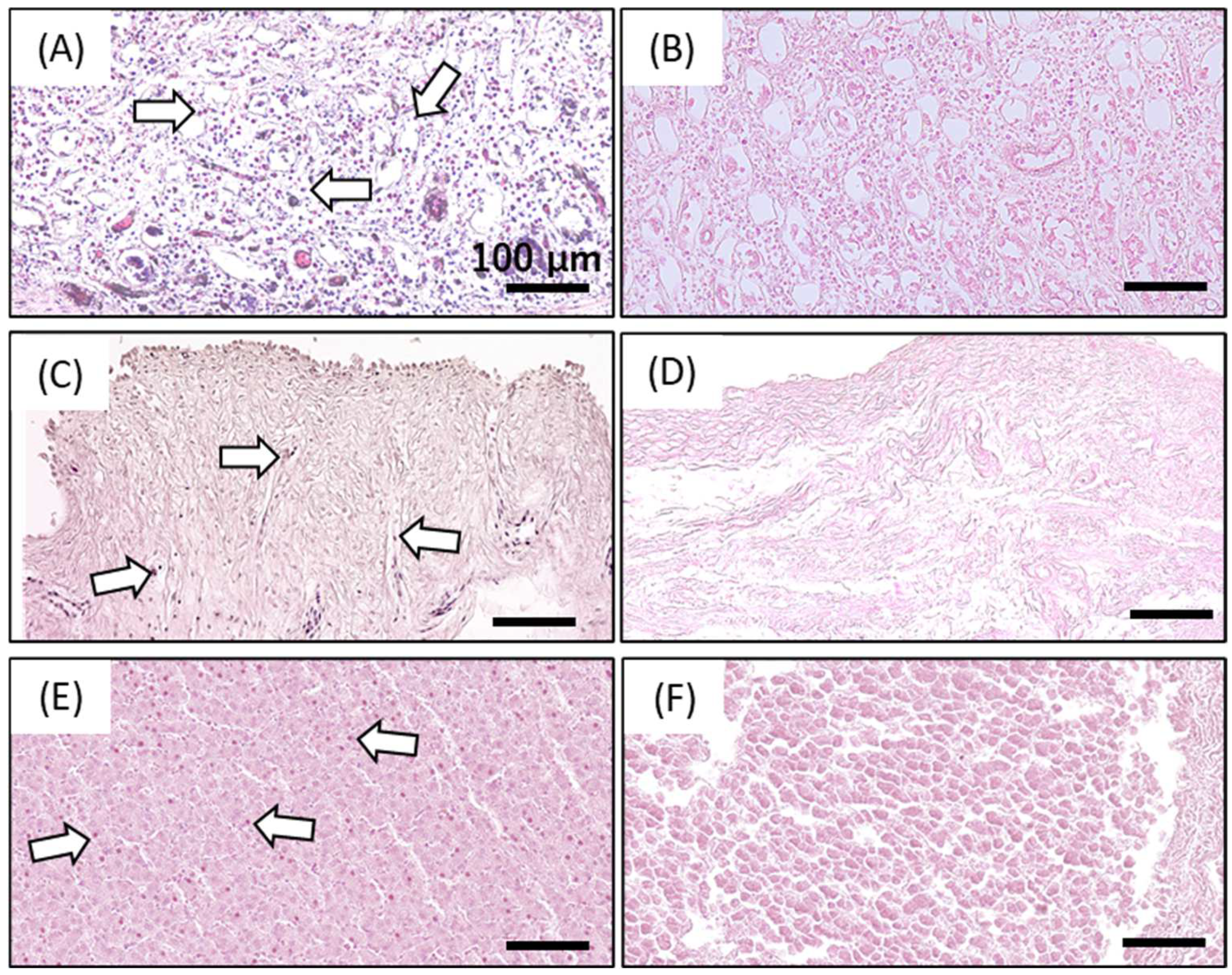
4.3. Determination of Decellularization Efficacy by Residual DNA Quantification and Hematoxylin and Eosin Staining
4.4. Isolation of Matrix-Bound Nanovesicles (MBVs)
4.5. Transmission Electron Microscopy (TEM) Observation of MBVs
4.6. MBV Size Analysis and Concentration by Nano Tracking Assay (NTA)
4.7. RNA Isolation and Analysis
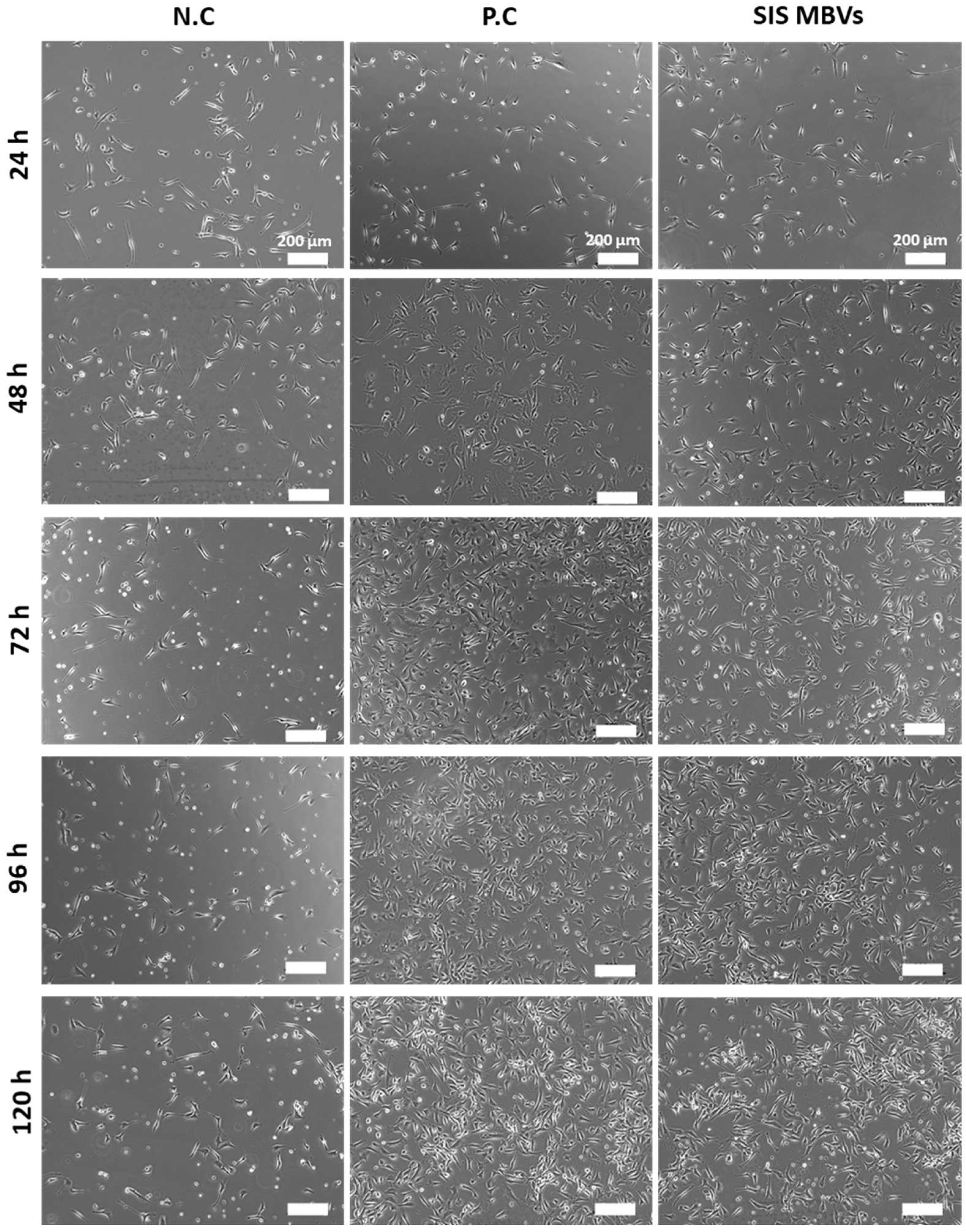
4.8. Cell Seeding and Growth Assay
4.9. Cell Observation and Measurement of Cell Proliferation
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valdoz, J.C.; Johnson, B.C.; Jacobs, D.J.; Franks, N.A.; Dodson, E.L.; Sanders, C.; Cribbs, C.G.; Van Ry, P.M. The ECM: To Scaffold, or Not to Scaffold, That Is the Question. Int. J. Mol. Sci. 2021, 22, 12690. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Agmon, G.; Christman, K.L. Controlling stem cell behavior with decellularized extracellular matrix scaffolds. Curr. Opin. Solid State Mater. Sci. 2016, 20, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, I.T.; Badylak, S.F. Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev. Dyn. 2015, 245, 351–360. [Google Scholar] [CrossRef]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Ge, H.; Tian, M.; Pei, Q.; Tan, F.; Pei, H. Extracellular Matrix Stiffness: New Areas Affecting Cell Metabolism. Front. Oncol. 2021, 11, 631991. [Google Scholar] [CrossRef]
- Reing, J.E.; Zhang, L.; Myers-Irvin, J.; Cordero, K.E.; Freytes, D.O.; Heber-Katz, E.; Bedelbaeva, K.; McIntosh, D.; Dewilde, A.; Braunhut, S.J.; et al. Degradation Products of Extracellular Matrix Affect Cell Migration and Proliferation. Tissue Eng. Part A 2009, 15, 605–614. [Google Scholar] [CrossRef]
- Maquart, F.; Bellon, G.; Pasco, S.; Monboisse, J. Matrikines in the regulation of extracellular matrix degradation. Biochimie 2005, 87, 353–360. [Google Scholar] [CrossRef]
- Davis, G.E.; Bayless, K.J.; Davis, M.J.; Meininger, G.A. Regulation of Tissue Injury Responses by the Exposure of Matricryptic Sites within Extracellular Matrix Molecules. Am. J. Pathol. 2000, 156, 1489–1498. [Google Scholar] [CrossRef]
- Huleihel, L.; Hussey, G.S.; Naranjo, J.D.; Zhang, L.; Dziki, J.L.; Turner, N.J.; Stolz, D.B.; Badylak, S.F. Matrix-bound nanovesicles within ECM bioscaffolds. Sci. Adv. 2016, 2, e1600502. [Google Scholar] [CrossRef]
- Huleihel, L.; Bartolacci, J.G.; Dziki, J.L.; Vorobyov, T.; Arnold, B.; Scarritt, M.E.; Pineda Molina, C.; Lopresti, S.T.; Brown, B.N.; Naranjo, J.D.; et al. Matrix-Bound Nanovesicles Recapitulate Extracellular Matrix Effects on Macrophage Phenotype. Tissue Eng. Part A 2017, 23, 1283–1294. [Google Scholar] [CrossRef]
- Hussey, G.S.; Dziki, J.L.; Lee, Y.C.; Bartolacci, J.G.; Behun, M.; Turnquist, H.R.; Badylak, S.F. Matrix bound nanovesicle-associated IL-33 activates a pro-remodelingmacrophage phenotype via a non-canonical, ST2-independent pathway. J. Immunol. Regen. Med. 2019, 3, 26–35. [Google Scholar]
- Funamoto, S.; Nam, K.; Kimura, T.; Murakoshi, A.; Hashimoto, Y.; Niwaya, K.; Kitamura, S.; Fujisato, T.; Kishida, A. The use of high-hydrostatic pressure treatment to decellularize blood vessels. Biomaterials 2010, 31, 3590–3595. [Google Scholar] [CrossRef]
- Kurokawa, S.; Hashimoto, Y.; Funamoto, S.; Murata, K.; Yamashita, A.; Yamazaki, K.; Ikeda, T.; Minatoya, K.; Kishida, A.; Masumoto, H. In vivo recellularization of xenogeneic vascular grafts decellularized with high hydrostatic pressure method in a porcine carotid arterial interpose model. PLoS ONE 2021, 16, e0254160. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Funamoto, S.; Sasaki, S.; Negishi, J.; Hattori, S.; Honda, T.; Kimura, T.; Kobayashi, H.; Kishida, A. Re-epithelialization and remodeling of decellularized corneal matrix in a rabbit corneal epithelial wound model. Mater. Sci. Eng. C 2019, 102, 238–246. [Google Scholar] [CrossRef]
- Tabuchi, M.; Negishi, J.; Yamashita, A.; Higami, T.; Kishida, A.; Funamoto, S. Effect of decellularized tissue powders on a rat model of acute myocardial infarction. Mater. Sci. Eng. C 2015, 56, 494–500. [Google Scholar] [CrossRef]
- Negishi, J.; Hashimoto, Y.; Yamashita, A.; Kimura, T.; Kishida, A.; Funamoto, S. Histological structure affects recellularization of decellularized arteries. Mater. Sci. Eng. C 2017, 70, 450–455. [Google Scholar] [CrossRef]
- Nakamura, N.; Ito, A.; Kimura, T.; Kishida, A. Extracellular Matrix Induces Periodontal Ligament Reconstruction In Vivo. Int. J. Mol. Sci. 2019, 20, 3277. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ohara, M.; Hashimoto, Y.; Nakamura, N.; Fujisato, T.; Kimura, T.; Kishida, A. In vitro evaluation of surface biological properties of decellularized aorta for cardiovascular use. J. Mater. Chem. B 2020, 8, 10977–10989. [Google Scholar] [CrossRef]
- Kobayashi, M.; Ohara, M.; Hashimoto, Y.; Nakamura, N.; Fujisato, T.; Kimura, T.; Kishida, A. Effect of luminal surface structure of decellularized aorta on thrombus formation and cell behavior. PLoS ONE 2021, 16, e0246221. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Funamoto, S.; Sasaki, S.; Honda, T.; Hattori, S.; Nam, K.; Kimura, T.; Mochizuki, M.; Fujisato, T.; Kobayashi, H.; et al. Preparation and characterization of decellularized cornea using high-hydrostatic pressurization for corneal tissue engineering. Biomaterials 2010, 31, 3941–3948. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Huang, Y.; Li, K.; Fan, Y.; Xie, H.; Li, X. Small intestinal submucosa: Superiority, limitations and solutions, and its potential to address bottlenecks in tissue repair. J. Mater. Chem. B 2019, 7, 5038–5055. [Google Scholar] [CrossRef] [PubMed]
- Parekh, A.; Mantle, B.; Banks, J.; Swarts, J.D.; Badylak, S.F.; Dohar, J.E.; Hebda, P.A. Repair of the tympanic membrane with urinary bladder matrix. Laryngoscope 2009, 119, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Revi, D.; Vineetha, V.P.; Muhamed, J.; Surendran, G.C.; Rajan, A.; Kumary, T.; Anilkumar, T.V. Wound healing potential of scaffolds prepared from porcine jejunum and urinary bladder by a non-detergent/enzymatic method. J. Biomater. Appl. 2014, 29, 1218–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Santoso, E.G.; Yoshida, K.; Hirota, Y.; Aizawa, M.; Yoshino, O.; Kishida, A.; Osuga, Y.; Saito, S.; Ushida, T.; Furukawa, K.S. Application of Detergents or High Hydrostatic Pressure as Decellularization Processes in Uterine Tissues and Their Subsequent Effects on In Vivo Uterine Regeneration in Murine Models. PLoS ONE 2014, 9, e103201. [Google Scholar] [CrossRef] [PubMed]
- Mahara, A.; Morimoto, N.; Sakuma, T.; Fujisato, T.; Yamaoka, T. Complete Cell Killing by Applying High Hydrostatic Pressure for Acellular Vascular Graft Preparation. BioMed Res. Int. 2014, 2014, 379607. [Google Scholar] [CrossRef]
- Yamamoto, K.; Zhang, X.; Inaoka, T.; Morimatsu, K.; Kimura, K.; Nakaura, Y. Bacterial Injury Induced by High Hydrostatic Pressure. Food Eng. Rev. 2021, 13, 442–453. [Google Scholar] [CrossRef]
- Silva, J.L.; Luan, P.; Glaser, M.; Voss, E.W.; Weber, G. Effects of hydrostatic pressure on a membrane-enveloped virus: High immunogenicity of the pressure-inactivated virus. J. Virol. 1992, 66, 2111–2117. [Google Scholar] [CrossRef]
- Kingsley, D.H.; Chen, H.; Hoover, D.G. Inactivation of selected picornaviruses by high hydrostatic pressure. Virus Res. 2004, 102, 221–224. [Google Scholar] [CrossRef]
- Quijano, L.M.; Naranjo, J.D.; El-Mossier, S.O.; Turner, N.J.; Molina, C.P.; Bartolacci, J.; Zhang, L.; White, L.; Li, H.; Badylak, S.F. Matrix-Bound Nanovesicles: The Effects of Isolation Method upon Yield, Purity, and Function. Tissue Eng. Part C Methods 2020, 26, 528–540. [Google Scholar] [CrossRef]
- Lee, J.-H.; Parthiban, P.; Jin, G.-Z.; Knowles, J.C.; Kim, H.-W. Materials roles for promoting angiogenesis in tissue regeneration. Prog. Mater. Sci. 2020, 117, 100732. [Google Scholar] [CrossRef]
- Li, S.-C.; Chan, W.-C.; Hu, L.-Y.; Lai, C.-H.; Hsu, C.-N.; Lin, W.-C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2010, 96, 1–9. [Google Scholar] [CrossRef]
- Pogue, A.I.; Clement, C.; Hill, J.M.; Lukiw, W.J. Evolution of microRNA (miRNA) Structure and Function in Plants and Animals: Relevance to Aging and Disease. J. Aging Sci. 2014, 2, 119. [Google Scholar]
- Kobayashi, M.; Kadota, J.; Hashimoto, Y.; Fujisato, T.; Nakamura, N.; Kimura, T.; Kishida, A. Elastic Modulus of ECM Hydrogels Derived from Decellularized Tissue Affects Capillary Network Formation in Endothelial Cells. Int. J. Mol. Sci. 2020, 21, 6304. [Google Scholar] [CrossRef]
- Bernard, M.P.; Chu, M.L.; Myers, J.C.; Ramirez, F.; Eikenberry, E.F.; Prockop, D.J. Nucleotide sequences of complementary deoxyribonucleic acids for the pro.alpha.1 chain of human type I procollagen. Statistical evaluation of structures that are conserved during evolution. Biochemistry 1983, 22, 5213–5223. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.-W.; Qiu, Y.; Dupee, D.; Noonan, M.; Lin, Y.-D.; Fisch, S.; Unno, K.; Sereti, K.-I.; Liao, R. MicroRNA-34a Plays a Key Role in Cardiac Repair and Regeneration Following Myocardial Infarction. Circ. Res. 2015, 117, 450–459. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA miR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Icli, B.; Wu, W.; Ozdemir, D.; Li, H.; Haemmig, S.; Liu, X.; Giatsidis, G.; Cheng, H.S.; Avci, S.N.; Kurt, M.; et al. MicroRNA-135a-3p regulates angiogenesis and tissue repair by targeting p38 signaling in endothelial cells. FASEB J. 2019, 33, 5599–5614. [Google Scholar] [CrossRef]
- Van Balkom, B.W.M.; de Jong, O.G.; Smits, M.; Brummelman, J.; den Ouden, K.; de Bree, P.M.; van Eijndhoven, M.A.; Pegtel, D.M.; Stoorvogel, W.; Würdinger, T.; et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 2013, 121, 3997–4006. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Piper, D.W.; Fenton, B.H. pH stability and activity curves of pepsin with special reference to their clinical importance. Gut 1965, 6, 506–508. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, M.; Ishida, N.; Hashimoto, Y.; Negishi, J.; Saga, H.; Sasaki, Y.; Akiyoshi, K.; Kimura, T.; Kishida, A. Extraction and Biological Evaluation of Matrix-Bound Nanovesicles (MBVs) from High-Hydrostatic Pressure-Decellularized Tissues. Int. J. Mol. Sci. 2022, 23, 8868. https://doi.org/10.3390/ijms23168868
Kobayashi M, Ishida N, Hashimoto Y, Negishi J, Saga H, Sasaki Y, Akiyoshi K, Kimura T, Kishida A. Extraction and Biological Evaluation of Matrix-Bound Nanovesicles (MBVs) from High-Hydrostatic Pressure-Decellularized Tissues. International Journal of Molecular Sciences. 2022; 23(16):8868. https://doi.org/10.3390/ijms23168868
Chicago/Turabian StyleKobayashi, Mako, Naoki Ishida, Yoshihide Hashimoto, Jun Negishi, Hideki Saga, Yoshihiro Sasaki, Kazunari Akiyoshi, Tsuyoshi Kimura, and Akio Kishida. 2022. "Extraction and Biological Evaluation of Matrix-Bound Nanovesicles (MBVs) from High-Hydrostatic Pressure-Decellularized Tissues" International Journal of Molecular Sciences 23, no. 16: 8868. https://doi.org/10.3390/ijms23168868
APA StyleKobayashi, M., Ishida, N., Hashimoto, Y., Negishi, J., Saga, H., Sasaki, Y., Akiyoshi, K., Kimura, T., & Kishida, A. (2022). Extraction and Biological Evaluation of Matrix-Bound Nanovesicles (MBVs) from High-Hydrostatic Pressure-Decellularized Tissues. International Journal of Molecular Sciences, 23(16), 8868. https://doi.org/10.3390/ijms23168868






