Distribution and Localization of Mahogunin Ring Finger 1 in the Mouse Central Nervous System
Abstract
:1. Introduction
2. Results
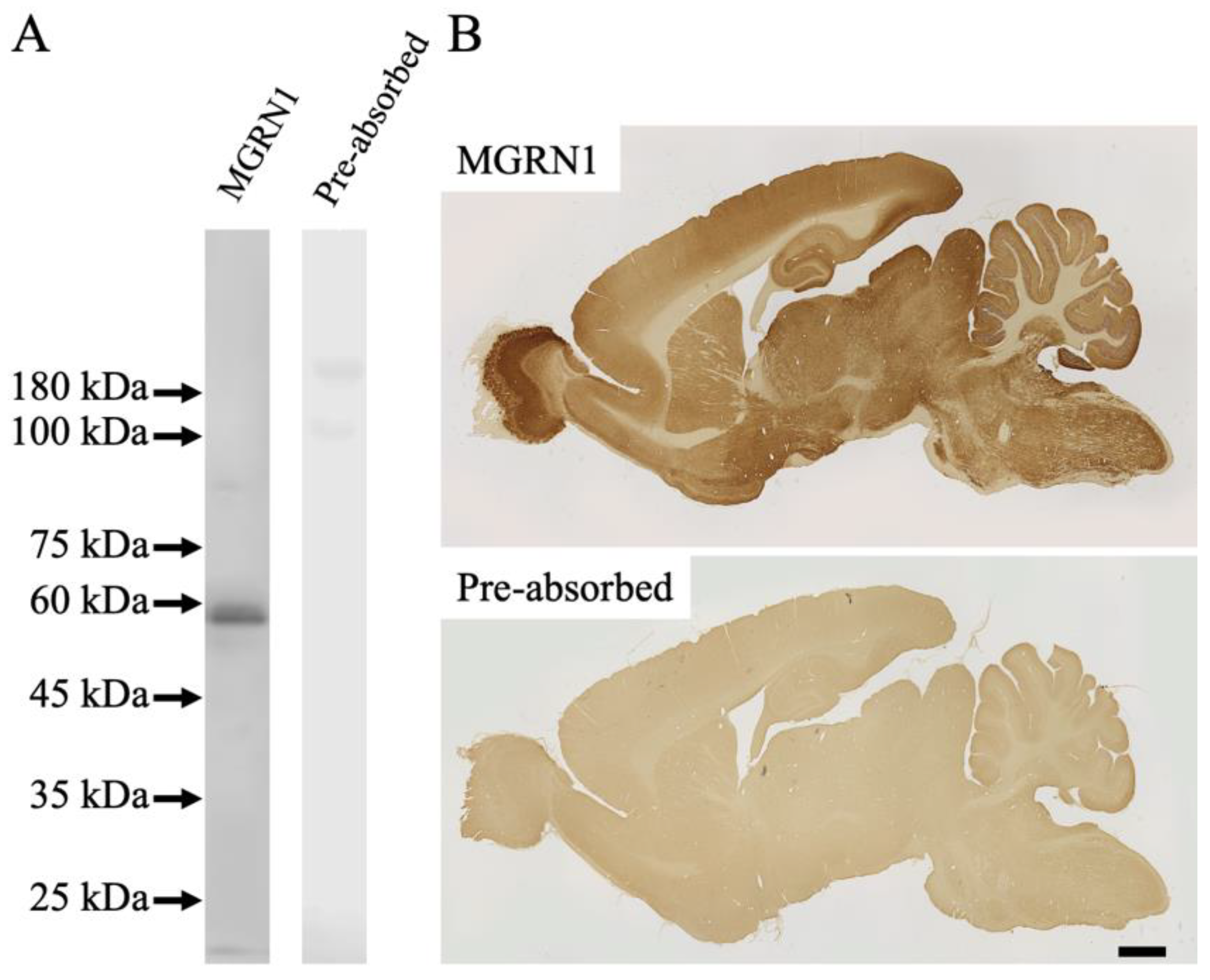
2.1. Specificity of Antibodies
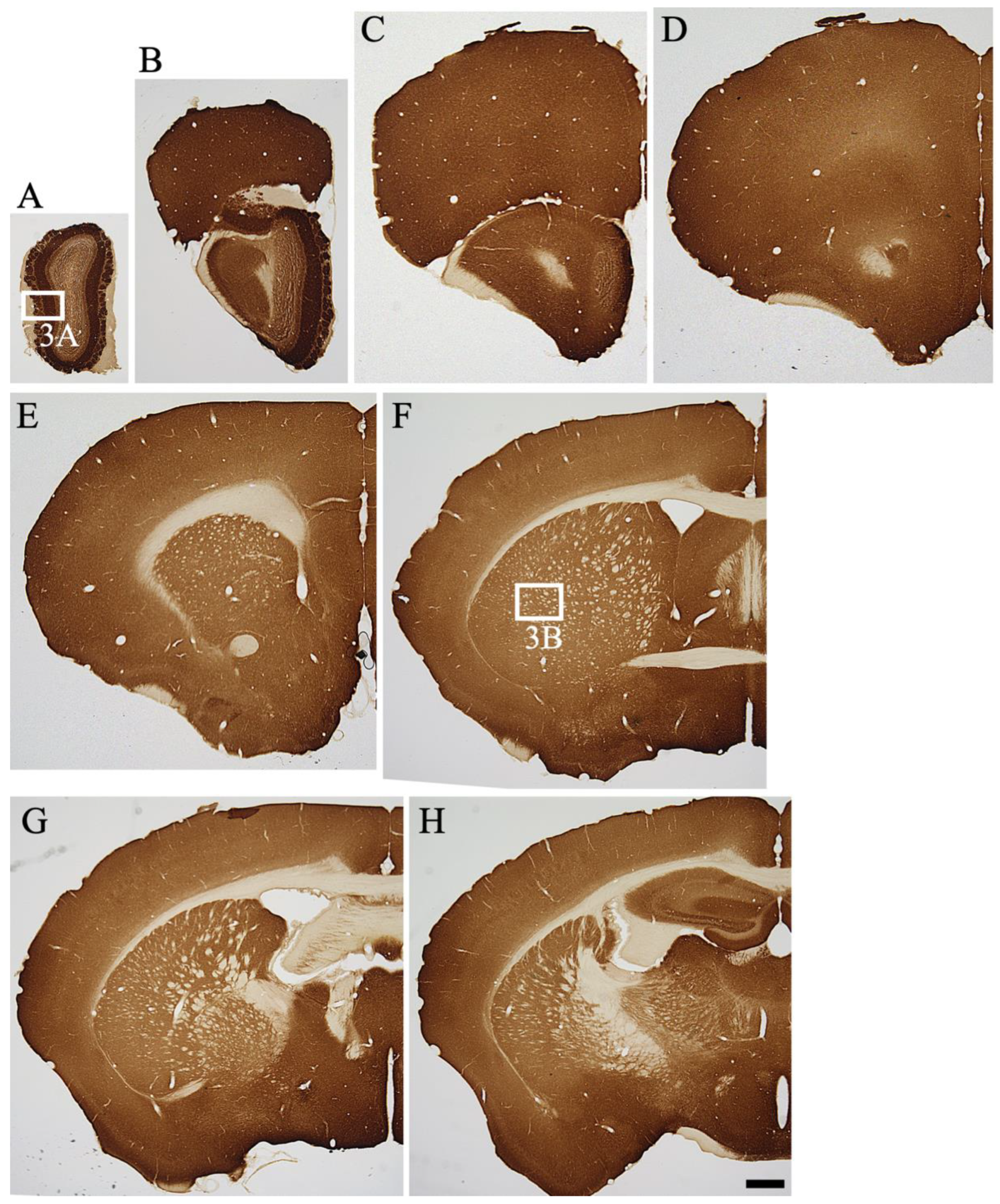
2.2. Overview of MGRN1 Distribution
2.3. Detailed Distribution of MGRN1-IR in the Mouse CNS
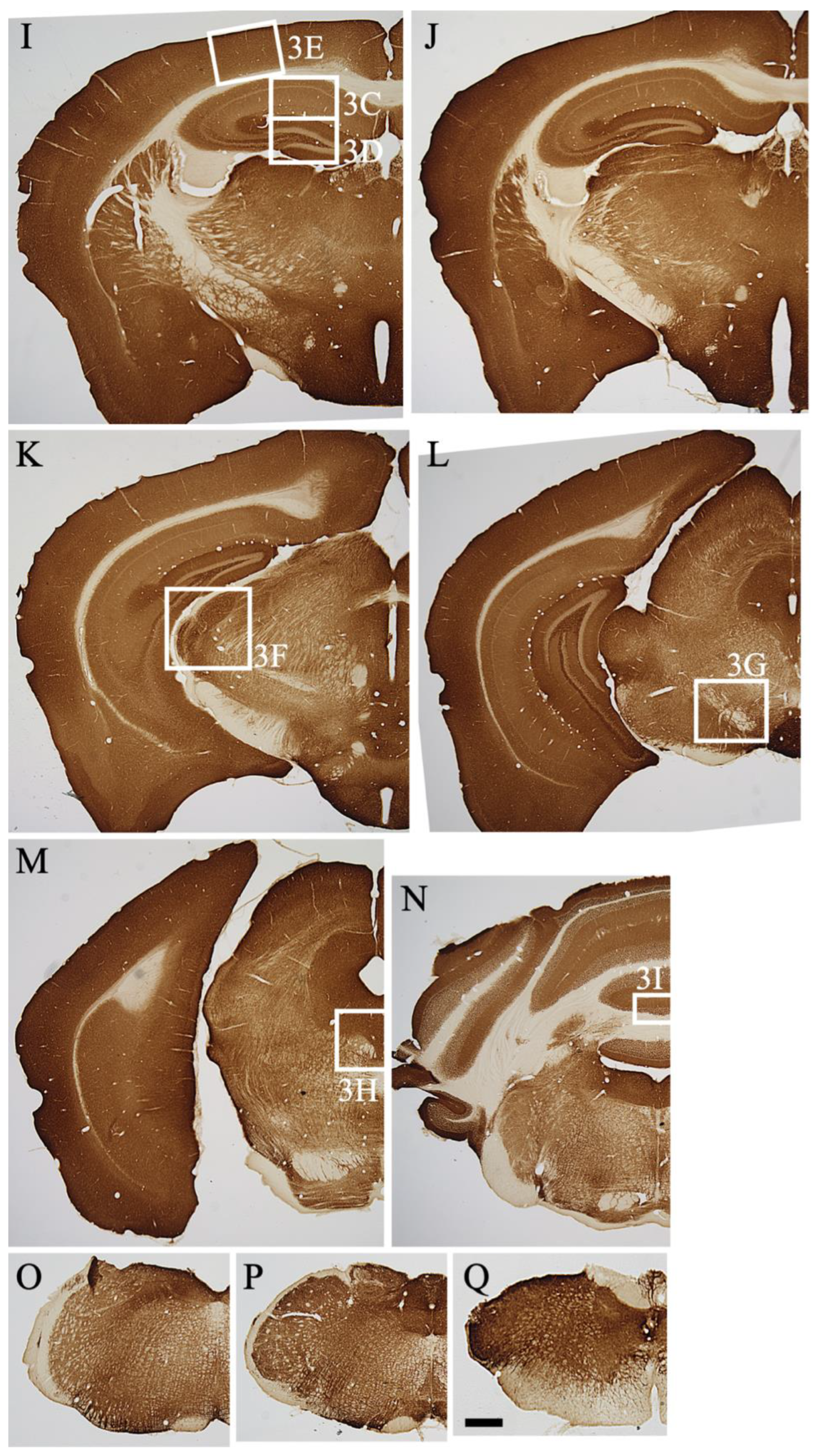
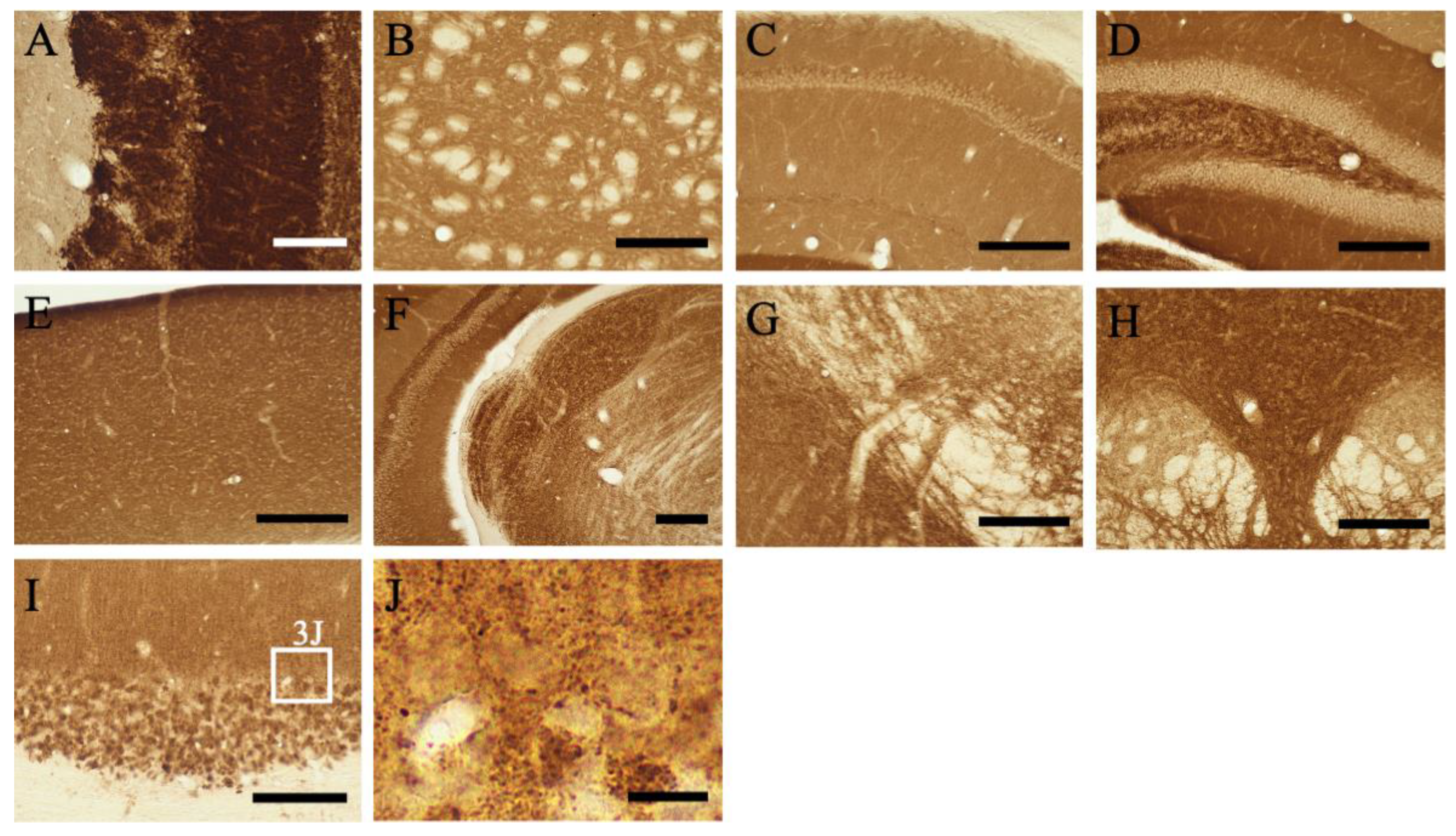
2.3.1. Telencephalon
2.3.2. Diencephalon
2.3.3. Mesencephalon
2.3.4. Pons and Medulla
2.3.5. Cerebellum
2.3.6. Spinal Cord
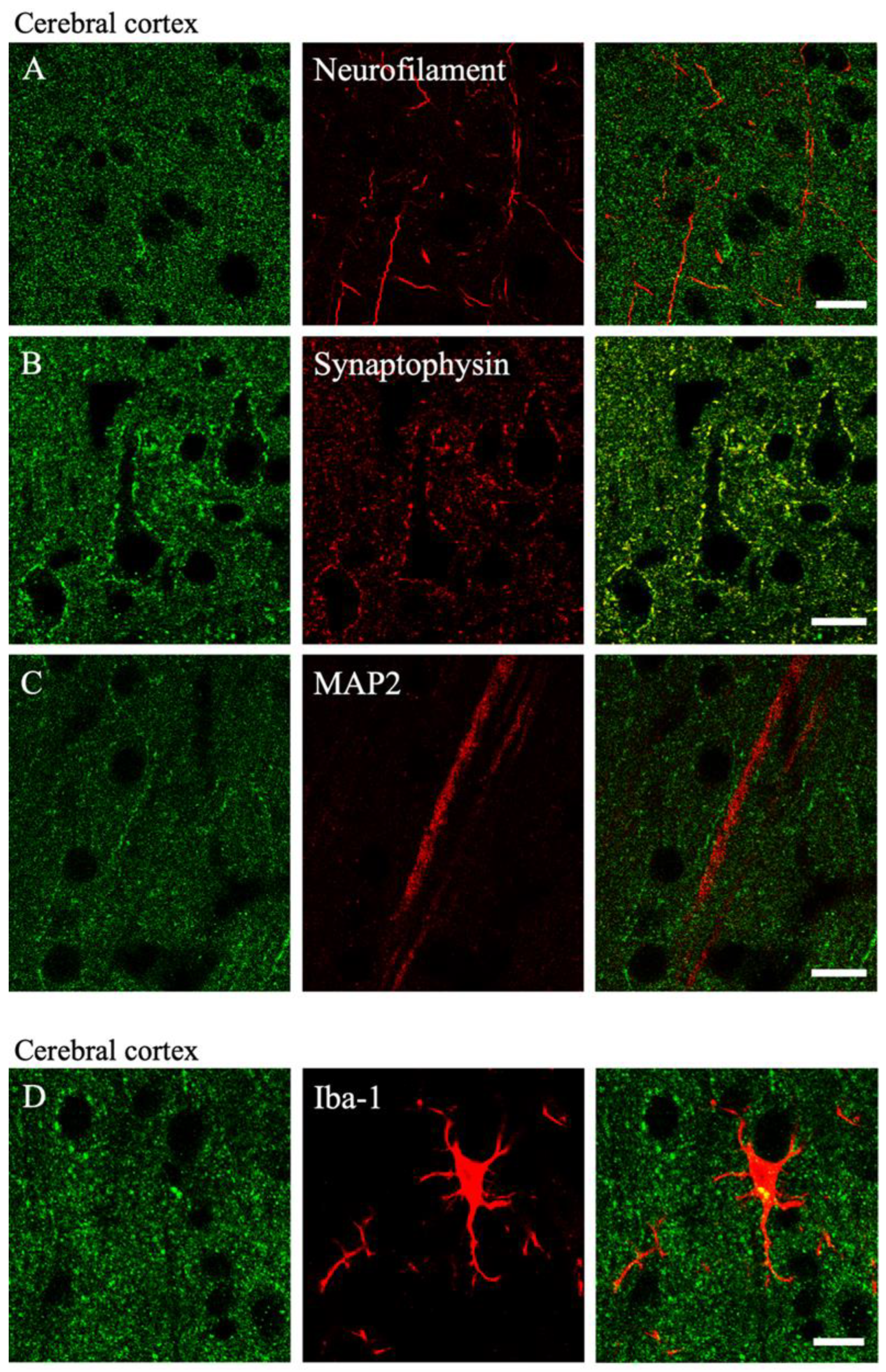
2.4. Colocalization of MGRN1
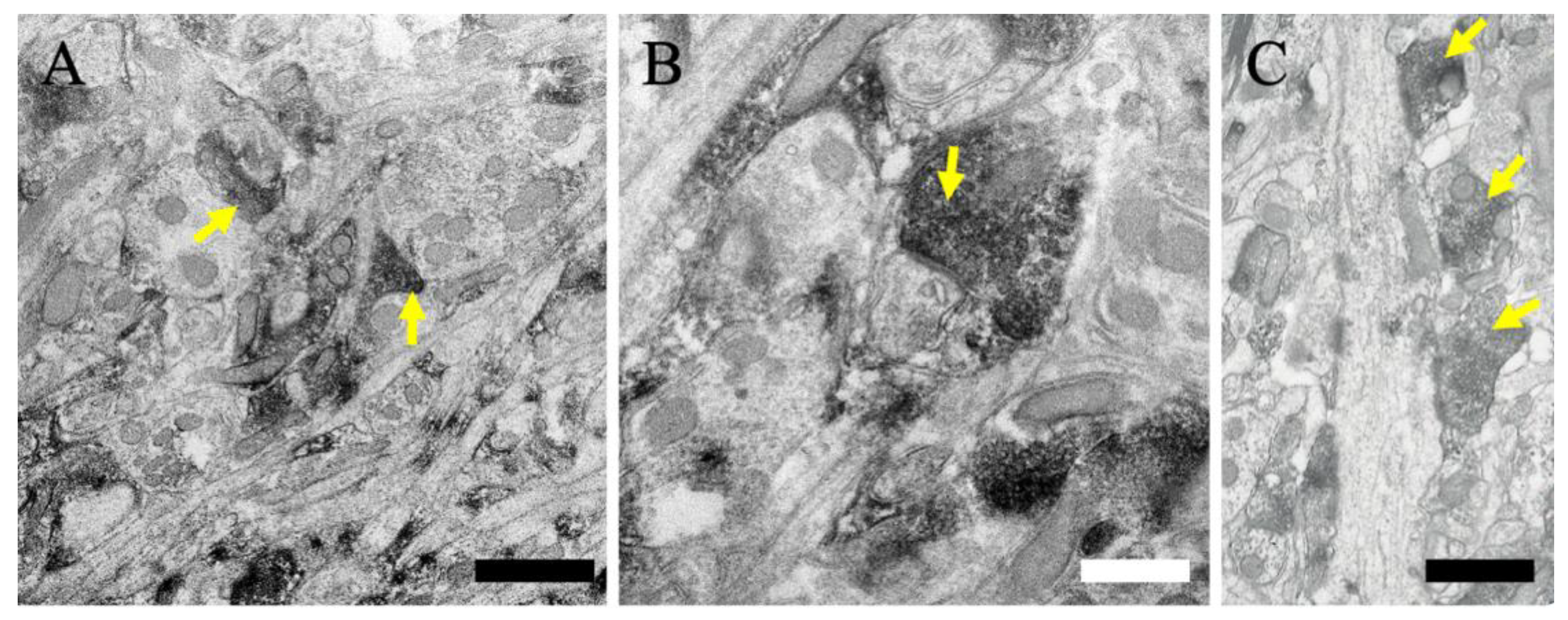
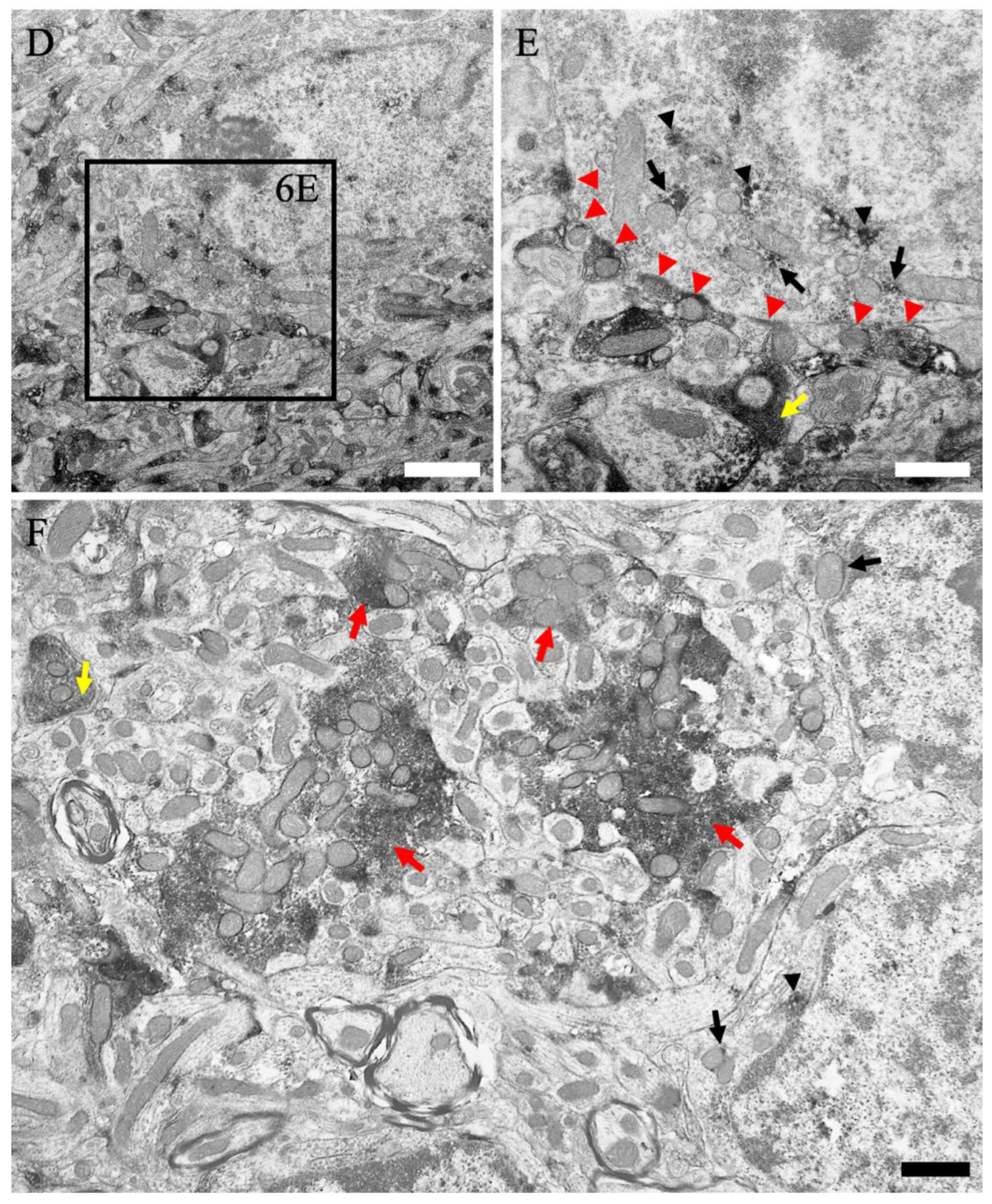
2.5. Subcellular Localization of MGRN1
3. Discussion
3.1. Antibody Specificity
3.2. Distribution of MGRN1
3.3. Spongiform Degeneration (Neuronal Vacuolation) and E3 Ubiquitin Ligase Activity
4. Materials and Methods
4.1. Animals
4.2. Production of the Anti-MGRN1 Antibody
4.3. Western Blotting
4.4. Immunoperoxidase Staining
4.5. Double Immunofluorescence Staining
4.6. Immunoelectron Microscopy
4.7. Image Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scotter, E.L.; Vance, C.; Nishimura, A.L.; Lee, Y.B.; Chen, H.J.; Urwin, H.; Sardone, V.; Mitchell, J.C.; Rogelj, B.; Rubinsztein, D.C.; et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 2014, 127, 1263–1278. [Google Scholar] [CrossRef]
- Lee, M.J.; Lee, J.H.; Rubinsztein, D.C. Tau degradation: The ubiquitin-proteasome system versus the autophagy-lysosome system. Prog. Neurobiol. 2013, 105, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2009, 584, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 2009, 5, 862–863. [Google Scholar] [CrossRef] [PubMed]
- Layfield, R.; Searle, M.S. Disruption of ubiquitin-mediated processes in diseases of the brain and bone. Biochem. Soc. Trans. 2008, 36, 469–471. [Google Scholar] [CrossRef]
- Layfield, R.; Lowe, J.; Bedford, L. The ubiquitin-proteasome system and neurodegenerative disorders. Essays Biochem. 2005, 41, 157–171. [Google Scholar] [CrossRef]
- Layfield, R.; Cavey, J.R.; Lowe, J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Res. Rev. 2003, 2, 343–356. [Google Scholar] [CrossRef]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, X.; Du, X.; Bi, M.; Ma, F.; Xie, J.; Jiang, H. The S-nitrosylation of parkin attenuated the ubiquitination of divalent metal transporter 1 in MPP+-treated SH-SY5Y cells. Sci. Rep. 2020, 10, 15542. [Google Scholar] [CrossRef]
- Kumar, M.; Acevedo-Cintron, J.; Jhaldiyal, A.; Wang, H.; Andrabi, S.A.; Eacker, S.; Karuppagounder, S.S.; Brahmachari, S.; Chen, R.; Kim, H.; et al. Defects in mitochondrial biogenesis drive mitochondrial alterations in PARKIN-deficient human dopamine neurons. Stem Cell Rep. 2020, 15, 629–645. [Google Scholar] [CrossRef]
- Mizuno, Y.; Hattori, N.; Kitada, T.; Matsumine, H.; Mori, H.; Shimura, H.; Kubo, S.; Kobayashi, H.; Asakawa, S.; Minoshima, S.; et al. Familial Parkinson’s disease. Alpha-synuclein and parkin. Adv. Neurol. 2001, 86, 13–21. [Google Scholar] [PubMed]
- Shimura, H.; Schlossmacher, M.G.; Hattori, N.; Frosch, M.P.; Trockenbacher, A.; Schneider, R.; Mizuno, Y.; Kosik, K.S.; Selkoe, D.J. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: Implications for Parkinson’s disease. Science 2001, 293, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Kishino, T.; Lalande, M.; Wagstaff, J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997, 15, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, T.; Sutcliffe, J.S.; Fang, P.; Galjaard, R.J.; Jiang, Y.H.; Benton, C.S.; Rommens, J.M.; Beaudet, A.L. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 1997, 15, 74–77. [Google Scholar] [CrossRef]
- Phan, L.K.; Chung, W.K.; Leibel, R.L. The mahoganoid mutation (Mgrn1md) improves insulin sensitivity in mice with mutations in the melanocortin signaling pathway independently of effects on adiposity. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E611–E620. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Eldridge, A.G.; Jackson, P.K.; Gunn, T.M.; Barsh, G.S. Accessory proteins for melanocortin signaling: Attractin and mahogunin. Ann. N. Y. Acad. Sci. 2003, 994, 288–298. [Google Scholar] [CrossRef]
- Mukherjee, R.; Bhattacharya, A.; Sau, A.; Basu, S.; Chakrabarti, S.; Chakrabarti, O. Calmodulin regulates MGRN1-GP78 interaction mediated ubiquitin proteasomal degradation system. FASEB J. 2019, 33, 1927–1945. [Google Scholar] [CrossRef]
- Mukherjee, R.; Chakrabarti, O. Ubiquitin-mediated regulation of the E3 ligase GP78 by MGRN1 in trans affects mitochondrial homeostasis. J. Cell Sci. 2016, 129, 757–773. [Google Scholar] [CrossRef]
- Mukherjee, R.; Majumder, P.; Chakrabarti, O. MGRN1-mediated ubiquitination of alpha-tubulin regulates microtubule dynamics and intracellular transport. Traffic 2017, 18, 791–807. [Google Scholar] [CrossRef]
- Phan, L.K.; Lin, F.; LeDuc, C.A.; Chung, W.K.; Leibel, R.L. The mouse mahoganoid coat color mutation disrupts a novel C3HC4 RING domain protein. J. Clin. Investig. 2002, 110, 1449–1459. [Google Scholar] [CrossRef]
- He, L.; Lu, X.Y.; Jolly, A.F.; Eldridge, A.G.; Watson, S.J.; Jackson, P.K.; Barsh, G.S.; Gunn, T.M. Spongiform degeneration in mahoganoid mutant mice. Science 2003, 299, 710–712. [Google Scholar] [CrossRef] [PubMed]
- Duke-Cohan, J.S.; Kim, J.H.; Azouz, A. Attractin: Cautionary tales for therapeutic intervention in molecules with pleiotropic functionality. J. Environ. Pathol. Toxicol. Oncol. 2004, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Duke-Cohan, J.S.; Tang, W.; Schlossman, S.F. Attractin: A cub-family protease involved in T cell-monocyte/macrophage interactions. Adv. Exp. Med. Biol. 2000, 477, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Duke-Cohan, J.S.; Gu, J.; McLaughlin, D.F.; Xu, Y.; Freeman, G.J.; Schlossman, S.F. Attractin (DPPT-L), a member of the CUB family of cell adhesion and guidance proteins, is secreted by activated human T lymphocytes and modulates immune cell interactions. Proc. Natl. Acad. Sci. USA 1998, 95, 11336–11341. [Google Scholar] [CrossRef] [PubMed]
- Kuramoto, T.; Kitada, K.; Inui, T.; Sasaki, Y.; Ito, K.; Hase, T.; Kawagachi, S.; Ogawa, Y.; Nakao, K.; Barsh, G.S.; et al. Attractin/mahogany/zitter plays a critical role in myelination of the central nervous system. Proc. Natl. Acad. Sci. USA 2001, 98, 559–564. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Gunn, T.M.; Bouley, D.M.; Lu, X.Y.; Watson, S.J.; Schlossman, S.F.; Duke-Cohan, J.S.; Barsh, G.S. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat. Genet. 2001, 27, 40–47. [Google Scholar] [CrossRef]
- Gunn, T.M.; Inui, T.; Kitada, K.; Ito, S.; Wakamatsu, K.; He, L.; Bouley, D.M.; Serikawa, T.; Barsh, G.S. Molecular and phenotypic analysis of Attractin mutant mice. Genetics 2001, 158, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Bronson, R.T.; Donahue, L.R.; Samples, R.; Kim, J.H.; Naggert, J.K. Mice with mutations in the mahogany gene Atrn have cerebral spongiform changes. J. Neuropathol. Exp. Neurol. 2001, 60, 724–730. [Google Scholar] [CrossRef]
- Lu, X.; Gunn, T.M.; Shieh, K.; Barsh, G.S.; Akil, H.; Watson, S.J. Distribution of Mahogany/Attractin mRNA in the rat central nervous system. FEBS Lett. 1999, 462, 101–107. [Google Scholar] [CrossRef]
- Nakadate, K.; Sakakibara, S.; Ueda, S. Attractin/mahogany protein expression in the rodent central nervous system. J. Comp. Neurol. 2008, 508, 94–111. [Google Scholar] [CrossRef]
- Chhangani, D.; Nukina, N.; Kurosawa, M.; Amanullah, A.; Joshi, V.; Upadhyay, A.; Mishra, A. Mahogunin ring finger 1 suppresses misfolded polyglutamine aggregation and cytotoxicity. Biochim. Biophys. Acta 2014, 1842, 1472–1484. [Google Scholar] [CrossRef]
- Ochikubo, F.; Gomi, H.; Kitamura, K.; Yoshikawa, Y.; Yamanouchi, K. Auditory brain-stem response in zitter rats with genetic spongiform encephalopathy. Electroencephalogr. Clin. Neurophysiol. 1992, 82, 145–151. [Google Scholar] [CrossRef]
- Ottersen, O.P.; Storm-Mathisen, J. Glutamate- and GABA-containing neurons in the mouse and rat brain, as demonstrated with a new immunocytochemical technique. J. Comp. Neurol. 1984, 229, 374–392. [Google Scholar] [CrossRef] [PubMed]
- Laszlo, L.; Lowe, J.; Self, T.; Kenward, N.; Landon, M.; McBride, T.; Farquhar, C.; McConnell, I.; Brown, J.; Hope, J.; et al. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J. Pathol. 1992, 166, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.A.; Masliah, E.; Terry, R.D.; Mirra, S.S. A neuropathological subset of Alzheimer’s disease with concomitant Lewy body disease and spongiform change. Acta Neuropathol. 1989, 78, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Artigas, J.; Niedobitek, F.; Grosse, G.; Heise, W.; Gosztonyi, G. Spongiform encephalopathy in AIDS dementia complex: Report of five cases. J. Acquir. Immune Defic. Syndr. 1989, 2, 374–381. [Google Scholar]
- Rossler, L.; Lemburg, S.; Weitkamper, A.; Thiels, C.; Hoffjan, S.; Nguyen, H.P.; Lucke, T.; Heyer, C.M. Canavan’s spongiform leukodystrophy (Aspartoacylase deficiency) with emphasis on sonographic features in infancy: Description of a case report and review of the literature. J. Ultrasound. 2022. [Google Scholar] [CrossRef]
- Baslow, M.H. Canavan’s spongiform leukodystrophy: A clinical anatomy of a genetic metabolic CNS disease. J. Mol. Neurosci. 2000, 15, 61–69. [Google Scholar] [CrossRef]
- Whatley, B.R.; Li, L.; Chin, L.S. The ubiquitin-proteasome system in spongiform degenerative disorders. Biochim. Biophys. Acta 2008, 1782, 700–712. [Google Scholar] [CrossRef]
- Sun, K.; Johnson, B.S.; Gunn, T.M. Mitochondrial dysfunction precedes neurodegeneration in mahogunin (Mgrn1) mutant mice. Neurobiol. Aging 2007, 28, 1840–1852. [Google Scholar] [CrossRef]
- Ehara, A.; Nakadate, K.; Sugimoto, H.; Yoshimoto, K.; Ueda, S. Role of neuronal nitric oxide synthase in slowly progressive dopaminergic neurodegeneration in the Zitter rat. Nitric Oxide 2018, 78, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Paz, J.; Yao, H.; Lim, H.S.; Lu, X.Y.; Zhang, W. The neuroprotective role of attractin in neurodegeneration. Neurobiol. Aging 2007, 28, 1446–1456. [Google Scholar] [CrossRef]
- Jen, H.I.; Lin, Z.Y.; Guo, J.X.; Lee, C.I. The Effects of Divalent cation-chelated prion fibrils on the immune response of EOC 13.31 microglia cells. Cells 2020, 9, 2285. [Google Scholar] [CrossRef] [PubMed]
- Majer, A.; Medina, S.J.; Sorensen, D.; Martin, M.J.; Frost, K.L.; Phillipson, C.; Manguiat, K.; Booth, S.A. The cell type resolved mouse transcriptome in neuron-enriched brain tissues from the hippocampus and cerebellum during prion disease. Sci. Rep. 2019, 9, 1099. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Nakadate, K.; Sakakibara, S.; Hirata, K.; Ueda, S. Expression of Iba1 protein in microglial cells of zitter mutant rat. Neurosci. Lett. 2007, 411, 26–31. [Google Scholar] [CrossRef]
- Streit, W.J.; Walter, S.A.; Pennell, N.A. Reactive microgliosis. Prog. Neurobiol. 1999, 57, 563–581. [Google Scholar] [CrossRef]
- Raivich, G.; Bohatschek, M.; Kloss, C.U.; Werner, A.; Jones, L.L.; Kreutzberg, G.W. Neuroglial activation repertoire in the injured brain: Graded response, molecular mechanisms and cues to physiological function. Brain Res. Rev. 1999, 30, 77–105. [Google Scholar] [CrossRef]
- Waxman, S.G. Axonal conduction and injury in multiple sclerosis: The role of sodium channels. Nat. Rev. Neurosci. 2006, 7, 932–941. [Google Scholar] [CrossRef]
- Li, J.; Baud, O.; Vartanian, T.; Volpe, J.J.; Rosenberg, P.A. Peroxynitrite generated by inducible nitric oxide synthase and NADPH oxidase mediates microglial toxicity to oligodendrocytes. Proc. Natl. Acad. Sci. USA 2005, 102, 9936–9941. [Google Scholar] [CrossRef]
- Bernardo, A.; Greco, A.; Levi, G.; Minghetti, L. Differential lipid peroxidation, Mn superoxide, and bcl-2 expression contribute to the maturation-dependent vulnerability of oligodendrocytes to oxidative stress. J. Neuropathol. Exp. Neurol. 2003, 62, 509–519. [Google Scholar] [CrossRef]
- Jiao, J.; Sun, K.; Walker, W.P.; Bagher, P.; Cota, C.D.; Gunn, T.M. Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim. Biophys. Acta 2009, 1792, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chakrabarti, O. Mahogunin regulates fusion between amphisomes/MVBs and lysosomes via ubiquitination of TSG101. Cell Death Dis. 2015, 6, e1970. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.P.; Oehler, A.; Edinger, A.L.; Wagner, K.U.; Gunn, T.M. Oligodendroglial deletion of ESCRT-I component TSG101 causes spongiform encephalopathy. Biol. Cell 2016, 108, 324–337. [Google Scholar] [CrossRef]
- Ding, W.X.; Yin, X.M. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012, 393, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.X.; Guo, F.; Ni, H.M.; Bockus, A.; Manley, S.; Stolz, D.B.; Eskelinen, E.L.; Jaeschke, H.; Yin, X.M. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J. Biol. Chem. 2012, 287, 42379–42388. [Google Scholar] [CrossRef] [PubMed]
- DiAntonio, A.; Hicke, L. Ubiquitin-dependent regulation of the synapse. Annu. Rev. Neurosci. 2004, 27, 223–246. [Google Scholar] [CrossRef]
- Waites, C.L.; Leal-Ortiz, S.A.; Okerlund, N.; Dalke, H.; Fejtova, A.; Altrock, W.D.; Gundelfinger, E.D.; Garner, C.C. Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation. EMBO J. 2013, 32, 954–969. [Google Scholar] [CrossRef]
- Hoffmann-Conaway, S.; Brockmann, M.M.; Schneider, K.; Annamneedi, A.; Rahman, K.A.; Bruns, C.; Textoris-Taube, K.; Trimbuch, T.; Smalla, K.H.; Rosenmund, C.; et al. Parkin contributes to synaptic vesicle autophagy in Bassoon-deficient mice. eLife 2020, 9, e56590. [Google Scholar] [CrossRef]
- Yumoto, T.; Nakadate, K.; Nakamura, Y.; Sugitani, Y.; Sugitani-Yoshida, R.; Ueda, S.; Sakakibara, S. Radmis, a novel mitotic spindle protein that functions in cell division of neural progenitors. PLoS ONE 2013, 8, e79895. [Google Scholar] [CrossRef] [PubMed]
- Nakadate, K.; Kamata, S. Severe acute hepatic dysfunction induced by ammonium acetate treatment results in choroid plexus swelling and ventricle enlargement in the brain. Int. J. Mol. Sci. 2022, 23, 2010. [Google Scholar] [CrossRef]
- Satoh, R.; Kawakami, K.; Nakadate, K. Effects of smart drugs on cholinergic system and non-neuronal acetylcholine in the mouse hippocampus: Histopathological approach. J. Clin. Med. 2022, 11, 3310. [Google Scholar] [CrossRef] [PubMed]
- Nakadate, K.; Imamura, K.; Watanabe, Y. Cellular and subcellular localization of alpha-1 adrenoceptors in the rat visual cortex. Neuroscience 2006, 141, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Paxinous, G.; Franklin, K. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]


| Brain Region | Expression | Brain Region | Expression | |||||
|---|---|---|---|---|---|---|---|---|
| Olfactory | Ventral posteromedial nucleus | ++ | ||||||
| Main olfactory bulb | Dorsal lateral geniculate nucleus | ++ | ||||||
| Glomerular layer | ++++ | Intergeniculate leaflet | ++ | |||||
| External plxiform layer | +++ | Medial geniculate nucleus | +++ | |||||
| Mitral layer | - | Mesencephalon | ||||||
| Internal granular layer | ++ | Superior colliculus | ||||||
| Accessory olfactory bulb | ++ | Superficial layer | +++ | |||||
| Vomeronasal nerve layer | + | Deep layer | ++ | |||||
| Cerebral cortex | Periaquedactal gray | +++ | ||||||
| Layer I–VI | ++ ~ +++ | Oculomotor nucleus | ++ | |||||
| Hippocampal formation | Red nucleus | +++ | ||||||
| Ammon’s horn (CA1–CA3) | Substantia nigra | +++ | ||||||
| Stratum oriens | ++ | Ventral tegmental area | ++ | |||||
| Pyramidal layer | ++ | Interpeduncular nucleus | +++ | |||||
| Stratum radiatum | + | Pons | ||||||
| Stratum lacnosum-moleculare | + | Dosal tegmental nucleus | +++ | |||||
| Dentate gyrus | Dosal raphe nucleus | +++ | ||||||
| Molecular layer | Outside half | +++ | Median raphe nucleus | ++ | ||||
| Intside half | +++ | Prabrachial nucleus | ++ | |||||
| Granular layer | ++ | Inferior colliculus | ++ | |||||
| Hillus | ++++ | Retrorubular area | ++ | |||||
| Basal forbrain and septal area | Pedunculo pontine nucleus | ++ | ||||||
| Bed nuclei of stria terminalis | ++ | Trapezoid body | +++ | |||||
| Claustrum | ++ | Locus coeruleus | ++ | |||||
| Basal ganglia | Motor trigeminal nucleus | ++ | ||||||
| Caudate putamen | + ~ ++ | Principal sensory trigeminal nucleus | ++ | |||||
| Globus pallidus | + | Medulla oblongata | ||||||
| Nucleus accumbens | Dorsal cochlear nucleus | +++ | ||||||
| Core | ++ | Ventral cochlear nucleus | +++ | |||||
| Shell | + ~ ++ | Medial vestibular nucleus | +++ | |||||
| Olfactory tubercle | + ~ +++ | other vestibular nucleus | ++ | |||||
| Island of Calleja | +++ | Spinal trigeminal nucleus | +++ | |||||
| Lateral septal nucleus | ++ | Facial nucleus | +++ | |||||
| Medial septal nucleus | ++ | Raphe pallidus | +++ | |||||
| Nucleus of lateral olfactry tract | +++ | Ambiguus nucleus | +++ | |||||
| Piriform cortex | ++++ | Nucleus of solitary tract | ++ | |||||
| Amygdaloid complex | Dorsal motor nucleus of vagus | +++ | ||||||
| Central amygdaloid nucleus | ++ | hypoglossal nucleus | +++ | |||||
| Basolateral amygdaloid nucleus | ++ | Inferior olive | ++ | |||||
| Medial amygdaloid nucleus | +++ | Externsl cuneate nucleus | +++ | |||||
| Hypothalamus | Cuneate nucleus | +++ | ||||||
| Medial preoptic area | +++ | Gracile nucleus | +++ | |||||
| Supraoptic nucleus | +++ | Cerebellum | ||||||
| Suprachiasmatic nucleus | +++ | Molecular layer | +++ | |||||
| Paraventricular nucleus | +++ | Purkinje cell layer | ++ | |||||
| Periventricular nucleus | +++ | Granular cell layer | +++ | |||||
| Lateral hypothalamic area | + | Cerebellar nuclei | ++ | |||||
| Dorsal hypothalamic area | ++ | Spinal cord | ||||||
| Arcuate nucleus | +++ | Dorsal horn | +++ | |||||
| Dorsomedial hyphothalamic nucleus | ++ | Ventral horn | +++ | |||||
| Ventromedical hypothalamic nucleus | +++ | Circum ventricular organ and related area | ||||||
| Zona incerta | + | Subfornical organ | +++ | |||||
| Medial mammillary nucleus | ++ | Median eminence | ++++ | |||||
| Caudal and posterior magnocellular nuclei | ++ | |||||||
| Epithalamus and thalamus | Subcommissural organ | +++ | ||||||
| Medial habenular nucleus | +++ | Area postrema | ++++ | |||||
| Lateral habenular nucleus | ++ | Rostral migratory stream | ++ | |||||
| Paraventricular thalamic nucleus | +++ | Ependyma | ++ | |||||
| Ventral posterolateral nucleus | ++ | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakadate, K.; Kawakami, K. Distribution and Localization of Mahogunin Ring Finger 1 in the Mouse Central Nervous System. Int. J. Mol. Sci. 2022, 23, 8956. https://doi.org/10.3390/ijms23168956
Nakadate K, Kawakami K. Distribution and Localization of Mahogunin Ring Finger 1 in the Mouse Central Nervous System. International Journal of Molecular Sciences. 2022; 23(16):8956. https://doi.org/10.3390/ijms23168956
Chicago/Turabian StyleNakadate, Kazuhiko, and Kiyoharu Kawakami. 2022. "Distribution and Localization of Mahogunin Ring Finger 1 in the Mouse Central Nervous System" International Journal of Molecular Sciences 23, no. 16: 8956. https://doi.org/10.3390/ijms23168956






