Transcriptome Analysis to Identify Genes Related to Flowering Reversion in Tomato
Abstract
1. Introduction
2. Results
2.1. Phenotypic Comparison between ‘116’ and ‘117’
2.2. Quality Assessment and Repeat Correlation Analysis of RNA-seq Data
2.3. Analysis of DEGs in Different Comparison Groups
2.4. GO and KEGG Enrichment Analysis of DEGs
2.5. Identification of Key Regulatory Genes for Flower Formation Reversal in Tomato
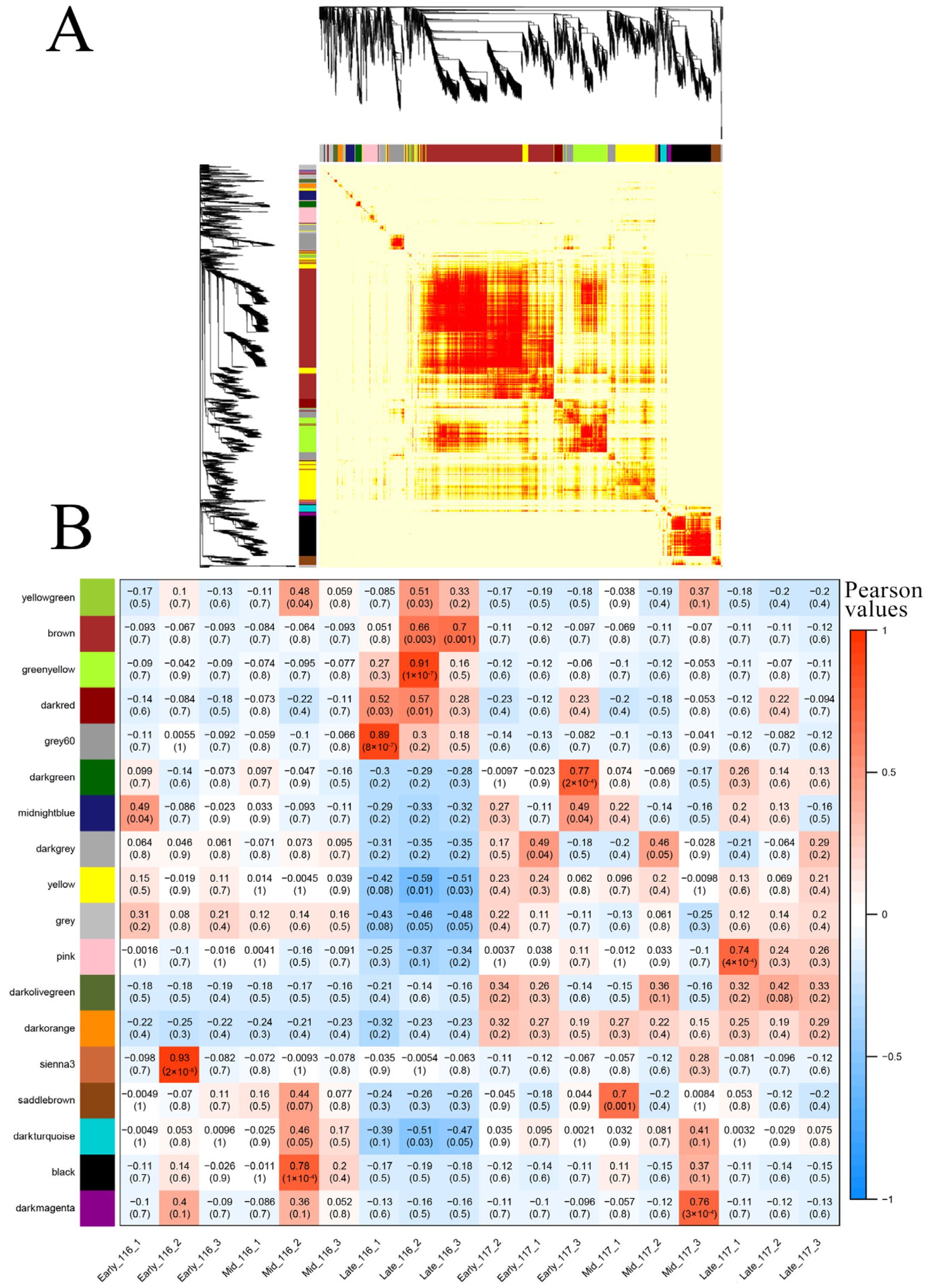
2.6. WGCNA of DEGs
2.7. Analysis of the Metabolic and Regulatory Pathways of DEGs
2.8. Validation of RNA-Seq Data by RT-qPCR
2.9. Measurements of GA3, IAA and 6-BA Hormones
2.10. Effects of Hormones on Flowering Reversion in Tomato
3. Discussion
3.1. Transcriptomic Analysis Yields 3223 DEGs Associated with Flowering Reversion
3.2. The Relationship between Floral Organ Development and Flowering Reversion
3.3. Effect of Plant Hormones on Flowering Reversion
3.4. Flowering Reversion Is the Result of Multiple Biological Processes
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. Microscopic Structure Analysis of Tomato Flowering Reversion
4.3. mRNA Library Construction and Sequencing
4.4. Mapping Reads and DEG Analysis
4.5. Gene Ontology (GO) Functional and KEGG Pathway Enrichment Analysis of DEGs
4.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.7. Analysis of MapMan Biological Functions of DEGs
4.8. Expression Profiles of Plant Flowering-Related Marker Genes
4.9. Real-Time qPCR Analysis
4.10. Measurements of Relevant Hormone Contents
4.11. Exogenous Hormone Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar] [PubMed]
- Goldberg, E.E.; Otto, S.P.; Vamosi, J.C.; Mayrose, I.; Sabath, N.; Ming, R.; Ashman, T.L. Macroevolutionary synthesis of flowering plant sexual systems. Evolution 2017, 71, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, A.; Sun, F.; Li, M.; Xu, K.; Zhang, C.; Liu, S.; Xi, Y. Using Transcriptome Analysis to Identify Genes Involved in Switchgrass Flower Reversion. Front. Plant Sci. 2018, 9, 1805. [Google Scholar] [CrossRef] [PubMed]
- MacAlister, C.A.; Park, S.J.; Jiang, K.; Marcel, F.; Bendahmane, A.; Izkovich, Y.; Eshed, Y.; Lippman, Z.B. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 2012, 44, 1393–1398. [Google Scholar] [CrossRef]
- Wang, G.; Köhler, C. Epigenetic processes in flowering plant reproduction. J. Exp. Bot. 2017, 68, 797–807. [Google Scholar] [CrossRef]
- Cho, L.H.; Yoon, J.; An, G. The control of flowering time by environmental factors. Plant J. 2017, 90, 708–719. [Google Scholar] [CrossRef]
- Li, L.; Li, X.; Liu, Y.; Liu, H. Flowering responses to light and temperature. Sci. China Life Sci. 2016, 59, 403–408. [Google Scholar] [CrossRef]
- Asbe, A.; Matsushita, S.C.; Gordon, S.; Kirkpatrick, H.E.; Madlung, A. Floral Reversion in Arabidopsis suecica Is Correlated with the Onset of Flowering and Meristem Transitioning. PLoS ONE 2015, 10, e0127897. [Google Scholar] [CrossRef]
- Olimpieri, I.; Mazzucato, A. Phenotypic and genetic characterization of the pistillate mutation in tomato. Theor. Appl. Genet. 2008, 118, 151–163. [Google Scholar] [CrossRef]
- McCullough, E.; Wright, K.M.; Alvarez, A.; Clark, C.P.; Rickoll, W.L.; Madlung, A. Photoperiod-dependent floral reversion in the natural allopolyploid Arabidopsis suecica. New Phytol. 2010, 186, 239–250. [Google Scholar] [CrossRef]
- Wang, K.; Tang, D.; Hong, L.; Xu, W.; Huang, J.; Li, M.; Gu, M.; Xue, Y.; Cheng, Z. DEP and AFO regulate reproductive habit in rice. PLoS Genet. 2010, 6, e1000818. [Google Scholar] [CrossRef] [PubMed]
- Washburn, C.F.; Thomas, J.F. Reversion of flowering in Glycine Max (Fabaceae). Am. J. Bot. 2000, 87, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Wang, L.; Liang, W.; Gai, Y.; Wang, X.; Chen, W. Floral reversion mechanism in longan (Dimocarpus longan Lour.) revealed by proteomic and anatomic analyses. J. Proteom. 2012, 75, 1099–1118. [Google Scholar] [CrossRef] [PubMed]
- Tooke, F.; Ordidge, M.; Chiurugwi, T.; Battey, N. Mechanisms and function of flower and inflorescence reversion. J. Exp. Bot. 2005, 56, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Jiang, B.; Ma, L.; Zhang, S.; Zhai, H.; Xu, X.; Hou, W.; Xia, Z.; Wu, C.; Sun, S.; et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation. New Phytol. 2018, 217, 1335–1345. [Google Scholar] [CrossRef]
- Xie, K.; Wang, Y.; Bai, X.; Ye, Z.; Zhang, C.; Sun, F.; Zhang, C.; Xi, Y. Overexpression of PvSTK1 gene from Switchgrass (Panicum virgatum L.) affects flowering time and development of floral organ in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 178, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, Y.; Zhang, Y.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Transcriptome Analysis of Flower Development and Mining of Genes Related to Flowering Time in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2021, 22, 8128. [Google Scholar] [CrossRef] [PubMed]
- Molinero-Rosales, N.; Latorre, A.; Jamilena, M.; Lozano, R. SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 2004, 218, 427–434. [Google Scholar] [CrossRef]
- Pnueli, L.; Carmel-Goren, L.; Hareven, D.; Gutfinger, T.; Alvarez, J.; Ganal, M.; Zamir, D.; Lifschitz, E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 1998, 125, 1979–1989. [Google Scholar] [CrossRef]
- Pnueli, L.; Gutfinger, T.; Hareven, D.; Ben-Naim, O.; Ron, N.; Adir, N.; Lifschitz, E. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell. 2001, 13, 2687–2702. [Google Scholar] [CrossRef]
- Lazaro, A.; Obeng-Hinneh, E.; Albani, M.C. Extended Vernalization Regulates Inflorescence Fate in Arabis alpina by Stably Silencing PERPETUAL FLOWERING1. Plant Physiol. 2018, 176, 2819–2833. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, H.; Chen, W.; Xie, D.; Zheng, S. Proteomic analysis of differentially expressed proteins in longan flowering reversion buds. Sci. Hortic. 2009, 122, 275–280. [Google Scholar] [CrossRef]
- Yu, S.; Galvão, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H.; Feng, Y.Q.; Wang, S.; Schmid, M.; Wang, J.W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.; Su, Y.H.; Zhao, X.Y.; Li, W.; Gao, X.Q.; Zhang, X.S. Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 2010, 450, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of auxin in regulating Arabidopsis flower development. Planta 2006, 223, 315–328. [Google Scholar] [CrossRef]
- You, X.; Zhu, S.; Zhang, W.; Zhang, J.; Wang, C.; Jing, R.; Chen, W.; Wu, H.; Cai, Y.; Feng, Z.; et al. OsPEX5 regulates rice spikelet development through modulating jasmonic acid biosynthesis. New Phytol. 2019, 224, 712–724. [Google Scholar] [CrossRef]
- Li, Z.; Ou, Y.; Zhang, Z.; Li, J.; He, Y. Brassinosteroid Signaling Recruits Histone 3 Lysine-27 Demethylation Activity to FLOWERING LOCUS C Chromatin to Inhibit the Floral Transition in Arabidopsis. Mol. Plant. 2018, 11, 1135–1146. [Google Scholar] [CrossRef]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef]
- Blázquez, M.A.; Ferrándiz, C.; Madueño, F.; Parcy, F. How floral meristems are built. Plant Mol. Biol. 2006, 60, 855–870. [Google Scholar] [CrossRef]
- Stewart, D.; Graciet, E.; Wellmer, F. Molecular and regulatory mechanisms controlling floral organ development. FEBS J. 2016, 283, 1823–1830. [Google Scholar] [CrossRef]
- Meir, Z.; Aviezer, I.; Chongloi, G.L.; Ben-Kiki, O.; Bronstein, R.; Mukamel, Z.; Keren-Shaul, H.; Jaitin, D.; Tal, L.; Shalev-Schlosser, G.; et al. Dissection of floral transition by single-meristem transcriptomes at high temporal resolution. Nat. Plants 2021, 7, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Szymkowiak, E.J.; Irish, E.E. Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell. 1999, 11, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Qi, S.; Touqeer, A.; Li, H.; Zhang, X.; Liu, X.; Wu, S. SlGT11 controls floral organ patterning and floral determinacy in tomato. BMC Plant Biol. 2020, 20, 562. [Google Scholar] [CrossRef]
- Matsoukas, I.G.; Massiah, A.J.; Thomas, B. Florigenic and antiflorigenic signaling in plants. Plant Cell Physiol. 2012, 53, 1827–1842. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Lee, H.T.; Jeong, H.H.; Park, J.H.; Kim, Y.C.; Lee, J.H.; Kim, J.K. The splicing factor 1-FLOWERING LOCUS M module spatially regulates temperature-dependent flowering by modulating FLOWERING LOCUS T and LEAFY expression. Plant Cell Rep. 2022, 41, 1603–1612. [Google Scholar] [CrossRef]
- Li, Z.; Zeng, S.; Li, Y.; Li, M.; Souer, E. Leaf-Like Sepals Induced by Ectopic Expression of a SHORT VEGETATIVE PHASE (SVP)-Like MADS-Box Gene from the Basal Eudicot Epimedium sagittatum. Front. Plant Sci. 2016, 7, 1461. [Google Scholar] [CrossRef]
- Coneva, V.; Zhu, T.; Colasanti, J. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J. Exp. Bot. 2007, 58, 3679–3693. [Google Scholar] [CrossRef]
- Wang, M.; Sun, S.; Wu, C.; Han, T.; Wang, Q. Isolation and characterization of the brassinosteroid receptor gene (GmBRI1) from Glycine max. Int. J. Mol. Sci. 2014, 15, 3871–3888. [Google Scholar] [CrossRef]
- Wang, M.; Xu, X.; Zhang, X.; Sun, S.; Wu, C.; Hou, W.; Wang, Q.; Han, T. Functional analysis of GmCPDs and investigation of their roles in flowering. PLoS ONE 2015, 10, e0118476. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ma, Q.; Yam, K.M.; Cheung, M.Y.; Xu, Y.; Han, T.; Lam, H.M.; Chong, K. In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 2006, 223, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Liu, Z.; Liu, Y.; Ou, L.; Deng, M.; Wang, J.; Song, J.; Ma, Y.; Chen, W.; Zhang, Z.; et al. Comparative transcriptome analysis between cytoplasmic male-sterile line and its maintainer during the floral bud development of pepper. Hortic. Plant J. 2020, 6, 89–98. [Google Scholar] [CrossRef]
- Battey, N.H.; Lyndon, R.F. Reversion of flowering. Bot. Rev. 1990, 56, 162–189. [Google Scholar] [CrossRef]
- Nie, J.; Shan, N.; Liu, H.; Yao, X.; Wang, Z.; Bai, R.; Guo, Y.; Duan, Y.; Wang, C.; Sui, X. Transcriptional control of local auxin distribution by the CsDFB1-CsPHB module regulates floral organogenesis in cucumber. Proc. Natl. Acad. Sci. USA 2021, 118, e2023942118. [Google Scholar] [CrossRef]
- Koh, K.W.; Lee, S.H.; Chen, H.K.; Chang, C.Y.; Chan, M.T. Phalaenopsis flowering locus VE regulates floral organ maturation. Plant Cell Rep. 2018, 37, 467–482. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Ito, T. Floral stem cells: From dynamic balance towards termination. Biochem. Soc. Trans. 2010, 38, 613–616. [Google Scholar] [CrossRef]
- Theissen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef]
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Thomson, B.; Zheng, B.; Wellmer, F. Floral Organogenesis: When Knowing Your ABCs Is not Enough. Plant Physiol. 2017, 173, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Xu, C.; Kwon, C.T.; Soyars, C.; Demesa-Arevalo, E.; Man, J.; Liu, L.; Lemmon, Z.H.; Jones, D.S.; Van, E.J.; et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 2019, 51, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Anfang, M.; Shani, E. Transport mechanisms of plant hormones. Curr. Opin. Plant Biol. 2021, 63, 102055. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Winter, C.M.; Wu, M.F.; Kanno, Y.; Yamaguchi, A.; Seo, M.; Wagner, D. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 2014, 344, 638–641. [Google Scholar] [CrossRef]
- Wang, Y.H.; Irving, H.R. Developing a model of plant hormone interactions. Plant Signal Behav. 2011, 6, 494–500. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2022, 4, 616–625. [Google Scholar] [CrossRef]
- Liang, Z.; Ma, Y.; Xu, T.; Cui, B.; Liu, Y.; Guo, Z.; Yang, D. Effects of abscisic acid, gibberellin, ethylene and their interactions on production of phenolic acids in salvia miltiorrhiza bunge hairy roots. PLoS ONE 2013, 8, e72806. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Boer, B.G.; Jofuku, K.D. Regulation of Arabidopsis flower development. Plant Cell. 1993, 5, 1183–1193. [Google Scholar] [PubMed][Green Version]
- Liu, L.; Farrona, S.; Klemme, S.; Turck, F.K. Post-fertilization expression of FLOWERING LOCUS T suppresses reproductive reversion. Front. Plant Sci. 2014, 5, 164. [Google Scholar] [CrossRef]
- Xu, W.; Tao, J.; Chen, M.; Dreni, L.; Luo, Z.; Hu, Y.; Liang, W.; Zhang, D. Interactions between FLORAL ORGAN NUMBER4 and floral homeotic genes in regulating rice flower development. J. Exp. Bot. 2017, 68, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Colasanti, J.; Yuan, Z.; Sundaresan, V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 1998, 93, 593–603. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, C.; Mu, J.; Zhang, H.; Yao, W.; Ding, X.; Ding, J.; Chang, Y. All-in-one sequencing: An improved library preparation method for cost-effective and high-throughput next-generation sequencing. Plant Methods 2020, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Lang, J.; Zhu, R.; Sun, X.; Zhu, S.; Li, T.; Shi, X.; Sun, Y.; Yang, Z.; Wang, W.; Bing, P.; et al. Evaluation of the MGISEQ-2000 Sequencing Platform for Illumina Target Capture Sequencing Libraries. Front. Genet. 2021, 12, 730519. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Neuparth, T.; Machado, A.M.; Montes, R.; Rodil, R.; Barros, S.; Alves, N.; Ruivo, R.; Castro, L.F.C.; Quintana, J.B.; Santos, M.M. Transcriptomic data on the transgenerational exposure of the keystone amphipod Gammarus locusta to simvastatin. Data Brief 2020, 32, 106248. [Google Scholar] [CrossRef]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef]
- Dang, Z.; Huang, L.; Jia, Y.; Lockhart, P.J.; Fong, Y.; Tian, Y. Identification of Genic SSRs Provide a Perspective for Studying Environmental Adaptation in the Endemic Shrub Tetraena mongolica. Genes 2020, 11, 322. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Bolger, M.; Schwacke, R.; Usadel, B. MapMan Visualization of RNA-Seq Data Using Mercator4 Functional Annotations. Methods Mol. Biol. 2021, 2354, 195–212. [Google Scholar]
- Sun, Y.; Yang, H.; Li, J. Transcriptome Analysis Reveals the Response Mechanism of Frl-Mediated Resistance to Fusarium oxysporum f. sp. radicis-lycopersici (FORL) Infection in Tomato. Int. J. Mol. Sci. 2022, 23, 7078. [Google Scholar] [PubMed]
- Li, Y.; Zhang, D.; Zhang, S.; Lou, Y.; An, X.; Jiang, Z.; Gao, Z. Transcriptome and miRNAome analysis reveals components regulating tissue differentiation of bamboo shoots. Plant Physiol. 2022, 188, 2182–2198. [Google Scholar] [CrossRef] [PubMed]











Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Yang, W.; Chen, J.; Chen, D.; Yang, H.; Xu, X. Transcriptome Analysis to Identify Genes Related to Flowering Reversion in Tomato. Int. J. Mol. Sci. 2022, 23, 8992. https://doi.org/10.3390/ijms23168992
Sun Y, Yang W, Chen J, Chen D, Yang H, Xu X. Transcriptome Analysis to Identify Genes Related to Flowering Reversion in Tomato. International Journal of Molecular Sciences. 2022; 23(16):8992. https://doi.org/10.3390/ijms23168992
Chicago/Turabian StyleSun, Yaoguang, Wenhui Yang, Jinxiu Chen, Dexia Chen, Huanhuan Yang, and Xiangyang Xu. 2022. "Transcriptome Analysis to Identify Genes Related to Flowering Reversion in Tomato" International Journal of Molecular Sciences 23, no. 16: 8992. https://doi.org/10.3390/ijms23168992
APA StyleSun, Y., Yang, W., Chen, J., Chen, D., Yang, H., & Xu, X. (2022). Transcriptome Analysis to Identify Genes Related to Flowering Reversion in Tomato. International Journal of Molecular Sciences, 23(16), 8992. https://doi.org/10.3390/ijms23168992





