Prokaryotic Na+/H+ Exchangers—Transport Mechanism and Essential Residues
Abstract
1. Introduction
2. Classification
2.1. The CPA Superfamily
2.2. The IT Superfamily
2.3. The Na+-Transporting Mrp Superfamily
3. Physiological Role
4. Structural Particularities
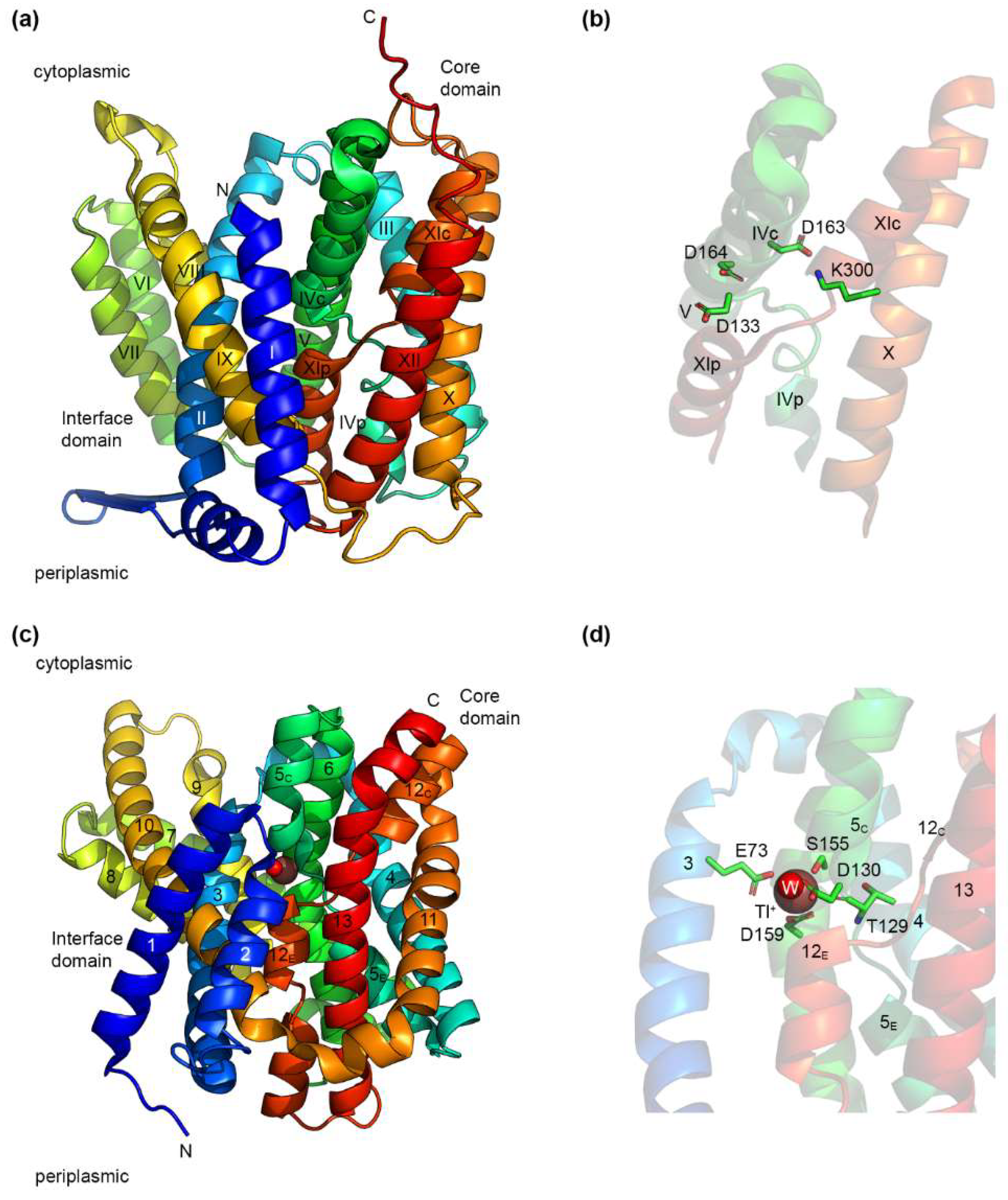
4.1. The NhaA Fold
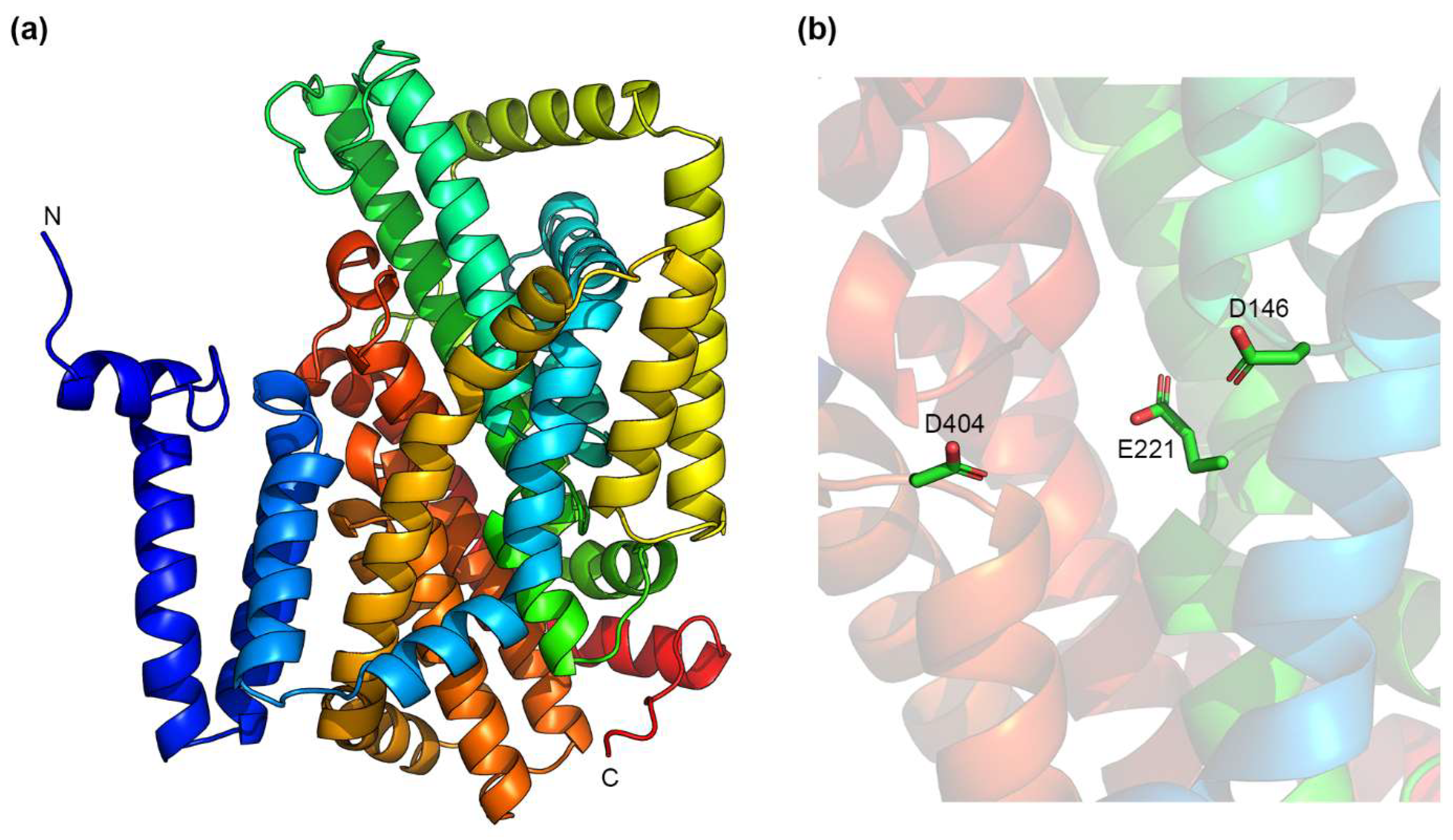
4.2. IT Na+/H+ Exchangers
4.3. Mrp Na+/H+ Exchangers
5. Essential Residues and the Substrate Binding Site(s)
5.1. The CPA Motif
5.2. The Substrate Binding Site of CPA Na+/H+ Exchangers
5.2.1. Asp164
5.2.2. Asp163 and Lys300
5.2.3. Asp133
5.2.4. Other Residues
5.3. “pH Sensor” Region
5.4. Essential Residues in IT Na+/H+ Exchangers
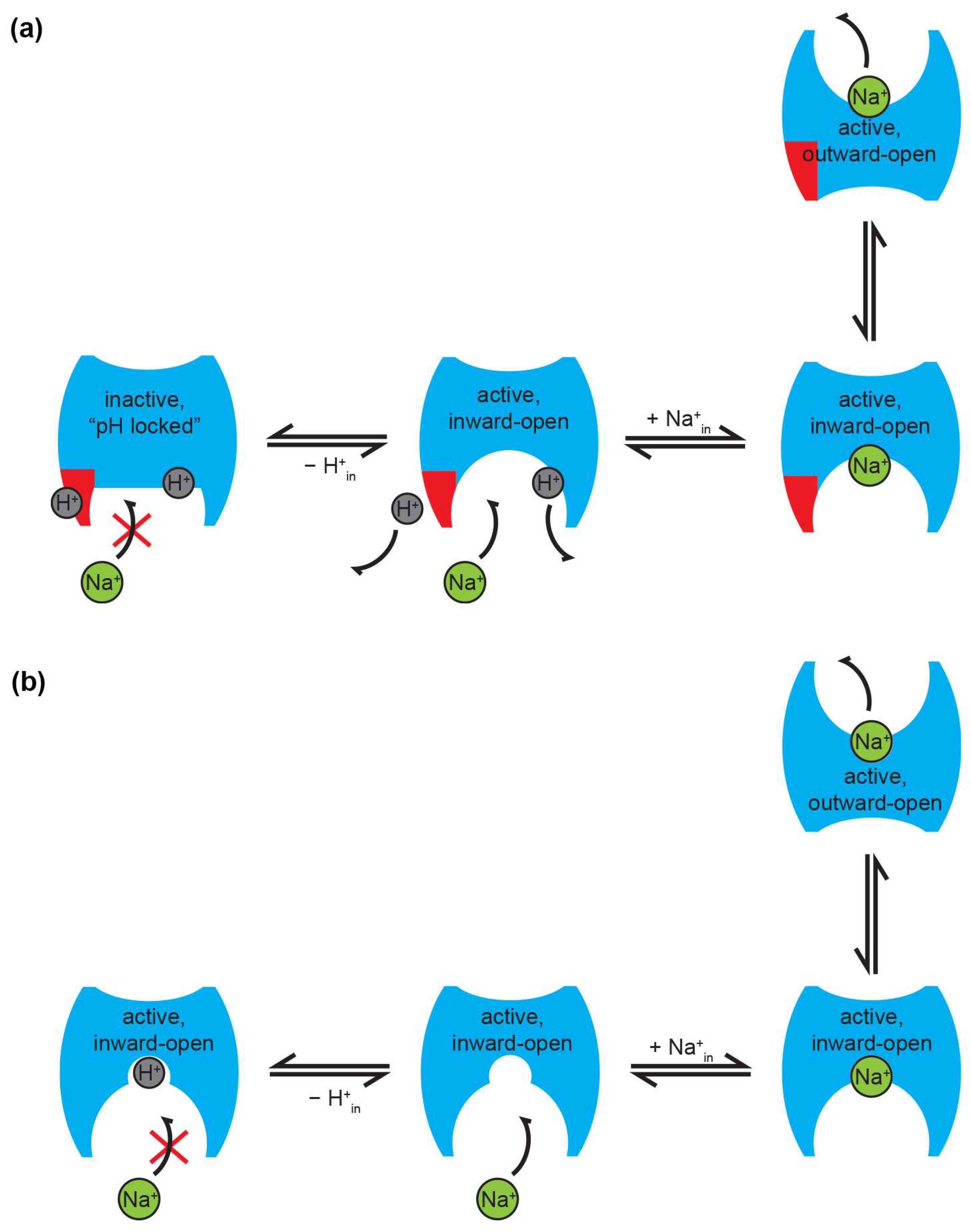
6. Transport Mechanism and Stoichiometry
6.1. pH Regulation of Na+/H+ Exchangers
6.2. Stoichiometry
7. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Krulwich, T.A.; Sachs, G.; Padan, E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 2011, 9, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Marshall, W.J. CHAPTER 5—Hydrogen ion homoeostasis and tissue oxygenation and their disorders. In Clinical Biochemistry: Metabolic and Clinical Aspects, 3rd ed.; Marshall, W.J., Lapsley, M., Day, A.P., Ayling, R.M., Eds.; Churchill Livingstone: London, UK, 2014; pp. 65–92. [Google Scholar]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline pH homeostasis in bacteria: New insights. Biochim. Biophys. Acta 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Slonczewski, J.L.; Fujisawa, M.; Dopson, M.; Krulwich, T.A. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv. Microb. Physiol. 2009, 55, 1–79, 317. [Google Scholar] [CrossRef]
- Murphy, E.; Eisner, D.A. Regulation of intracellular and mitochondrial sodium in health and disease. Circ. Res. 2009, 104, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Zacchia, M.; Abategiovanni, M.L.; Stratigis, S.; Capasso, G. Potassium: From Physiology to Clinical Implications. Kidney Dis. 2016, 2, 72–79. [Google Scholar] [CrossRef]
- Gao, A.Y.L.; Lourdin-De Filippis, E.; Orlowski, J.; McKinney, R.A. Roles of Endomembrane Alkali Cation/Proton Exchangers in Synaptic Function and Neurodevelopmental Disorders. Front. Physiol. 2022, 13, 892196. [Google Scholar] [CrossRef]
- Padan, E.; Landau, M. Sodium-Proton (Na(+)/H(+)) Antiporters: Properties and Roles in Health and Disease. Met. Ions Life Sci. 2016, 16, 391–458. [Google Scholar] [CrossRef]
- Pedersen, S.F.; Counillon, L. The SLC9A-C Mammalian Na(+)/H(+) Exchanger Family: Molecules, Mechanisms, and Physiology. Physiol. Rev. 2019, 99, 2015–2113. [Google Scholar] [CrossRef]
- Fuster, D.G.; Alexander, R.T. Traditional and emerging roles for the SLC9 Na+/H+ exchangers. Pflugers Arch. 2014, 466, 61–76. [Google Scholar] [CrossRef]
- Donowitz, M.; Ming Tse, C.; Fuster, D. SLC9/NHE gene family, a plasma membrane and organellar family of Na(+)/H(+) exchangers. Mol. Aspects Med. 2013, 34, 236–251. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.R.; Chen, M.; Deng, S.L.; Hao, X.X.; Wang, X.X.; Liu, Y.X. Sodium-hydrogen exchanger NHA1 and NHA2 control sperm motility and male fertility. Cell Death Dis. 2016, 7, e2152. [Google Scholar] [CrossRef]
- Kondapalli, K.C.; Todd Alexander, R.; Pluznick, J.L.; Rao, R. NHA2 is expressed in distal nephron and regulated by dietary sodium. J. Physiol. Biochem. 2017, 73, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Deisl, C.; Simonin, A.; Anderegg, M.; Albano, G.; Kovacs, G.; Ackermann, D.; Moch, H.; Dolci, W.; Thorens, B.; Hediger, M.A.; et al. Sodium/hydrogen exchanger NHA2 is critical for insulin secretion in beta-cells. Proc. Natl. Acad. Sci. USA 2013, 110, 10004–10009. [Google Scholar] [CrossRef]
- Cavarocchi, E.; Whitfield, M.; Chargui, A.; Stouvenel, L.; Lores, P.; Coutton, C.; Arnoult, C.; Santulli, P.; Patrat, C.; Thierry-Mieg, N.; et al. The sodium/proton exchanger SLC9C1 (sNHE) is essential for human sperm motility and fertility. Clin. Genet. 2021, 99, 684–693. [Google Scholar] [CrossRef]
- Orlowski, J.; Grinstein, S. Na+/H+ exchangers. Compr. Physiol. 2011, 1, 2083–2100. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ghishan, F.K.; Kiela, P.R. SLC9 Gene Family: Function, Expression, and Regulation. Compr. Physiol. 2018, 8, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Anderegg, M.A.; Gyimesi, G.; Ho, T.M.; Hediger, M.A.; Fuster, D.G. The Less Well-Known Little Brothers: The SLC9B/NHA Sodium Proton Exchanger Subfamily-Structure, Function, Regulation and Potential Drug-Target Approaches. Front. Physiol. 2022, 13, 898508. [Google Scholar] [CrossRef] [PubMed]
- Landau, M.; Herz, K.; Padan, E.; Ben-Tal, N. Model structure of the Na+/H+ exchanger 1 (NHE1): Functional and clinical implications. J. Biol. Chem. 2007, 282, 37854–37863. [Google Scholar] [CrossRef] [PubMed]
- Padan, E. Functional and structural dynamics of NhaA, a prototype for Na(+) and H(+) antiporters, which are responsible for Na(+) and H(+) homeostasis in cells. Biochim. Biophys. Acta 2014, 1837, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Minato, Y.; Ghosh, A.; Faulkner, W.J.; Lind, E.J.; Schesser Bartra, S.; Plano, G.V.; Jarrett, C.O.; Hinnebusch, B.J.; Winogrodzki, J.; Dibrov, P.; et al. Na+/H+ antiport is essential for Yersinia pestis virulence. Infect. Immun. 2013, 81, 3163–3172. [Google Scholar] [CrossRef]
- Karpel, R.; Alon, T.; Glaser, G.; Schuldiner, S.; Padan, E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J. Biol. Chem. 1991, 266, 21753–21759. [Google Scholar] [CrossRef]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef] [PubMed]
- Hunte, C.; Screpanti, E.; Venturi, M.; Rimon, A.; Padan, E.; Michel, H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 2005, 435, 1197–1202. [Google Scholar] [CrossRef]
- Lee, C.; Kang, H.J.; von Ballmoos, C.; Newstead, S.; Uzdavinys, P.; Dotson, D.L.; Iwata, S.; Beckstein, O.; Cameron, A.D.; Drew, D. A two-domain elevator mechanism for sodium/proton antiport. Nature 2013, 501, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Paulino, C.; Wohlert, D.; Kapotova, E.; Yildiz, O.; Kuhlbrandt, W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. eLife 2014, 3, e03583. [Google Scholar] [CrossRef] [PubMed]
- Wohlert, D.; Kuhlbrandt, W.; Yildiz, O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. eLife 2014, 3, e03579. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Gao, Y.; Ilie, A.; Kim, D.; Boucher, A.; Li, B.; Zhang, X.C.; Orlowski, J.; Zhao, Y. Structure and mechanism of the human NHE1-CHP1 complex. Nat. Commun. 2021, 12, 3474. [Google Scholar] [CrossRef]
- Matsuoka, R.; Fudim, R.; Jung, S.; Zhang, C.; Bazzone, A.; Chatzikyriakidou, Y.; Robinson, C.V.; Nomura, N.; Iwata, S.; Landreh, M.; et al. Structure, mechanism and lipid-mediated remodeling of the mammalian Na(+)/H(+) exchanger NHA2. Nat. Struct. Mol. Biol. 2022, 29, 108–120. [Google Scholar] [CrossRef]
- Dwivedi, M. Site-directed mutations reflecting functional and structural properties of Ec-NhaA. Biochimie 2021, 180, 79–89. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Tamang, D.G.; Vastermark, A. The transporter classification database. Nucleic Acids Res. 2014, 42, D251–D258. [Google Scholar] [CrossRef]
- Bruford, E.A.; Lush, M.J.; Wright, M.W.; Sneddon, T.P.; Povey, S.; Birney, E. The HGNC Database in 2008: A resource for the human genome. Nucleic Acids Res. 2008, 36, D445–D448. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Tran, C.V.; Barabote, R.D. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Shao, L.; Xu, T.; Zheng, X.; Shao, D.; Zhang, H.; Chen, H.; Zhang, Z.; Yan, M.; Abdel-Motaal, H.; Jiang, J. A novel three-TMH Na(+)/H(+) antiporter and the functional role of its oligomerization. J. Mol. Biol. 2021, 433, 166730. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Masrati, G.; Dwivedi, M.; Rimon, A.; Gluck-Margolin, Y.; Kessel, A.; Ashkenazy, H.; Mayrose, I.; Padan, E.; Ben-Tal, N. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat. Commun. 2018, 9, 4205. [Google Scholar] [CrossRef]
- Mourin, M.; Wai, A.; O’Neil, J.; Hausner, G.; Dibrov, P. Physiological, Structural, and Functional Analysis of the Paralogous Cation-Proton Antiporters of NhaP Type from Vibrio cholerae. Int. J. Mol. Sci. 2019, 20, 2572. [Google Scholar] [CrossRef]
- Xiang, M.; Feng, M.; Muend, S.; Rao, R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc. Natl. Acad. Sci. USA 2007, 104, 18677–18681. [Google Scholar] [CrossRef]
- Padan, E.; Michel, H. NhaA: A Unique Structural Fold of Secondary Active Transporters. Isr. J. Chem. 2015, 55, 1233–1239. [Google Scholar] [CrossRef]
- Prakash, S.; Cooper, G.; Singhi, S.; Saier, M.H., Jr. The ion transporter superfamily. Biochim. Biophys. Acta 2003, 1618, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Pinner, E.; Padan, E.; Schuldiner, S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J. Biol. Chem. 1992, 267, 11064–11068. [Google Scholar] [CrossRef]
- Pinner, E.; Kotler, Y.; Padan, E.; Schuldiner, S. Physiological role of nhaB, a specific Na+/H+ antiporter in Escherichia coli. J. Biol. Chem. 1993, 268, 1729–1734. [Google Scholar] [CrossRef]
- Pinner, E.; Padan, E.; Schuldiner, S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J. Biol. Chem. 1994, 269, 26274–26279. [Google Scholar] [CrossRef]
- Ito, M.; Guffanti, A.A.; Zemsky, J.; Ivey, D.M.; Krulwich, T.A. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 1997, 179, 3851–3857. [Google Scholar] [CrossRef] [PubMed]
- Haja, D.K.; Adams, M.W.W. pH Homeostasis and Sodium Ion Pumping by Multiple Resistance and pH Antiporters in Pyrococcus furiosus. Front. Microbiol. 2021, 12, 712104. [Google Scholar] [CrossRef]
- Nozaki, K.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim. Biophys. Acta 1998, 1369, 213–220. [Google Scholar] [CrossRef]
- Dzioba, J.; Ostroumov, E.; Winogrodzki, A.; Dibrov, P. Cloning, functional expression in Escherichia coli and primary characterization of a new Na+/H+ antiporter, NhaD, of Vibrio cholerae. Mol. Cell. Biochem. 2002, 229, 119–124. [Google Scholar] [CrossRef]
- Wang, Y.; Song, N.; Yang, L.; Abdel-Motaal, H.; Zhang, R.; Zhang, Z.; Meng, F.; Jiang, J. A novel NhaD-type Na(+)/H(+) antiporter from the moderate halophile and alkaliphile Halomonas alkaliphila. Can. J. Microbiol. 2017, 63, 596–607. [Google Scholar] [CrossRef]
- Melo, A.M.; Felix, N.A.; Carita, J.N.; Saraiva, L.M.; Teixeira, M. The Na+/H+ antiporter of the thermohalophilic bacterium Rhodothermus marinus. Biochem. Biophys. Res. Commun. 2006, 348, 1011–1017. [Google Scholar] [CrossRef]
- Sousa, P.M.; Videira, M.A.; Vorburger, T.; Silva, S.T.; Moir, J.W.; Steuber, J.; Melo, A.M. The novel NhaE-type Na(+)/H (+) antiporter of the pathogenic bacterium Neisseria meningitidis. Arch. Microbiol. 2013, 195, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Morino, M.; Krulwich, T.A. Mrp Antiporters Have Important Roles in Diverse Bacteria and Archaea. Front. Microbiol. 2017, 8, 2325. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Hino, M.; Kitada, M.; Horikoshi, K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J. Bacteriol. 1990, 172, 7282–7283. [Google Scholar] [CrossRef]
- Hamamoto, T.; Hashimoto, M.; Hino, M.; Kitada, M.; Seto, Y.; Kudo, T.; Horikoshi, K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 1994, 14, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Imaizumi, R.; Igarashi, K.; Kobayashi, H. Escherichia coli is able to grow with negligible sodium ion extrusion activity at alkaline pH. J. Bacteriol. 1992, 174, 7743–7749. [Google Scholar] [CrossRef][Green Version]
- Padan, E.; Maisler, N.; Taglicht, D.; Karpel, R.; Schuldiner, S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s). J. Biol. Chem. 1989, 264, 20297–20302. [Google Scholar] [CrossRef]
- Taglicht, D.; Padan, E.; Schuldiner, S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J. Biol. Chem. 1993, 268, 5382–5387. [Google Scholar] [CrossRef]
- Padan, E.; Venturi, M.; Gerchman, Y.; Dover, N. Na(+)/H(+) antiporters. Biochim. Biophys. Acta 2001, 1505, 144–157. [Google Scholar] [CrossRef]
- Herz, K.; Vimont, S.; Padan, E.; Berche, P. Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of Vibrio cholerae in a saline environment. J. Bacteriol. 2003, 185, 1236–1244. [Google Scholar] [CrossRef]
- Kuroda, T.; Mizushima, T.; Tsuchiya, T. Physiological roles of three Na+/H+ antiporters in the halophilic bacterium Vibrio parahaemolyticus. Microbiol. Immunol. 2005, 49, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Foreman, S.; Ferrara, K.; Hreha, T.N.; Duran-Pinedo, A.E.; Frias-Lopez, J.; Barquera, B. Genetic and Biochemical Characterization of the Na(+)/H(+) Antiporters of Pseudomonas aeruginosa. J. Bacteriol. 2021, 203, e0028421. [Google Scholar] [CrossRef] [PubMed]
- Schubiger, C.B.; Hoang, K.H.T.; Hase, C.C. Sodium antiporters of Pseudomonas aeruginosa in challenging conditions: Effects on growth, biofilm formation, and swarming motility. J. Genet. Eng. Biotechnol. 2020, 18, 4. [Google Scholar] [CrossRef]
- Kuroda, T.; Fujita, N.; Utsugi, J.; Kuroda, M.; Mizushima, T.; Tsuchiya, T. A major Li(+) extrusion system NhaB of Pseudomonas aeruginosa: Comparison with the major Na(+) extrusion system NhaP. Microbiol. Immunol. 2004, 48, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.W.; Illias, R.M.; Mahadi, N.M.; Najimudin, N. Expression of the Na+/H+ antiporter gene (g1-nhaC) of alkaliphilic Bacillus sp. G1 in Escherichia coli. FEMS Microbiol. Lett. 2007, 276, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Guffanti, A.A.; Ito, M.; Krulwich, T.A. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J. Biol. Chem. 2000, 275, 30287–30292. [Google Scholar] [CrossRef] [PubMed]
- Dzioba-Winogrodzki, J.; Winogrodzki, O.; Krulwich, T.A.; Boin, M.A.; Hase, C.C.; Dibrov, P. The Vibrio cholerae Mrp system: Cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J. Mol. Microbiol. Biotechnol. 2009, 16, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Kosono, S.; Ohashi, Y.; Kawamura, F.; Kitada, M.; Kudo, T. Function of a principal Na(+)/H(+) antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 2000, 182, 898–904. [Google Scholar] [CrossRef]
- Kosono, S.; Haga, K.; Tomizawa, R.; Kajiyama, Y.; Hatano, K.; Takeda, S.; Wakai, Y.; Hino, M.; Kudo, T. Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: Its involvement in pathogenesis. J. Bacteriol. 2005, 187, 5242–5248. [Google Scholar] [CrossRef]
- Mager, T.; Rimon, A.; Padan, E.; Fendler, K. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: An electrophysiological study. J. Biol. Chem. 2011, 286, 23570–23581. [Google Scholar] [CrossRef] [PubMed]
- Calinescu, O.; Fendler, K. A universal mechanism for transport and regulation of CPA sodium proton exchangers. Biol. Chem. 2015, 396, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Warnau, J.; Wohlert, D.; Okazaki, K.I.; Yildiz, O.; Gamiz-Hernandez, A.P.; Kaila, V.R.I.; Kuhlbrandt, W.; Hummer, G. Ion Binding and Selectivity of the Na+/H+ Antiporter MjNhaP1 from Experiment and Simulation. J. Phys. Chem. B 2020, 124, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Calinescu, O.; Linder, M.; Wohlert, D.; Yildiz, O.; Kuhlbrandt, W.; Fendler, K. Electrogenic Cation Binding in the Electroneutral Na+/H+ Antiporter of Pyrococcus abyssi. J. Biol. Chem. 2016, 291, 26786–26793. [Google Scholar] [CrossRef]
- Vinothkumar, K.R.; Smits, S.H.; Kuhlbrandt, W. pH-induced structural change in a sodium/proton antiporter from Methanococcus jannaschii. EMBO J. 2005, 24, 2720–2729. [Google Scholar] [CrossRef] [PubMed]
- Mourin, M.; Schubiger, C.B.; Resch, C.T.; Hase, C.C.; Dibrov, P. Physiology of the Vc-NhaP paralogous group of cation-proton antiporters in Vibrio cholerae. Mol. Cell. Biochem. 2017, 428, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Taglicht, D.; Padan, E.; Schuldiner, S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J. Biol. Chem. 1991, 266, 11289–11294. [Google Scholar] [CrossRef]
- Furrer, E.M.; Ronchetti, M.F.; Verrey, F.; Pos, K.M. Functional characterization of a NapA Na+/H+ antiporter from Thermus thermophilus. FEBS Lett. 2007, 581, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Guffanti, A.A.; Oudega, B.; Krulwich, T.A. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 1999, 181, 2394–2402. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yashiro, S.; Dotson, D.L.; Uzdavinys, P.; Iwata, S.; Sansom, M.S.; von Ballmoos, C.; Beckstein, O.; Drew, D.; Cameron, A.D. Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J. Gen. Physiol. 2014, 144, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.J.; Iwata, S.; Cameron, A.D.; Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 2011, 478, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Levin, E.J.; Pan, Y.; McCoy, J.G.; Sharma, R.; Kloss, B.; Bruni, R.; Quick, M.; Zhou, M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 2014, 505, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Quick, M.; Dwivedi, M.; Padan, E. Insight into the direct interaction of Na+ with NhaA and mechanistic implications. Sci. Rep. 2021, 11, 7045. [Google Scholar] [CrossRef] [PubMed]
- Rimon, A.; Tzubery, T.; Padan, E. Monomers of the NhaA Na+/H+ antiporter of Escherichia coli are fully functional yet dimers are beneficial under extreme stress conditions at alkaline pH in the presence of Na+ or Li+. J. Biol. Chem. 2007, 282, 26810–26821. [Google Scholar] [CrossRef]
- Padan, E.; Danieli, T.; Keren, Y.; Alkoby, D.; Masrati, G.; Haliloglu, T.; Ben-Tal, N.; Rimon, A. NhaA antiporter functions using 10 helices, and an additional 2 contribute to assembly/stability. Proc. Natl. Acad. Sci. USA 2015, 112, E5575–E5582. [Google Scholar] [CrossRef] [PubMed]
- Herz, K.; Rimon, A.; Jeschke, G.; Padan, E. Beta-sheet-dependent dimerization is essential for the stability of NhaA Na+/H+ antiporter. J. Biol. Chem. 2009, 284, 6337–6347. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.I.; Wohlert, D.; Warnau, J.; Jung, H.; Yildiz, O.; Kuhlbrandt, W.; Hummer, G. Mechanism of the electroneutral sodium/proton antiporter PaNhaP from transition-path shooting. Nat. Commun. 2019, 10, 1742. [Google Scholar] [CrossRef] [PubMed]
- Masrati, G.; Mondal, R.; Rimon, A.; Kessel, A.; Padan, E.; Lindahl, E.; Ben-Tal, N. An angular motion of a conserved four-helix bundle facilitates alternating access transport in the TtNapA and EcNhaA transporters. Proc. Natl. Acad. Sci. USA 2020, 117, 31850–31860. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Patino-Ruiz, M.; Ganea, C.; Fendler, K.; Calinescu, O. Competition is the basis of the transport mechanism of the NhaB Na+/H+ exchanger from Klebsiella pneumoniae. PLoS ONE 2017, 12, e0182293. [Google Scholar] [CrossRef]
- Patino-Ruiz, M.; Fendler, K.; Calinescu, O. Mutation of two key aspartate residues alters stoichiometry of the NhaB Na+/H+ exchanger from Klebsiella pneumoniae. Sci. Rep. 2019, 9, 15390. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy protein structure prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Mancusso, R.; Gregorio, G.G.; Liu, Q.; Wang, D.N. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature 2012, 491, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.; Li, Y.; Wang, R.; Zhang, Y.; Wang, W.; Liu, Y.; Shang, Y.; Su, D.; Wang, W.; Yang, C. The pH sensor and ion binding of NhaD Na+/H+ antiporter from IT superfamily. Mol. Microbiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Morino, M.; Suzuki, T.; Ito, M.; Krulwich, T.A. Purification and functional reconstitution of a seven-subunit mrp-type na+/h+ antiporter. J. Bacteriol. 2014, 196, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.; Sazanov, L. Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter. eLife 2020, 9, e59407. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Nie, Y.; Wang, X.; Zhao, Y.; Zhang, X.C.; Wu, X.L. Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump. Proc. Natl. Acad. Sci. USA 2020, 117, 31166–31176. [Google Scholar] [CrossRef] [PubMed]
- Morino, M.; Natsui, S.; Swartz, T.H.; Krulwich, T.A.; Ito, M. Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J. Bacteriol. 2008, 190, 4162–4172. [Google Scholar] [CrossRef] [PubMed]
- Swartz, T.H.; Ito, M.; Hicks, D.B.; Nuqui, M.; Guffanti, A.A.; Krulwich, T.A. The Mrp Na+/H+ antiporter increases the activity of the malate:quinone oxidoreductase of an Escherichia coli respiratory mutant. J. Bacteriol. 2005, 187, 388–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ito, M.; Guffanti, A.A.; Wang, W.; Krulwich, T.A. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J. Bacteriol. 2000, 182, 5663–5670. [Google Scholar] [CrossRef] [PubMed]
- Hellmer, J.; Teubner, A.; Zeilinger, C. Conserved arginine and aspartate residues are critical for function of MjNhaP1, a Na+/H+ antiporter of M. jannaschii. FEBS Lett. 2003, 542, 32–36. [Google Scholar] [CrossRef]
- Padan, E.; Kozachkov, L.; Herz, K.; Rimon, A. NhaA crystal structure: Functional-structural insights. J. Exp. Biol. 2009, 212, 1593–1603. [Google Scholar] [CrossRef]
- Calinescu, O.; Paulino, C.; Kuhlbrandt, W.; Fendler, K. Keeping it simple, transport mechanism and pH regulation in Na+/H+ exchangers. J. Biol. Chem. 2014, 289, 13168–13176. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Rimon, A.; Kozachkov-Magrisso, L.; Friedler, A.; Padan, E. Revealing the ligand binding site of NhaA Na+/H+ antiporter and its pH dependence. J. Biol. Chem. 2012, 287, 38150–38157. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.M. Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Calinescu, O.; Danner, E.; Bohm, M.; Hunte, C.; Fendler, K. Species differences in bacterial NhaA Na+/H+ exchangers. FEBS Lett. 2014, 588, 3111–3116. [Google Scholar] [CrossRef]
- Inoue, H.; Noumi, T.; Tsuchiya, T.; Kanazawa, H. Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+ antiporter (NhaA) from Escherichia coli. FEBS Lett. 1995, 363, 264–268. [Google Scholar] [CrossRef]
- Tsuboi, Y.; Inoue, H.; Nakamura, N.; Kanazawa, H. Identification of membrane domains of the Na+/H+ antiporter (NhaA) protein from Helicobacter pylori required for ion transport and pH sensing. J. Biol. Chem. 2003, 278, 21467–21473. [Google Scholar] [CrossRef]
- Kuwabara, N.; Inoue, H.; Tsuboi, Y.; Mitsui, K.; Matsushita, M.; Kanazawa, H. Structure-function relationship of the fifth transmembrane domain in the Na+/H+ antiporter of Helicobacter pylori: Topology and function of the residues, including two consecutive essential aspartate residues. Biochemistry 2006, 45, 14834–14842. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, W.; Dotson, D.L.; Beckstein, O.; Shen, J. Mechanism of pH-dependent activation of the sodium-proton antiporter NhaA. Nat. Commun. 2016, 7, 12940. [Google Scholar] [CrossRef] [PubMed]
- Olkhova, E.; Hunte, C.; Screpanti, E.; Padan, E.; Michel, H. Multiconformation continuum electrostatics analysis of the NhaA Na+/H+ antiporter of Escherichia coli with functional implications. Proc. Natl. Acad. Sci. USA 2006, 103, 2629–2634. [Google Scholar] [CrossRef]
- Arkin, I.T.; Xu, H.; Jensen, M.O.; Arbely, E.; Bennett, E.R.; Bowers, K.J.; Chow, E.; Dror, R.O.; Eastwood, M.P.; Flitman-Tene, R.; et al. Mechanism of Na+/H+ antiporting. Science 2007, 317, 799–803. [Google Scholar] [CrossRef]
- Olkhova, E.; Kozachkov, L.; Padan, E.; Michel, H. Combined computational and biochemical study reveals the importance of electrostatic interactions between the “pH sensor” and the cation binding site of the sodium/proton antiporter NhaA of Escherichia coli. Proteins 2009, 76, 548–559. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, M.; Sukenik, S.; Friedler, A.; Padan, E. The Ec-NhaA antiporter switches from antagonistic to synergistic antiport upon a single point mutation. Sci. Rep. 2016, 6, 23339. [Google Scholar] [CrossRef] [PubMed]
- Uzdavinys, P.; Coincon, M.; Nji, E.; Ndi, M.; Winkelmann, I.; von Ballmoos, C.; Drew, D. Dissecting the proton transport pathway in electrogenic Na+/H+ antiporters. Proc. Natl. Acad. Sci. USA 2017, 114, E1101–E1110. [Google Scholar] [CrossRef]
- Patino-Ruiz, M.; Dwivedi, M.; Calinescu, O.; Karabel, M.; Padan, E.; Fendler, K. Replacement of Lys-300 with a glutamine in the NhaA Na+/H+ antiporter of Escherichia coli yields a functional electrogenic transporter. J. Biol. Chem. 2019, 294, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.A.; Huang, Y.; Beckstein, O.; Shen, J. Alternative proton-binding site and long-distance coupling in Escherichia coli sodium-proton antiporter NhaA. Proc. Natl. Acad. Sci. USA 2020, 117, 25517–25522. [Google Scholar] [CrossRef]
- Galili, L.; Rothman, A.; Kozachkov, L.; Rimon, A.; Padan, E. Trans membrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na+/H+ antiporter of Escherichia coli. Biochemistry 2002, 41, 609–617. [Google Scholar] [CrossRef]
- Mondal, R.; Rimon, A.; Masrati, G.; Ben-Tal, N.; Friedler, A.; Padan, E. Towards Molecular Understanding of the pH Dependence Characterizing NhaA of Which Structural Fold is Shared by Other Transporters. J. Mol. Biol. 2021, 433, 167156. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.; Paulino, C.; Hizlan, D.; Vonck, J.; Yildiz, O.; Kuhlbrandt, W. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 2011, 30, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Gerchman, Y.; Rimon, A.; Padan, E. A pH-dependent conformational change of NhaA Na(+)/H(+) antiporter of Escherichia coli involves loop VIII-IX, plays a role in the pH response of the protein, and is maintained by the pure protein in dodecyl maltoside. J. Biol. Chem. 1999, 274, 24617–24624. [Google Scholar] [CrossRef] [PubMed]
- Tzubery, T.; Rimon, A.; Padan, E. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J. Biol. Chem. 2004, 279, 3265–3272. [Google Scholar] [CrossRef] [PubMed]
- Tzubery, T.; Rimon, A.; Padan, E. Structure-based functional study reveals multiple roles of transmembrane segment IX and loop VIII-IX in NhaA Na+/H+ antiporter of Escherichia coli at physiological pH. J. Biol. Chem. 2008, 283, 15975–15987. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Fujisaki, Y.; Enomoto, H.; Nakayama, Y.; Takabe, T.; Yamaguchi, N.; Uozumi, N. Residue aspartate-147 from the third transmembrane region of Na(+)/H(+) antiporter NhaB of Vibrio alginolyticus plays a role in its activity. J. Bacteriol. 2001, 183, 5762–5767. [Google Scholar] [CrossRef] [PubMed]
- Habibian, R.; Dzioba, J.; Barrett, J.; Galperin, M.Y.; Loewen, P.C.; Dibrov, P. Functional analysis of conserved polar residues in Vc-NhaD, Na+/H+ antiporter of Vibrio cholerae. J. Biol. Chem. 2005, 280, 39637–39643. [Google Scholar] [CrossRef] [PubMed]
- Ostroumov, E.; Dzioba, J.; Loewen, P.C.; Dibrov, P. Asp(344) and Thr(345) are critical for cation exchange mediated by NhaD, Na(+)/H(+) antiporter of Vibrio cholerae. Biochim. Biophys. Acta 2002, 1564, 99–106. [Google Scholar] [CrossRef]
- Yang, Z.; Meng, Y.; Zhao, Q.; Cheng, B.; Xu, P.; Yang, C. Critical Functions of Region 1-67 and Helix XIII in Retaining the Active Structure of NhaD Antiporter in Halomonas sp. Y2. Front. Microbiol. 2018, 9, 831. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Yang, Z.; Cheng, B.; Nie, X.; Li, S.; Yin, H.; Xu, P.; Yang, C. Functional Interaction between the N and C Termini of NhaD Antiporters from Halomonas sp. Strain Y2. J. Bacteriol. 2017, 199, e00302-17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheng, B.; Meng, Y.; Cui, Y.; Li, C.; Tao, F.; Yin, H.; Yang, C.; Xu, P. Alkaline Response of a Halotolerant Alkaliphilic Halomonas Strain and Functional Diversity of Its Na+(K+)/H+ Antiporters. J. Biol. Chem. 2016, 291, 26056–26065. [Google Scholar] [CrossRef]
- Nie, R.; Stark, S.; Symersky, J.; Kaplan, R.S.; Lu, M. Structure and function of the divalent anion/Na(+) symporter from Vibrio cholerae and a humanized variant. Nat. Commun. 2017, 8, 15009. [Google Scholar] [CrossRef] [PubMed]
- Jardetzky, O. Simple allosteric model for membrane pumps. Nature 1966, 211, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Kozachkov, L.; Herz, K.; Padan, E. Functional and structural interactions of the transmembrane domain X of NhaA, Na+/H+ antiporter of Escherichia coli, at physiological pH. Biochemistry 2007, 46, 2419–2430. [Google Scholar] [CrossRef] [PubMed]
- Olkhova, E.; Padan, E.; Michel, H. The influence of protonation states on the dynamics of the NhaA antiporter from Escherichia coli. Biophys. J. 2007, 92, 3784–3791. [Google Scholar] [CrossRef] [PubMed]
- Lentes, C.J.; Mir, S.H.; Boehm, M.; Ganea, C.; Fendler, K.; Hunte, C. Molecular characterization of the Na+/H+-antiporter NhaA from Salmonella Typhimurium. PLoS ONE 2014, 9, e101575. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calinescu, O.; Dwivedi, M.; Patino-Ruiz, M.; Padan, E.; Fendler, K. Lysine 300 is essential for stability but not for electrogenic transport of the Escherichia coli NhaA Na(+)/H(+) antiporter. J. Biol. Chem. 2017, 292, 7932–7941. [Google Scholar] [CrossRef] [PubMed]
| Superfamily | Transporter | H+:Na+ Stoichiometry | Physiological Role | Organism | References |
|---|---|---|---|---|---|
| CPA | NhaP | 1:1 | pH regulation, H+ export | M. jannaschii | [74] |
| 1:1 | Resistance to K+ at alkaline pH, K+/H+ exchange | V. cholerae | [75] | ||
| NhaA | 2:1 | Salt resistance, Na+ (Li+) export | E. coli | [76] | |
| NapA | 2:1 | Salt resistance | T. thermophilus | [77] | |
| IT | NhaB | 3:2 | pH homeostasis at neutral pH conditions and low concentrations of Na+ | E. coli | [44] |
| NhaC | NA (electrogenic) | Salt tolerance and pH homeostasis at alkaline conditions | B. firmus | [46] | |
| NhaD | NA | Salt tolerance and pH homeostasis at alkaline conditions | V. cholerae | [49] | |
| NhaE | NA | Salt tolerance at neutral pH | R. marinus | [51] | |
| Mrp | Mrp | NA (electrogenic) | Salt resistance, pH homeostasis | B. subtilis | [78] |
| NA (electrogenic) | Salt resistance, bile acid export | V. cholerae | [67] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patiño-Ruiz, M.; Ganea, C.; Călinescu, O. Prokaryotic Na+/H+ Exchangers—Transport Mechanism and Essential Residues. Int. J. Mol. Sci. 2022, 23, 9156. https://doi.org/10.3390/ijms23169156
Patiño-Ruiz M, Ganea C, Călinescu O. Prokaryotic Na+/H+ Exchangers—Transport Mechanism and Essential Residues. International Journal of Molecular Sciences. 2022; 23(16):9156. https://doi.org/10.3390/ijms23169156
Chicago/Turabian StylePatiño-Ruiz, Miyer, Constanța Ganea, and Octavian Călinescu. 2022. "Prokaryotic Na+/H+ Exchangers—Transport Mechanism and Essential Residues" International Journal of Molecular Sciences 23, no. 16: 9156. https://doi.org/10.3390/ijms23169156
APA StylePatiño-Ruiz, M., Ganea, C., & Călinescu, O. (2022). Prokaryotic Na+/H+ Exchangers—Transport Mechanism and Essential Residues. International Journal of Molecular Sciences, 23(16), 9156. https://doi.org/10.3390/ijms23169156






