The Reduced Graphene Oxide (rGO) Induces Apoptosis, Autophagy and Cell Cycle Arrest in Breast Cancer Cells
Abstract
:1. Introduction
2. Results
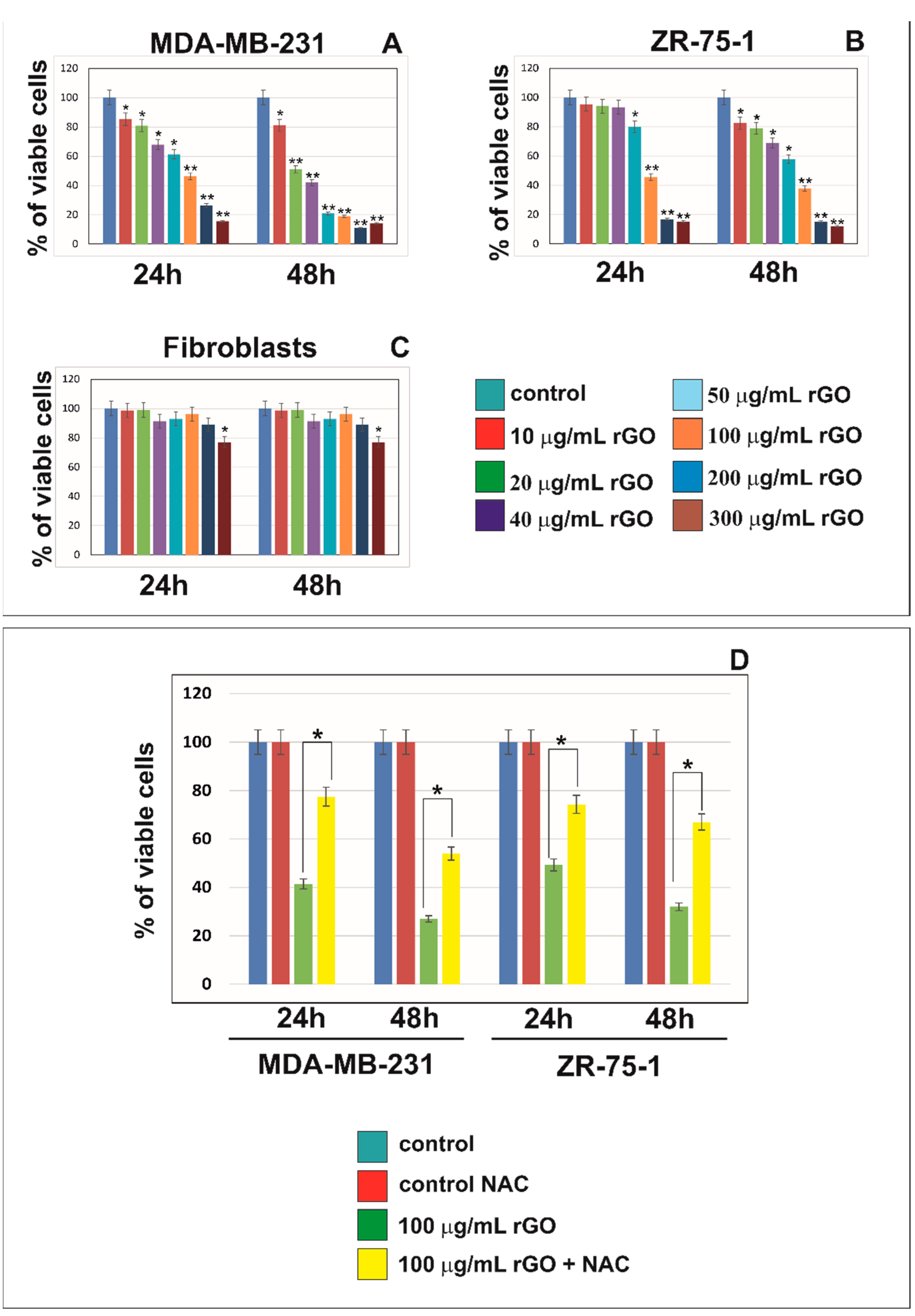
2.1. rGO Induced Cytotoxicity of MDA-MB-231 and ZR-75-1 Cells in Dose-Dependent Manner
2.2. rGO Induces S Phase Arrest in Breast Cancer Cells
2.3. rGO Induced Apoptosis in MDA-MB-231 and ZR-75-1 Cells
2.4. The Effect of rGO on Mitochondria Dysfunction in MDA-MB-231 and ZR-75-1 Cells
2.5. The Effect of rGO on Apoptosis Markers
2.6. The Effect of rGO on Autophagy Markers
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Cultures and Exposure to rGO
4.3. Cell Viability
4.4. Cell Cycle Analysis
4.5. Fluorescent Microscopy
4.6. Mitochondrial Membrane Potential (∆Ψm) Analysis
4.7. Caspase-9 Enzymatic Activity Assay
4.8. Caspase-3 Assay
4.9. Western Blot Analysis
4.10. Chemiluminescence Detection
4.11. Total Protein Content in Cells
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AKT | serine/threonine kinase |
| AMPK | 5′ AMP-activated protein kinase |
| APAF-1 | apoptotic protease activating factor 1 |
| ATG | autophagy-related protein |
| BAX | Bcl-2-associated X protein |
| BCL-2 | B-cell CLL/lymphoma 2 |
| BCL-xL | B-cell lymphoma-extra large |
| BIM | Bcl-2-like protein |
| JC-1 | 5,5,6,6-tetrachloro-1,1,3,3-tetraethylbenzimidazolcarbocyanine iodide |
| LC3 | microtubule-associated protein 1A/1B-light chain |
| MMP | mitochondrial membrane potential |
| MPTP | mitochondrial permeability transition pores |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NAC | N-acetylcysteine |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| PARP | poly(ADP-ribose) polymerase-1 |
| PI3K | phosphoinositide 3-kinases |
| rGO | reduced graphene oxide |
| ROS | reactive oxygen species |
References
- Hiroki, N.; Young, H.K. Cancer prevention from the perspective of global cancer burden patterns. J. Thorac. Dis. 2017, 9, 448–451. [Google Scholar]
- Thun, M.J.; DeLancey, J.O.; Center, M.M.; Jemal, A.; Ward, E.M. The global burden of cancer: Priorities for prevention. Carcinogenesis 2010, 31, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Castelli, G.; Pelosi, E. Breast Cancer: A Molecularly Heterogenous Disease Needing Subtype-Specific Treatments. Med. Sci. 2020, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Tuasha, N.; Petros, B. Heterogeneity of Tumors in Breast Cancer: Implications and Prospects for Prognosis and Therapeutics. Scientifica 2020, 2020, 4736091. [Google Scholar] [CrossRef]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef]
- Ou, L.; Lin, S.; Song, B.; Liu, J.; Lai, R.; Shao, L. The mechanisms of graphene-based materials-induced programmed cell death: A review of apoptosis, autophagy, and programmed necrosis. Int. J. Nanomed. 2017, 12, 6633–6646. [Google Scholar] [CrossRef]
- Yang, K.; Feng, L.; Liu, Z. Stimuli responsive drug delivery systems based on nano-graphene for cancer therapy. Adv. Drug Deliv. Rev. 2016, 105, 228–241. [Google Scholar] [CrossRef]
- Ristic, B.; Harhaji-Trajkovic, L.; Dakic, M.B.I.; Mijatovic, S.; Trajkovic, V. Modulation of Cancer Cell Autophagic Responses by Graphene-Based Nanomaterials: Molecular Mechanisms and Therapeutic Implications. Cancers 2021, 13, 4145. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, J.; Gao, G.; Sheng, Z.; Cui, H.; Cai, L. Indocyanine green-loaded polydopamine-reduced graphene oxide nanocomposites with amplifying photoacoustic and photothermal effects for cancer theranostics. Theranostics 2016, 6, 1043–1052. [Google Scholar] [CrossRef]
- Battogtokh, G.; Ko, Y.T. Graphene oxide-incorporated pH-responsive folate-albumin-photosensitizer nanocomplex as image-guided dual therapeutics. J. Control. Release 2016, 234, 10–20. [Google Scholar] [CrossRef]
- Teixeira, S.R.; Lloyd, C.; Yao, S. Polyaniline-graphene based α-amylase biosensor with a linear dynamic range in excess of 6 orders of magnitude. Biosens. Bioelectron. 2016, 85, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Shirai, A.; Henares, T.G.; Sueyoshi, K.; Endo, T.; Hisamoto, H. Fast and single-step immunoassay based on fluorescence quenching within a square glass capillary immobilizing graphene oxide-antibody conjugate and fluorescently labelled antibody. Analyst 2016, 141, 3389–3394. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, X.; Huang, P. Graphene-based nanomaterials for bioimaging. Adv. Drug Deliv. Rev. 2016, 105, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Neethirajan, S.A. Microfluidic biosensor using graphene oxide and aptamer-functionalized quantum dots for peanut allergen detection. Biosens. Bioelectron. 2016, 85, 649–656. [Google Scholar] [CrossRef]
- Ali Tahir, A.; Ullah, H.; Sudhagar, P.; Asri Mat Teridi, M.; Devadoss, A.; Sundaram, S. The application of graphene and its derivatives to energy conversion, storage, and environmental and biosensing devices. Chem. Rec. 2016, 16, 1591–1634. [Google Scholar] [CrossRef]
- McCallion, C.; Burthem, J.; Rees-Unwin, K.; Golovanov, A.; Pluen, A. Graphene in therapeutics delivery: Problems, solutions and future opportunities. Eur. J. Pharm. Biopharm. 2016, 104, 235–250. [Google Scholar] [CrossRef]
- Liu, H.; Li, T.; Liu, Y.; Qin, G.; Wang, X.; Chen, T. Glucose-reduced graphene oxide with excellent biocompatibility and photothermal efficiency as well as drug loading. Nanoscale Res. Lett. 2016, 11, 1–10. [Google Scholar] [CrossRef]
- Szczepaniak, J.; Strojny, B.; Chwalibog, E.S.; Jaworski, S.; Jagiello, J.; Winkowska, M.; Szmidt, M.; Wierzbicki, M.; Sosnowska, M.; Balaban, J.; et al. Effects of Reduced Graphene Oxides on Apoptosis and Cell Cycle of Glioblastoma Multiforme. Int. J. Mol. Sci. 2018, 19, 3939. [Google Scholar] [CrossRef]
- Hua, S.; Yang, L.; Gao, T.; Jiang, P.; Jiang, F.; Liu, Y. Graphene Quantum Dots Induce Autophagy and Reveal Protection Against Hydrogen Peroxide-Induced Oxidative Stress Injury. ACS Appl. Bio Mater. 2019, 2, 5760–5768. [Google Scholar] [CrossRef]
- Wang, J.; Wang, P.; He, Y.; Liu, X.; Wang, S.; Ma, C.; Tian, X.; Wang, J.; Wu, X. Graphene oxide inhibits cell migration and invasion by destroying actin cytoskeleton in cervical cancer cells. Aging 2020, 12, 17625–17633. [Google Scholar] [CrossRef]
- Yiru, Q.; Zhi-Wei, Z.; Shu-Ting, P.; Zhi-Xu, H.; Xueji, Z.; Jia-Xuan, Q.; Wei, D.; Tianxin, Y.; Shu-Feng, Z. Graphene quantum dots induce apoptosis, autophagy, and inflammatory response via p38 mitogen-activated protein kinase and nuclear factor-kB mediated signaling pathways in activated THP-1 macrophages. Toxicology 2015, 327, 62–76. [Google Scholar]
- Krętowski, R.; Jabłońska-Trypuć, A.; Cechowska-Pasko, M. The Preliminary Study on the Proapoptotic Effect of Reduced Graphene Oxide in Breast Cancer Cell Lines. Int. J. Mol. Sci. 2021, 22, 12593. [Google Scholar] [CrossRef] [PubMed]
- Minfei, S.; Yang, M.; Sangita, S. Role of the Crosstalk between Autophagy and Apoptosis in Cancer. J. Oncol. 2013, 2013, 102735. [Google Scholar] [CrossRef]
- Gump, J.M.; Thorburn, A. Autophagy and apoptosis- what’s the connection? Trends Cell Biol. 2011, 21, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS ONE 2009, 4, e6146. [Google Scholar] [CrossRef]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 1–24. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, J.H. An in vitro evaluation of graphene oxide reduced by Gonoderma spp. In human breast cancer cells (MDA-MB-231). Int. J. Nanomed. 2014, 9, 1783–1797. [Google Scholar] [CrossRef]
- Mendes, R.G.; Koch, B.; Bachmatiuk, A.; Ma, X.; Sanchez, S.; Damm, C.; Schmidt, O.G.; Gemming, T.; Eckert, J.; Rummeli, M.H. A size dependent evaluation of the cytotoxicity and uptake of nanographene oxide. J. Mater. Chem. B 2015, 3, 2522–2529. [Google Scholar] [CrossRef]
- Wu, J.; Yang, R.; Zhang, L.; Fan, Z.; Liu, S. Cytotoxicity effect of graphene oxide on human MDA-MB-231 cells. Toxicol. Mech. Methods 2015, 25, 312–319. [Google Scholar] [CrossRef]
- Alsaedi, I.I.J.; Taqi, Z.J.; Abdul Hussien, A.M.; Sulaiman, G.M.; Jabir, M.S. Graphene nanoparticles induces apoptosis in MCF-7 cells through mitochondrial damage and NF-KB pathway. Mater. Res. Express 2019, 6, 095413. [Google Scholar] [CrossRef]
- Jaworski, S.; Sawosz, E.; Kutwin, K.; Wierzbicki, M.; Hinzmann, M.; Grodzik, M.; Winnicka, A.; Lipińska, L.; Włodyga, K.; Chwalibog, A. In vitro and in vivo effects of graphene oxide and reduced graphene oxide on glioblastoma. Int. J. Nanomed. 2015, 10, 1585–1596. [Google Scholar]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiełło, J.; Koziński, R.; Wierzbicki, M.; Grodzik, M.; Lipińska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar]
- Markovic, Z.M.; Harhaji-Trajkovic, L.M.; Todorovic-Markovic, B.M.; Kepić, D.P.; Arsikin, K.M.; Jovanović, S.P.; Pantovic, A.C.; Dramićanin, M.D.; Trajkovic, V.S. In vitro comparison of the photothermal anticancer activity of graphene nanoparticles and carbon nanotubes. Biomaterials 2011, 32, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Liu, J.; Wu, J.; Yin, Q.; Liang, H.; Chen, A.; Shao, L. Graphene oxide and reduced graphene oxide induce neural pheochromocytoma-derived PC12 cell lines apoptosis and cell cycle alterations via the ERK signaling pathway. Int. J. Nanomed. 2017, 12, 5501–5510. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, Z.; Qu, L. Graphene-Based Fibers: Recent Advances in Preparation and Application. Adv. Mater. 2020, 32, e1901979. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Tang, T.; Zhao, L.; Yang, Z.; Liu, Z.; Gu, J.; Bai, B.; Liu, J.; Xu, J.; Yang, H. Mechanisms of oxidative stress, apoptosis, and autophagy involved in graphene oxide nanomaterial anti-osteosarcoma effect. Int. J. Nanomed. 2018, 17, 2907–2919. [Google Scholar] [CrossRef]
- Krętowski, R.; Borzym-Kluczyk, M.; Stypułkowska, A.; Brańska-Januszewska, J.; Ostrowska, H.; Cechowska-Pasko, M. Low glucose dependent decrease of apoptosis and induction of autophagy in breast cancer MCF-7 cells. Mol. Cell. Biochem. 2016, 417, 35–47. [Google Scholar] [CrossRef]
- Lammel, T.; Boisseaux, P.; Fernandez-Cruz, M.L.; Navas, J.M. Internalization and cytotoxicity of graphene oxide and carboxyl graphene nanoplatelets in the human hepatocellular carcinoma cell line Hep G2. Part. Fibre Toxicol. 2013, 10, 1–21. [Google Scholar] [CrossRef]
- Zhang, B.; Wei, P.; Zhou, Z.; Wei, T. Interactions of graphene with mammalian cells: Molecular mechanisms and biomedical insights. Adv. Drug Deliv. Rev. 2016, 105, 145–162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, B.; Zheng, J.; Yu, M.; Zhou, T.; Zhao, K.; Jia, Y.; Gao, X.; Chen, C.; Wei, T. The inhibition of migration and invasion of cancer cells by graphene via the impairment of mitochondrial respiration. Biomaterials 2014, 35, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Ghosh, C.; Basu, S. Polymer conjugated graphene-oxide nanoparticles impair nuclear DNA and Topoisomerase I in cancer. Nanoscale Adv. 2019, 1, 4965–4971. [Google Scholar] [CrossRef]
- Jiang, J.-H.; Pi, J.; Jin, H.; Cai, J.-Y. Functional graphene oxide as cancer-targeted drug delivery system to selectively induce oesophageal cancer cell apoptosis. Artif. Cells Nanomed. Biotechnol. 2018, 46, 297–307. [Google Scholar] [CrossRef]
- Abbaspour Babaei, M.; Zaman Huri, H.; Kamalidehghan, B.; Yeap, S.K.; Ahmadipour, F. Apoptotic induction and inhibition of NF-κB signaling pathway in human prostatic cancer PC3 cells by natural compound 2,2’-oxybis (4-allyl-1-methoxybenzene), biseugenol B, from Litsea costalis: An in vitro study. OncoTargets Ther. 2017, 10, 277–294. [Google Scholar] [CrossRef]
- Bajpai, V.K.; Khan, I.; Shukla, S.; Kang, S.M.; Aziz, F.; Tripathi, K.M.; Saini, D.; Cho, H.J.; Heo, N.S.; Sonkar, S.K.; et al. Multifunctional N-P-doped carbon dots for regulation of apoptosis and autophagy in B16F10 melanoma cancer cells and in vitro imaging applications. Theranostic 2020, 10, 7841–7856. [Google Scholar] [CrossRef]
- Kusaczuk, M.; Krętowski, R.; Bartoszewicz, M.; Cechowska-Pasko, M. Phenylbutyrate—A pan-HDAC inhibitor—Suppresses proliferation of glioblastoma LN-229 cell line. Tumor Biol. 2016, 37, 931–942. [Google Scholar] [CrossRef]
- Banerjee, P.P.; Bandyopadhyay, A.; Mondal, P.; Mondal, M.K.; Chowdhury, P.; Chakraborty, A.; Sudarshan, M.; Bhattacharya, S.; Chattopadhyay, A. Cytotoxic effect of graphene oxide-functionalized gold nanoparticles in human breast cancer cell lines. Nucleus 2019, 62, 243–250. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J. Funcjonalized graphene oxide triggers cell cycle checkpoint control through both the ATM and the ATR signaling pathway. Carbon 2018, 129, 495–503. [Google Scholar] [CrossRef]
- Hashemi, E.; Akhavan, O.; Shaamsara, M.; Majd, S.A.; Sanati, M.H.; Joupari, M.D.; Farmany, A. Graphene oxide negatively regulates cell cycle in embryonic fibroblasts cells. Int. J. Nanomed. 2020, 15, 6201–6209. [Google Scholar] [CrossRef]
- Krętowski, R.; Kusaczuk, M.; Naumowicz, M.; Kotyńska, J.; Szynaka, B.; Cechowska-Pasko, M. The Effects of Silica Nanoparticles on Apoptosis and Autophagy of Glioblastoma Cell Lines. Nanomaterials 2017, 7, 230. [Google Scholar] [CrossRef] [PubMed]
- Bjorkoy, G.; Lamark, T.; Pankiv, S.; Overvatn, A.; Brech, A.; Johansen, T. Monitoring Autophagic Degradation of p62/SQSTM1. Methods Enzymol. 2009, 452, 181–197. [Google Scholar] [PubMed]
- Zhang, L.; Ouyang, S.; Zhang, H.; Qiu, M.; Dai, Y.; Wang, Y.; Ou, J. Graphene oxide induces dose-dependent lung injury in rats by regulating autophagy. Exp. Ther. Med. 2021, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, J.; DeGraff, W.G.; Gazdar, A.F.; Minna, J.D.; Mitchell, J.B. Evaluation of a tetrazolium-based semiautomated colorimetric assay: Assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krętowski, R.; Cechowska-Pasko, M. The Reduced Graphene Oxide (rGO) Induces Apoptosis, Autophagy and Cell Cycle Arrest in Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 9285. https://doi.org/10.3390/ijms23169285
Krętowski R, Cechowska-Pasko M. The Reduced Graphene Oxide (rGO) Induces Apoptosis, Autophagy and Cell Cycle Arrest in Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(16):9285. https://doi.org/10.3390/ijms23169285
Chicago/Turabian StyleKrętowski, Rafał, and Marzanna Cechowska-Pasko. 2022. "The Reduced Graphene Oxide (rGO) Induces Apoptosis, Autophagy and Cell Cycle Arrest in Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 16: 9285. https://doi.org/10.3390/ijms23169285
APA StyleKrętowski, R., & Cechowska-Pasko, M. (2022). The Reduced Graphene Oxide (rGO) Induces Apoptosis, Autophagy and Cell Cycle Arrest in Breast Cancer Cells. International Journal of Molecular Sciences, 23(16), 9285. https://doi.org/10.3390/ijms23169285





