Isolation and Characterization of an LBD Transcription Factor CsLBD39 from Tea Plant (Camellia sinensis) and Its Roles in Modulating Nitrate Content by Regulating Nitrate-Metabolism-Related Genes
Abstract
1. Introduction
2. Results
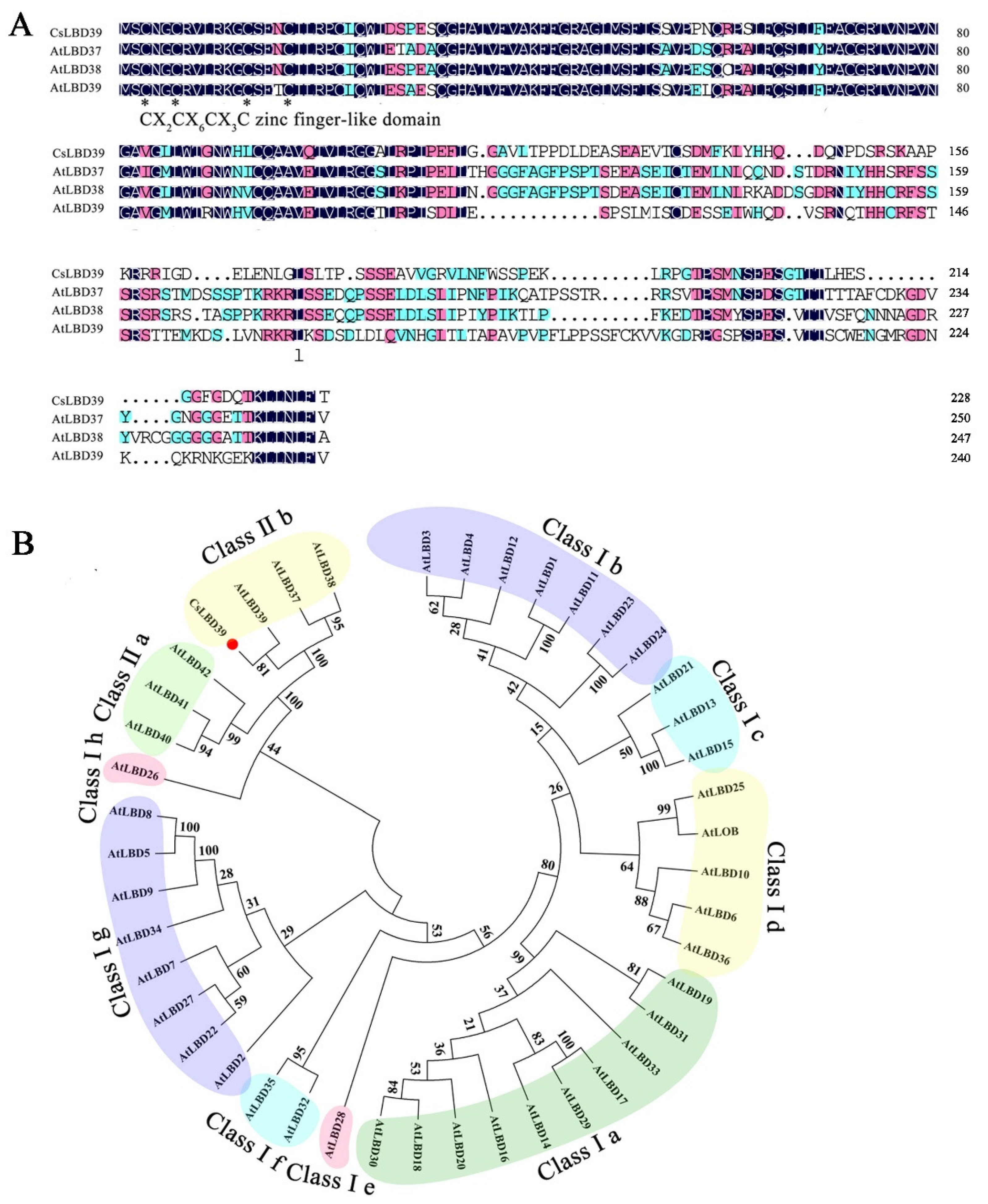
2.1. Sequence and Phylogenetic Tree Analysis of the CsLBD39
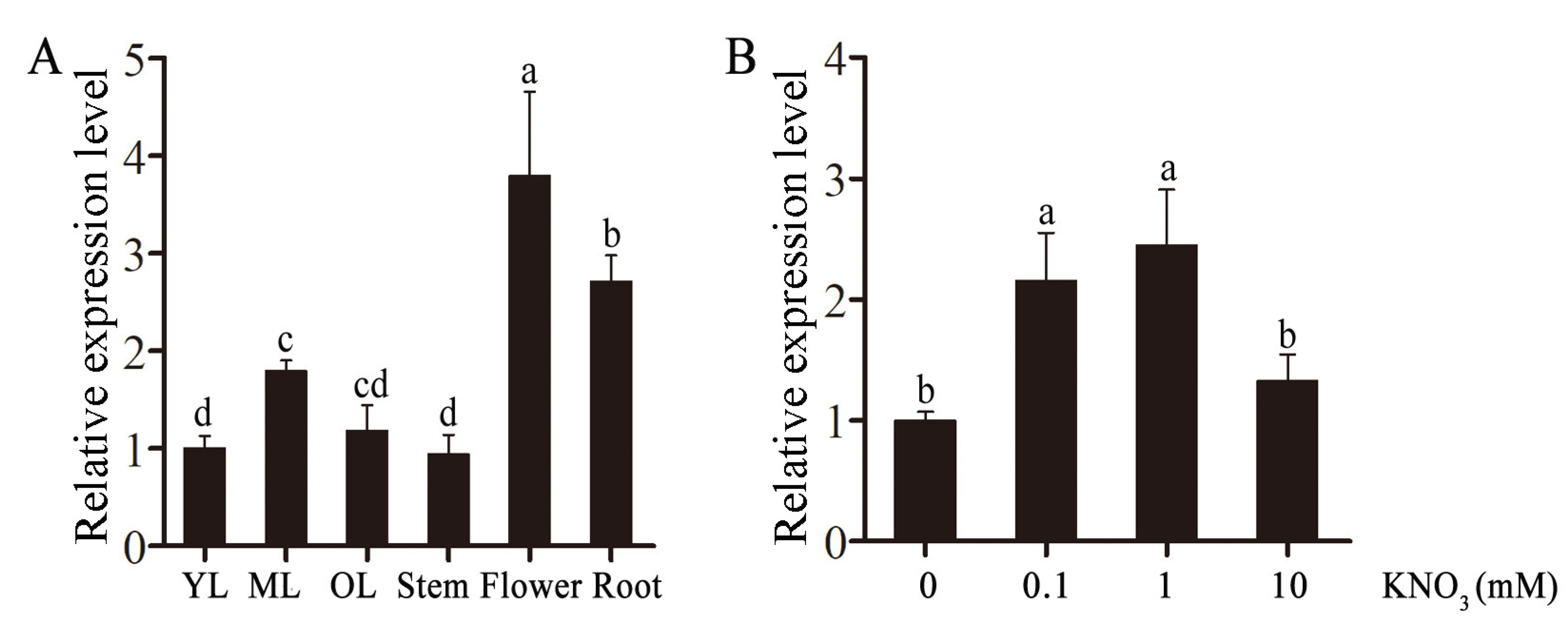
2.2. Relative Expression Level of CsLBD39 in Tea Plant
2.3. Subcellular Localization and Transcriptional Activation Activity Analysis of CsLBD39
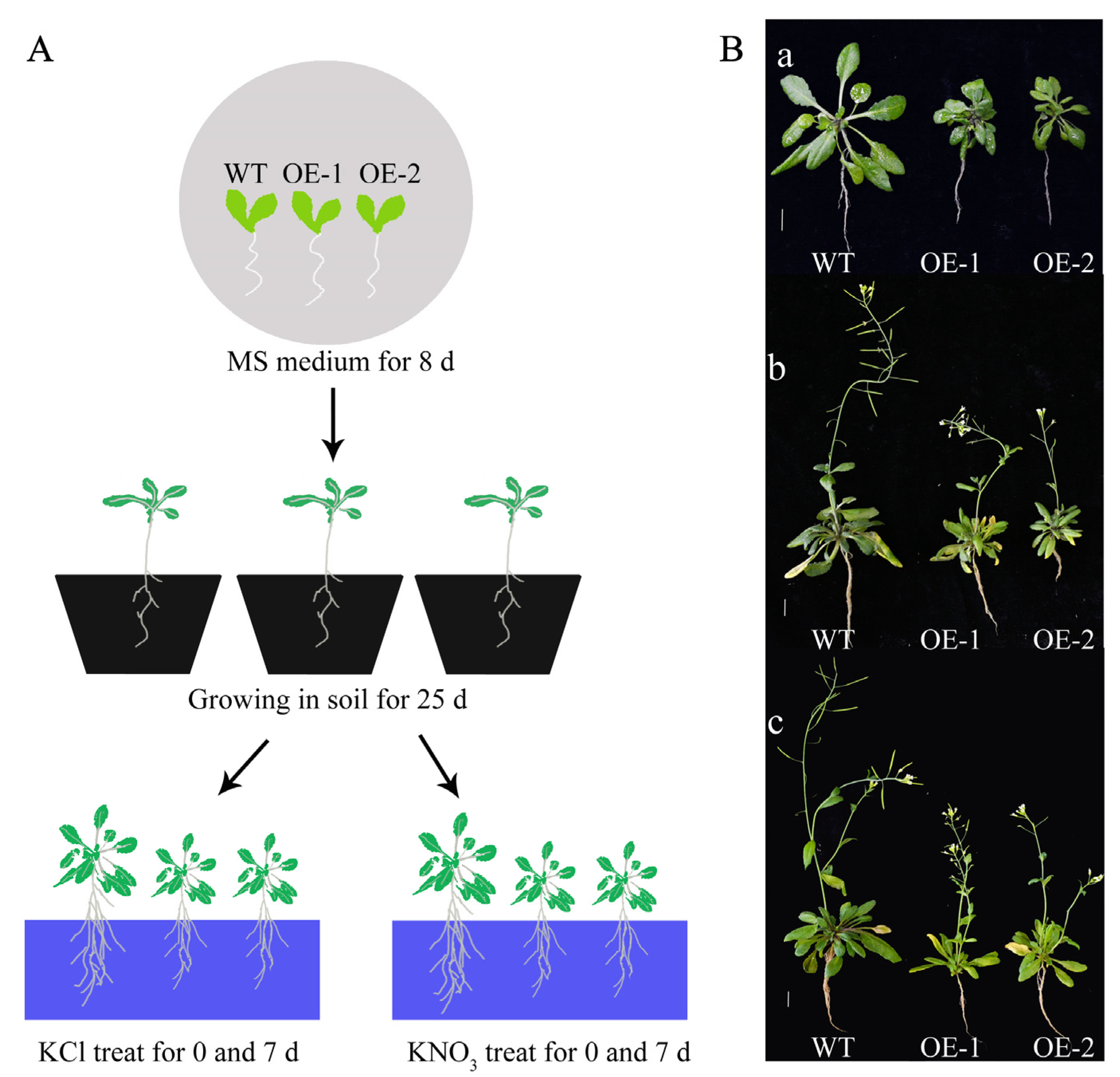
2.4. Overexpression of CsLBD39 in Arabidopsis
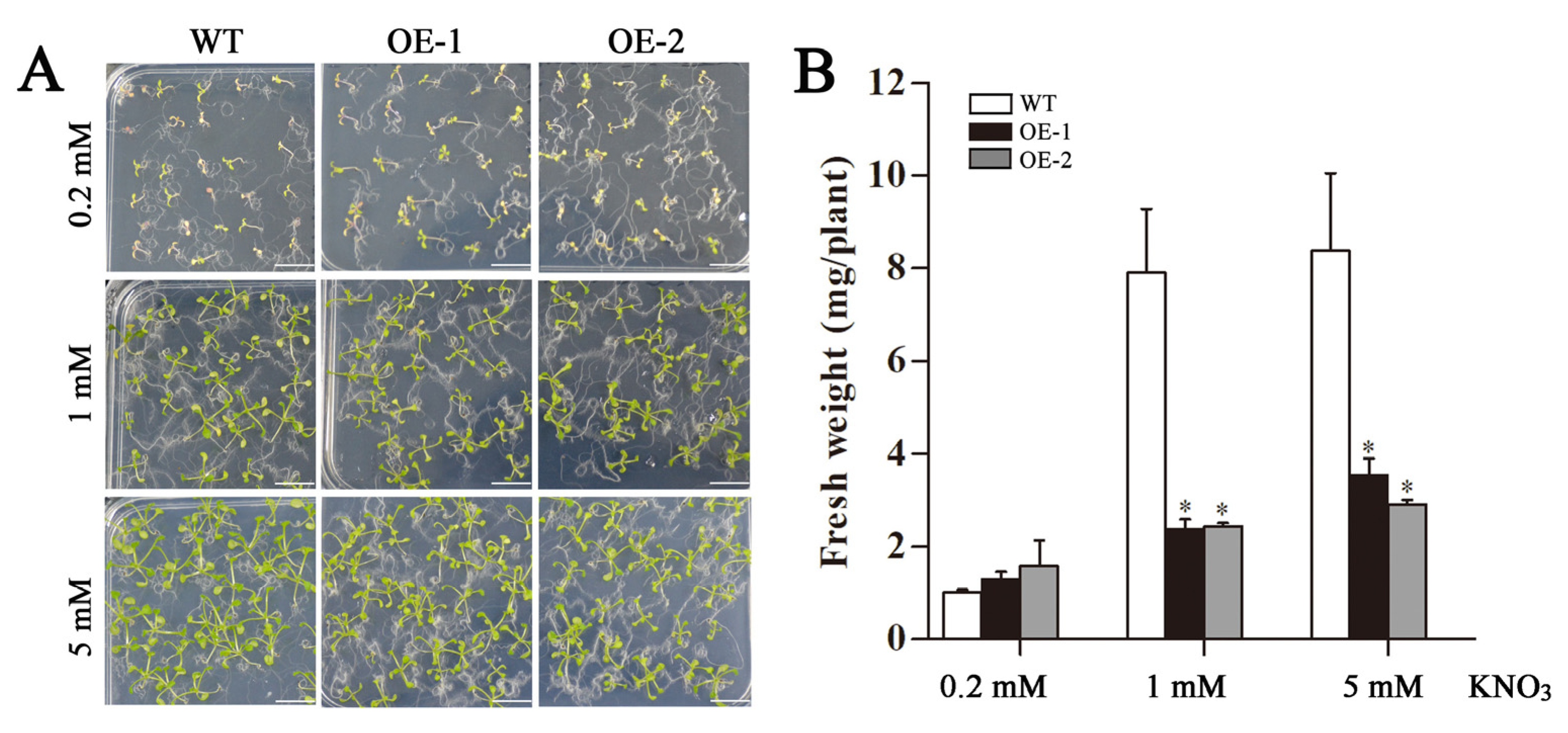
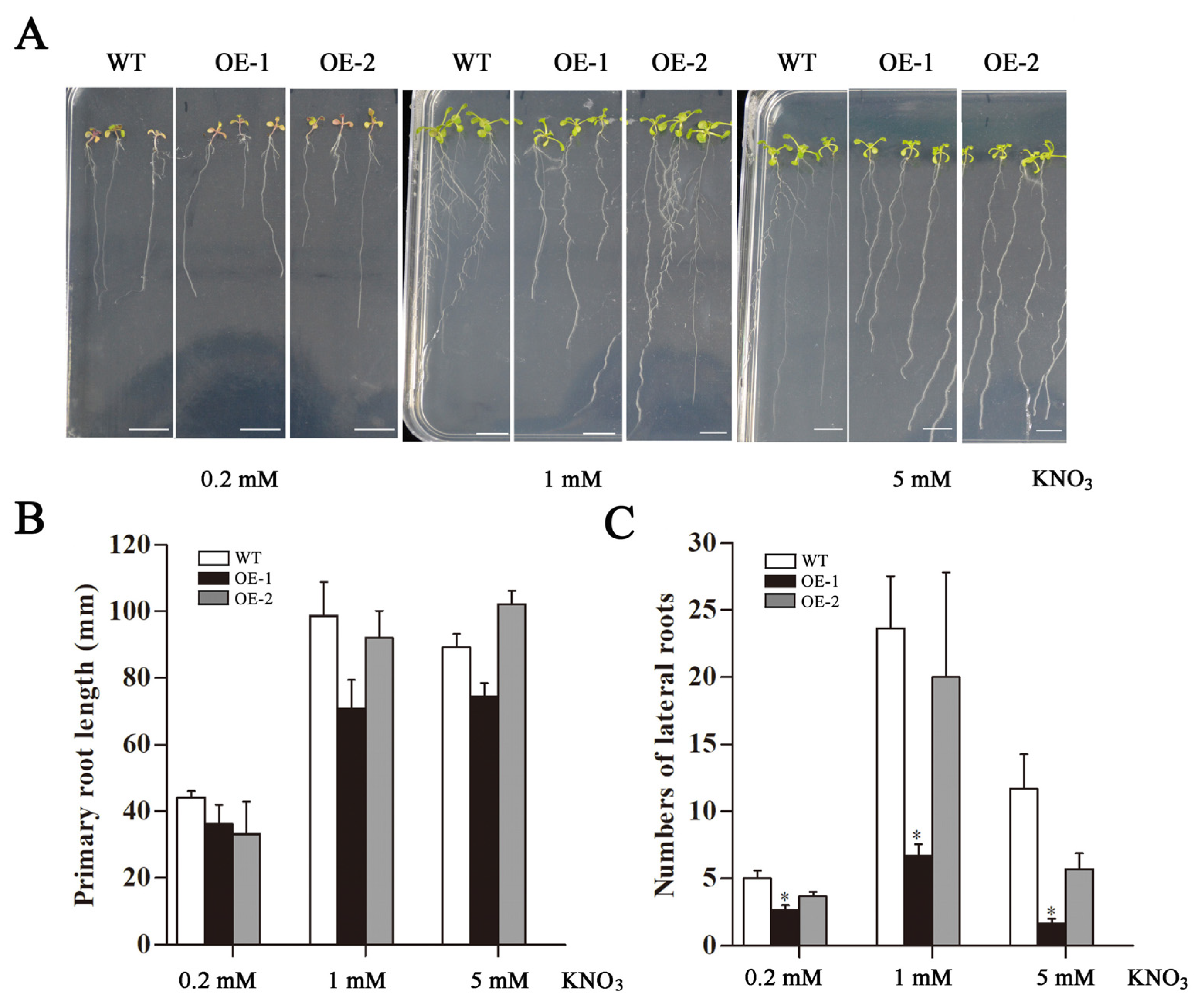
2.5. Changes in Fresh Weight and Roots of Transgenic Arabidopsis Overexpressing CsLBD39
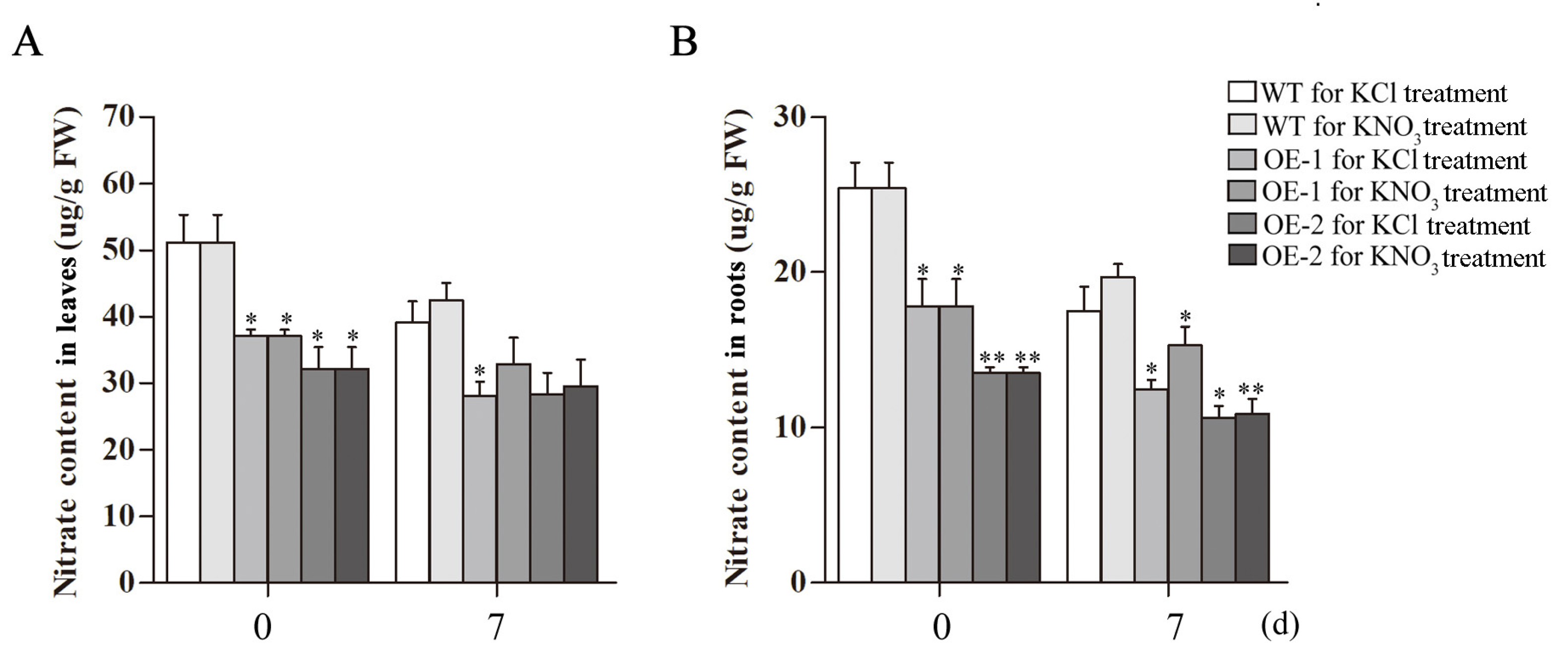
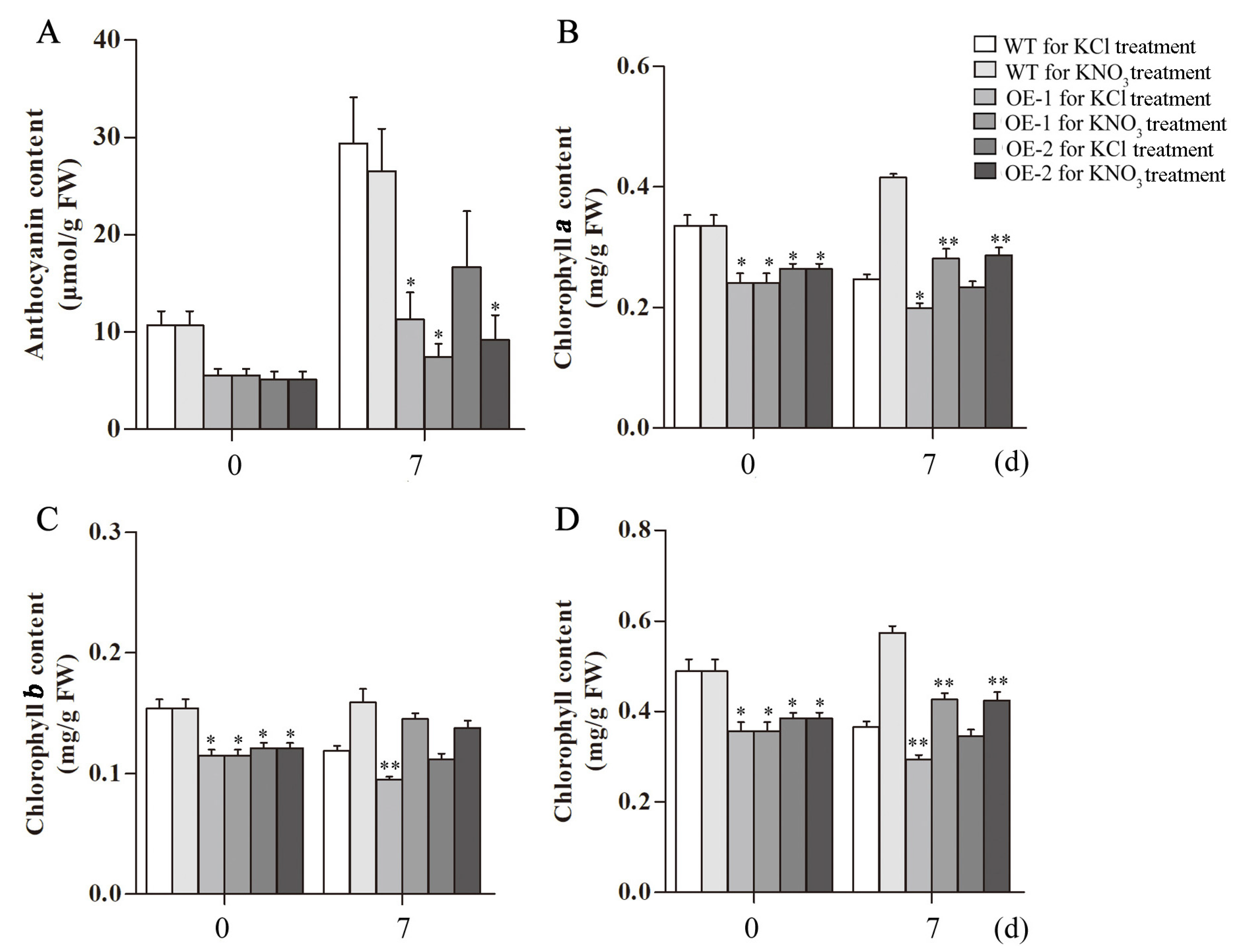
2.6. Analysis of Nitrate, Anthocyanin and Chlorophyll Contents in Transgenic Arabidopsis Overexpressing CsLBD39 Gene
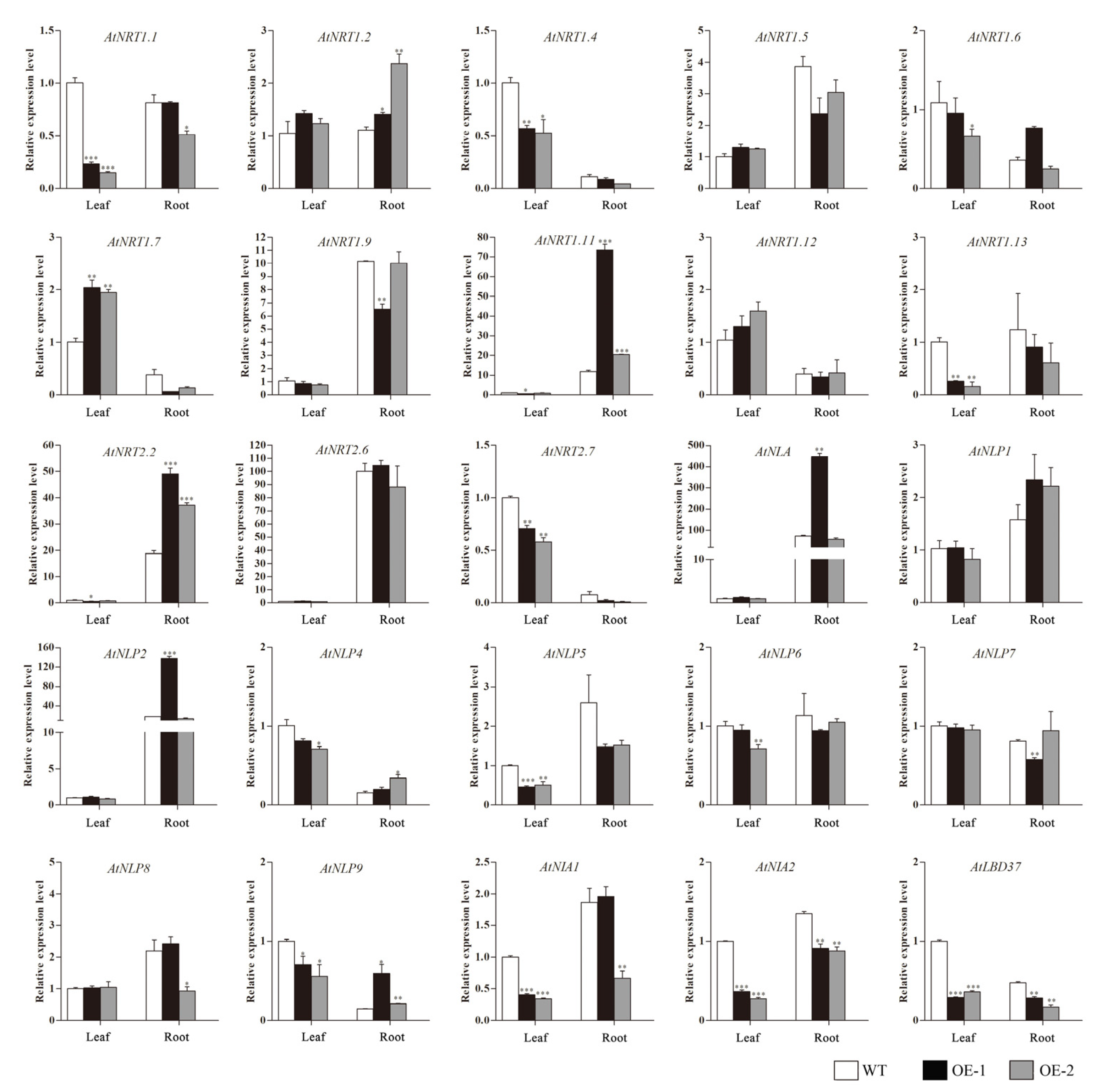
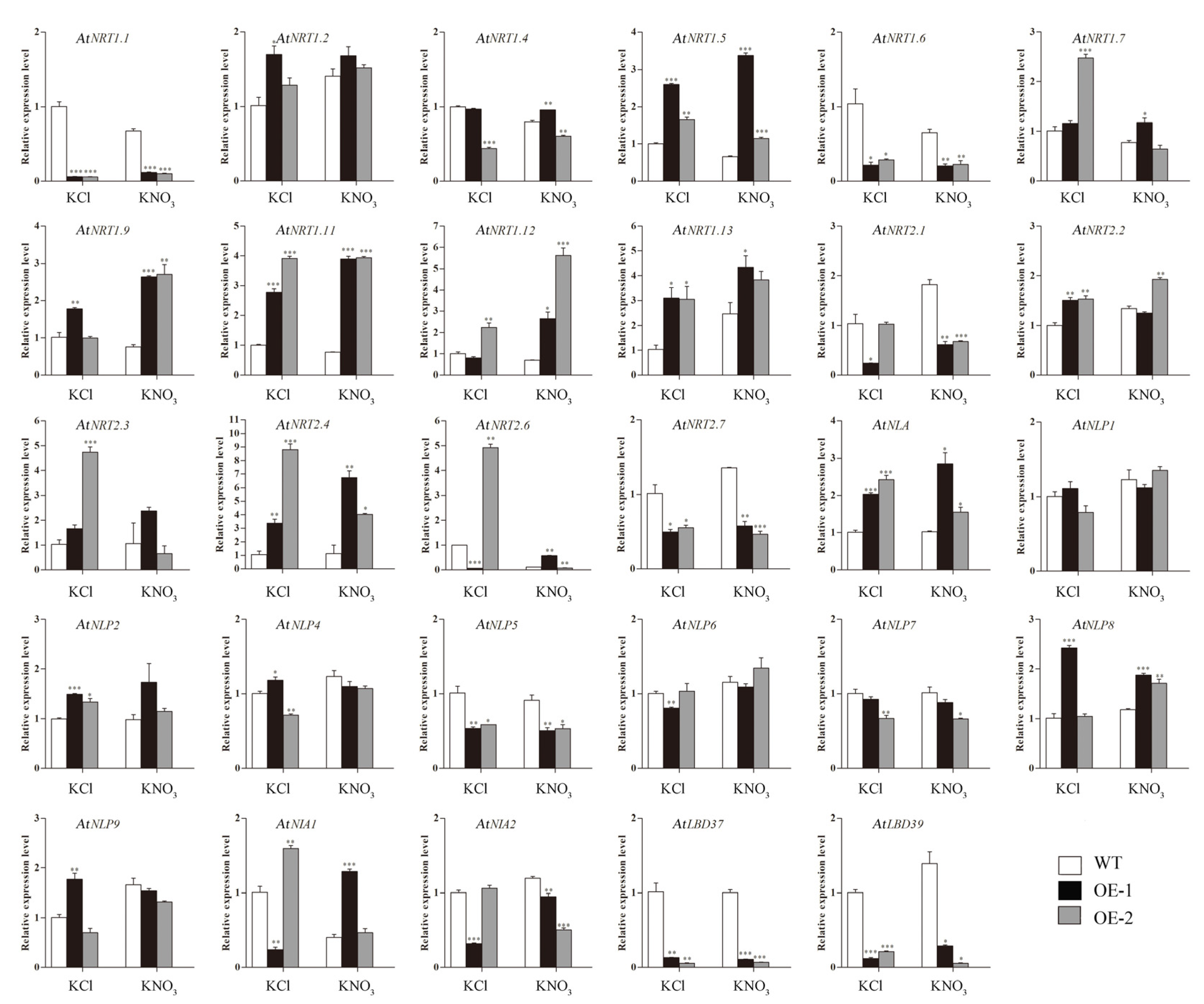
2.7. Expression Analysis of Nitrate Uptake and Transport-Related Genes in Transgenic Arabidopsis Plants Overexpressing CsLBD39 Gene
2.8. Analysis of Nitrate, Anthocyanins and Chlorophyll Contents in Transgenic Arabidopsis Overexpressing CsLBD39 under Nitrate Treatment
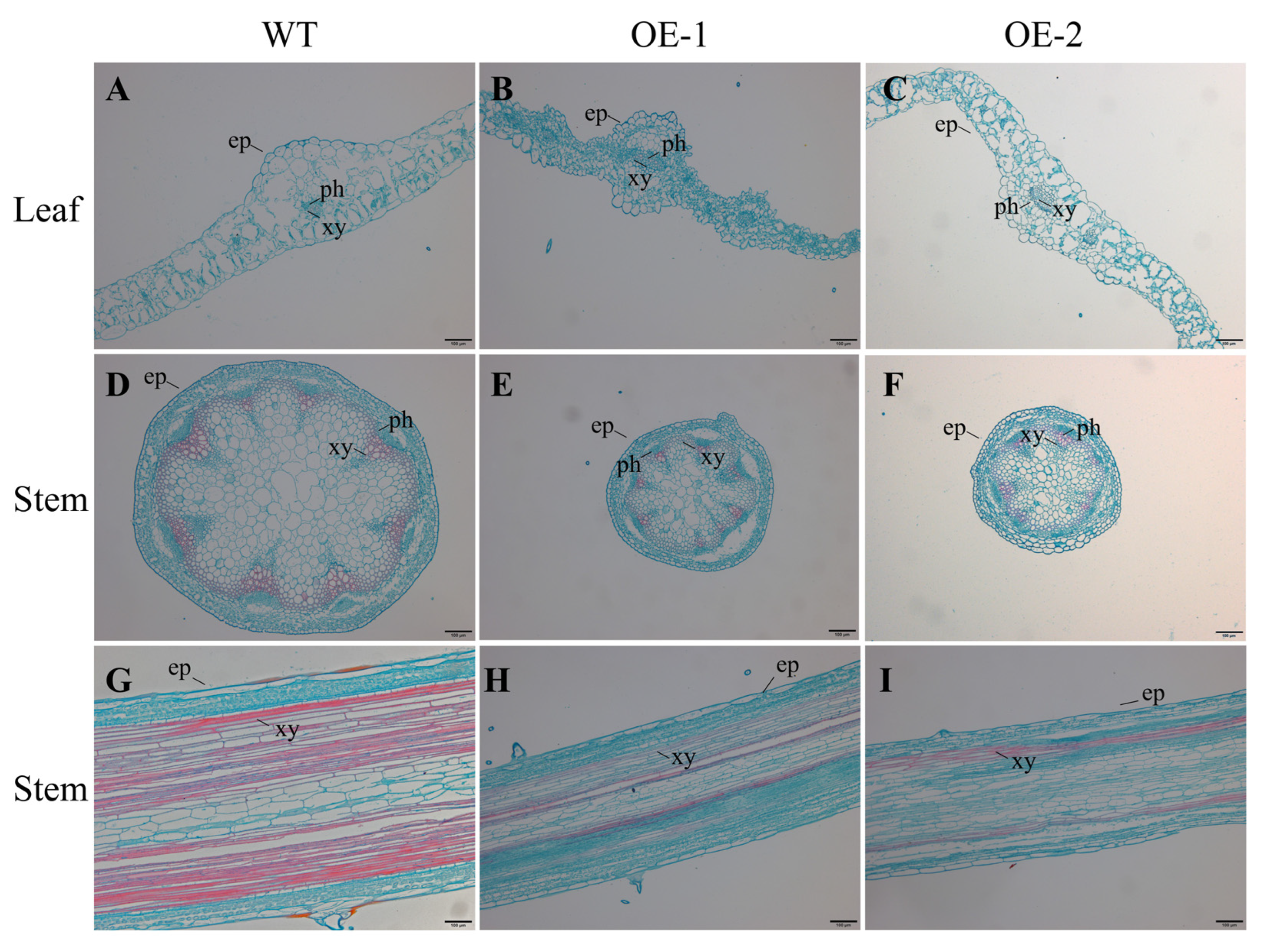
2.9. Cytological Observation on Leaves and Stems of Transgenic Arabidopsis
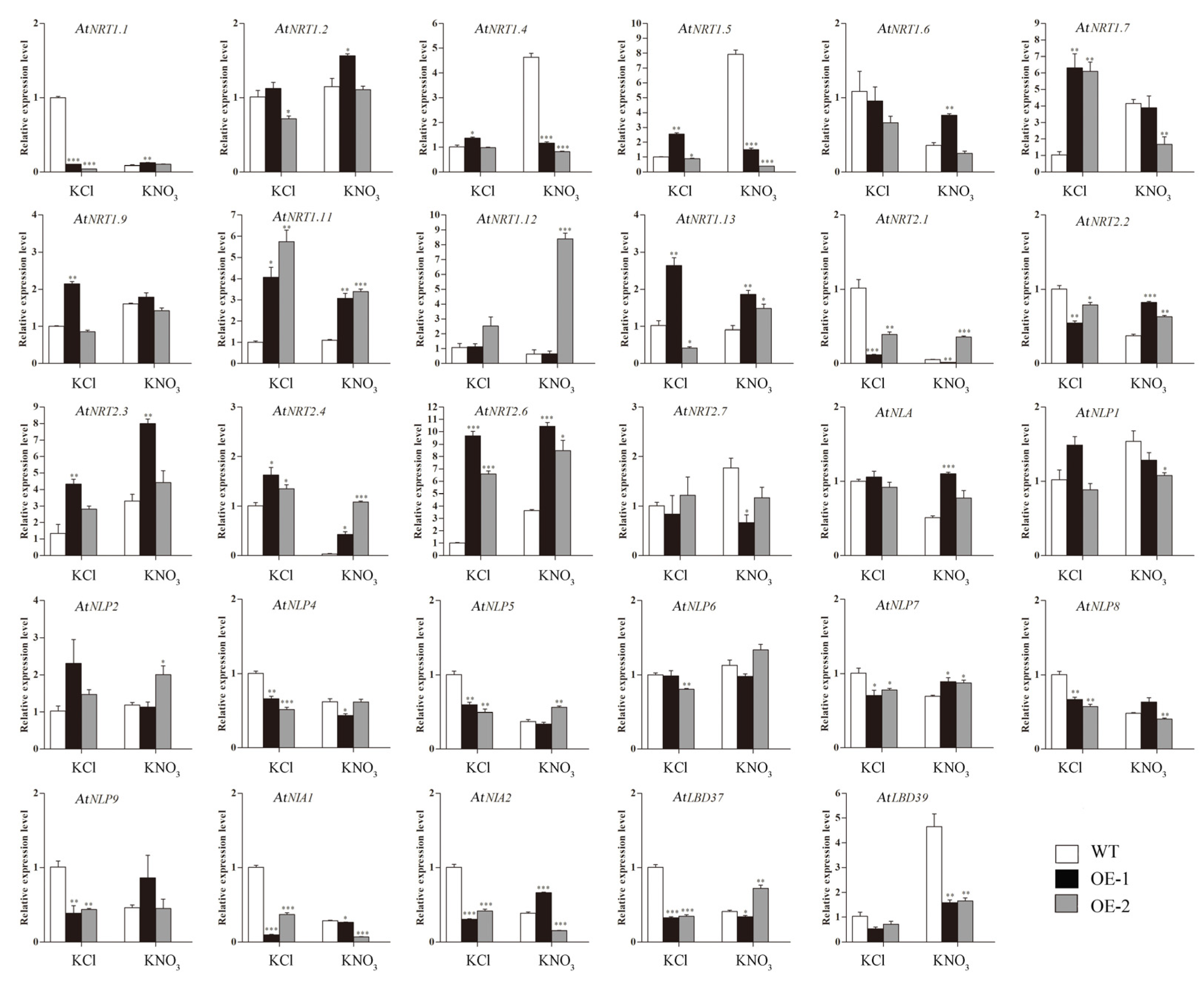
2.10. The Expression Analysis of Nitrate Uptake and Transport-Related Genes in Transgenic Arabidopsis Plants Overexpressing CsLBD39 under Nitrate Treatment
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions
4.2. RNA Extraction and cDNA Synthesis
4.3. Isolation and Bioinformatics Analysis of CsLBD39
4.4. Subcellular Localization of CsLBD39
4.5. Transcriptional Activation Activity Analysis of CsLBD39
4.6. Overexpression Plasmid Construction and Transformation
4.7. Nitrate Treatment Conditions in Arabidopsis
4.8. Measurement of the Nitrate Content
4.9. Determination of Chlorophyll
4.10. Determination of Anthocyanins
4.11. Histochemical Staining
4.12. Gene Expression Analysis
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, N.; Wang, R.; Zhao, L.; Zhang, C.; Wang, Y. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 2016, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Li, H.; Liu, J.X.; Wang, Y.; Zhuang, J. Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis). Hortic. Res. 2020, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar] [PubMed]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Walch-Liu, P.; Forde, B.G. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J. 2008, 54, 820–828. [Google Scholar] [CrossRef]
- Stitt, M. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 1999, 2, 178–186. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Bi, Y.M.; Wang, R.L.; Zhu, T.; Rothstein, S.J. Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genom. 2007, 8, 281. [Google Scholar] [CrossRef]
- Wang, R.; Xing, X.; Crawford, N. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 2007, 145, 1735–1745. [Google Scholar] [CrossRef]
- Yu, L.H.; Wu, J.; Tang, H.; Yuan, Y.; Wang, S.M.; Wang, Y.P.; Zhu, Q.S.; Li, S.G.; Xiang, C.B. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 2016, 6, 27795. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef] [PubMed]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.M.S.; Springer, P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef]
- Li, H.H.; Liu, X.; An, J.P.; Hao, Y.J.; Wang, X.F.; You, C.X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Org. 2017, 130, 47–59. [Google Scholar] [CrossRef]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.J.; Kim, N.Y.; Lee, S.H.; Kim, J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013, 73, 212–224. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013, 54, 1600–1611. [Google Scholar] [CrossRef]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.W.; Wu, Z.J.; Wang, Y.X.; Teng, R.M.; Zhuang, J. Differentially expressed protein and gene analysis revealed the effects of temperature on changes in ascorbic acid metabolism in harvested tea leaves. Hortic. Res. 2018, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.X.; Liu, Z.W.; Wu, Z.J.; Li, H.; Wang, W.L.; Cui, X.; Zhuang, J. Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 2018, 8, 3949. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Li, X.H.; Ratcliffe, R.G.; Ruan, J.Y. Characterization of ammonium and nitrate uptake and assimilation in roots of tea plants. Russ. J. Plant Physiol. 2012, 60, 91–99. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, L.; Bai, P.; Wei, K.; Cheng, H. Identification of regulatory networks and hub genes controlling nitrogen uptake in tea plants [Camellia sinensis (L.) O. Kuntze. J. Agric. Food Chem. 2020, 68, 2445–2456. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Tsai, Y.S.; Lin, Y.S.; Chiu, T.Y.; Hsiung, C.C.; Lee, M.I.; Simpson, J.C.; Hsu, C.N. Boosting multiclass learning with repeating codes and weak detectors for protein subcellular localization. Bioinformatics 2007, 23, 3374–3381. [Google Scholar] [CrossRef][Green Version]
- Miller, A.J.; Cramer, M. Root nitrogen acquisition and assimilation. Plant Soil 2005, 274, 1–36. [Google Scholar] [CrossRef]
- Poitout, A.; Crabos, A.; Petřík, I.; Novák, O.; Krouk, G.; Lacombe, B.T.; Ruffel, S. Responses to systemic nitrogen signaling in Arabidopsis roots involve trans-zeatin in shoots. Plant Cell 2018, 30, 1243–1257. [Google Scholar] [CrossRef]
- Hirel, B.; Bertin, P.; Quillere, I.; Bourdoncle, W.; Attagnant, C.; Dellay, C.; Gouy, A.; Cadiou, S.; Retailliau, C.; Falque, M.; et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001, 125, 1258–1270. [Google Scholar] [CrossRef]
- Jukanti, A.K.; Fischer, A.M. A high-grain protein content locus on barley (Hordeum vulgare) chromosome 6 is associated with increased flag leaf proteolysis and nitrogen remobilization. Physiol Plantarum. 2008, 132, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sun, Q.; Wang, K.; Du, Q.; Li, W.X. Nitrogen limitation adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol. 2017, 214, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Chalfun-Junior, A.; Franken, J.; Mes, J.J.; Marsch-Martinez, N.; Pereira, A.; Angenent, G.C. ASYMMETRIC LEAVES2-LIKE1 gene a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 2005, 57, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef]
- Feng, Z.; Sun, X.; Wang, G.; Liu, H.; Zhu, J. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot. 2012, 110, 1–10. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Busov, V. Boundary genes in regulation and evolution of secondary growth. Plant Signal. Behav. 2011, 6, 688–690. [Google Scholar] [CrossRef][Green Version]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Stecher, G.; Peterson, N.; Kumar, S.; Nei, M. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Feng, K.; Duan, A.Q.; Li, H.; Yang, Q.Q.; Xu, Z.S.; Xiong, A.S. Isolation, purification and characterization of an ascorbate peroxidase from celery and overexpression of the AgAPX1 gene enhanced ascorbate content and drought tolerance in Arabidopsis. BMC Plant Biol. 2019, 19, 488. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, Y.; Khadr, A.; Wang, Y.-H.; Xu, Z.-S.; Xiong, A.-S. DcDREB1A, a DREB-binding transcription factor from Daucus carota, enhances drought tolerance in transgenic Arabidopsis thaliana and modulates lignin levels by regulating lignin-biosynthesis-related genes. Environ. Exp. Bot. 2020, 169, 103896. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum. 1962, 15, 473. [Google Scholar] [CrossRef]
- Yu, X.; Hu, S.; He, C.; Zhou, J.; Qu, F.; Ai, Z.; Chen, Y.; Ni, D. Chlorophyll metabolism in postharvest tea (Camellia sinensis L.) leaves: Variations in color values, chlorophyll derivatives, and gene expression levels under different withering treatments. J Agric. Food Chem. 2019, 67, 10624–10636. [Google Scholar] [CrossRef]
- Ren, Y.R.; Zhao, Q.; Yang, Y.Y.; Zhang, T.E.; Wang, X.F.; You, C.X.; Hao, Y.J. The apple 14-3-3 protein MdGRF11 interacts with the BTB protein MdBT2 to regulate nitrate deficiency-induced anthocyanin accumulation. Hortic. Res. 2021, 8, 22. [Google Scholar] [CrossRef]
- Han, M.H.; Yang, N.; Wan, Q.W.; Teng, R.M.; Duan, A.Q.; Wang, Y.H.; Zhuang, J. Exogenous melatonin positively regulates lignin biosynthesis in Camellia sinensis. Int. J. Biol. Macromol. 2021, 179, 485–499. [Google Scholar] [CrossRef]
- Liu, J.X.; Jiang, Q.; Tao, J.P.; Feng, K.; Li, T.; Duan, A.Q.; Wang, H.; Xu, Z.S.; Liu, H.; Xiong, A.S. Integrative genome, transcriptome, microRNA, and degradome analysis of water dropwort (Oenanthe javanica) in response to water stress. Hortic. Res. 2021, 8, 262. [Google Scholar] [CrossRef]
- Wu, Z.J.; Tian, C.; Jiang, Q.; Li, X.H.; Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 2016, 6, 19748. [Google Scholar] [CrossRef]
- Lu, K.; Li, T.; Jian, H.; Wei, C.; Rui, Z.; Liu, M.; Yu, M.; Fan, Y.; Ma, J.; Wei, S. qPrimerDB: A thermodynamics-based gene-specific qPCR primer database for 147 organisms. Nucleic Acids Res. 2018, 46, D1229–D1236. [Google Scholar] [CrossRef] [PubMed]
- Menz, J.; Li, Z.; Schulze, W.X.; Ludewig, U. Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 2016, 88, 717–734. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, R.-M.; Yang, N.; Li, J.-W.; Liu, C.-F.; Chen, Y.; Li, T.; Wang, Y.-H.; Xiong, A.-S.; Zhuang, J. Isolation and Characterization of an LBD Transcription Factor CsLBD39 from Tea Plant (Camellia sinensis) and Its Roles in Modulating Nitrate Content by Regulating Nitrate-Metabolism-Related Genes. Int. J. Mol. Sci. 2022, 23, 9294. https://doi.org/10.3390/ijms23169294
Teng R-M, Yang N, Li J-W, Liu C-F, Chen Y, Li T, Wang Y-H, Xiong A-S, Zhuang J. Isolation and Characterization of an LBD Transcription Factor CsLBD39 from Tea Plant (Camellia sinensis) and Its Roles in Modulating Nitrate Content by Regulating Nitrate-Metabolism-Related Genes. International Journal of Molecular Sciences. 2022; 23(16):9294. https://doi.org/10.3390/ijms23169294
Chicago/Turabian StyleTeng, Rui-Min, Ni Yang, Jing-Wen Li, Chun-Fang Liu, Yi Chen, Tong Li, Ya-Hui Wang, Ai-Sheng Xiong, and Jing Zhuang. 2022. "Isolation and Characterization of an LBD Transcription Factor CsLBD39 from Tea Plant (Camellia sinensis) and Its Roles in Modulating Nitrate Content by Regulating Nitrate-Metabolism-Related Genes" International Journal of Molecular Sciences 23, no. 16: 9294. https://doi.org/10.3390/ijms23169294
APA StyleTeng, R.-M., Yang, N., Li, J.-W., Liu, C.-F., Chen, Y., Li, T., Wang, Y.-H., Xiong, A.-S., & Zhuang, J. (2022). Isolation and Characterization of an LBD Transcription Factor CsLBD39 from Tea Plant (Camellia sinensis) and Its Roles in Modulating Nitrate Content by Regulating Nitrate-Metabolism-Related Genes. International Journal of Molecular Sciences, 23(16), 9294. https://doi.org/10.3390/ijms23169294







