Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability
Abstract
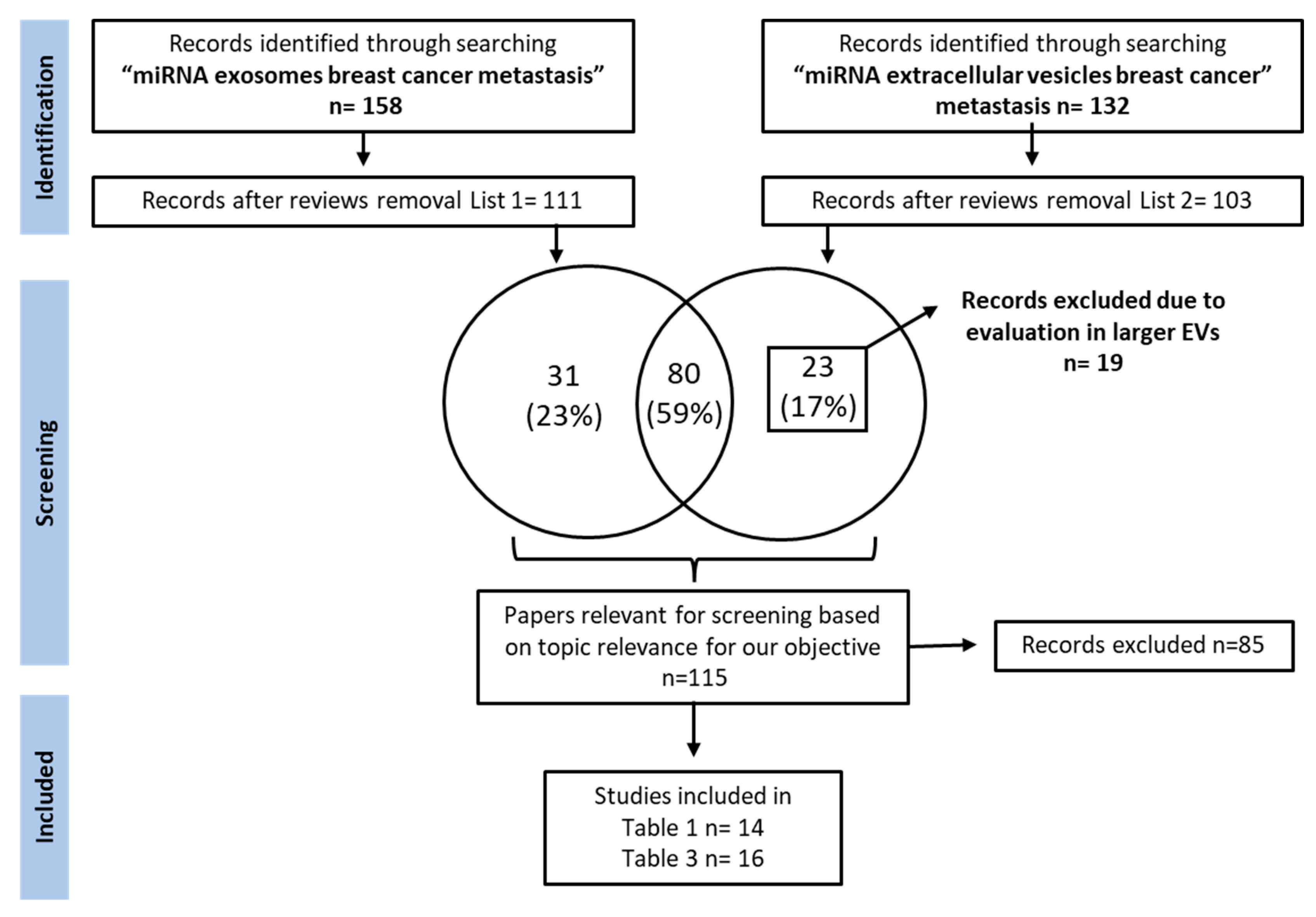
:1. Introduction
2. Clinically Validated Exosomal miRNAs as Biomarkers for Breast Cancer Metastasis by Site
2.1. Exosomal miRNAs Associated with Lymph Node Metastasis
2.2. MiRNAs Associated with Bone Metastasis
2.3. MiRNAs Associated with Brain Metastasis
2.4. MiRNAs Associated with Distant Metastasis, without Organ Specificity
| Nr crt | miR | Function | Metastatic Site | Number of Cases | Clinical Significance | Biological Function | Target Gene | Refs. |
|---|---|---|---|---|---|---|---|---|
| 1 | miR-363-5p | Tumor-suppressor miR | Lymph node | 10 BC (6 LNM+, 4 LNM−) and 10 healthy controls | Significantly associated with breast cancer (p = 0.047) and axillary lymph node metastasis (p = 0.019). High LMN detection power with an AUC of 0.958 in patients’ samples and an AUC of 0.733 in TCGA dataset. OS prediction (HR = 0.63, 95% CI 0.45–0.89; p = 0.0075, log-rank test). | Suppresses migration, invasion, proliferation, and colony formation in vitro. | PDGFB | [41] |
| 2 | miR-370-3p | Oncomir | Lymph node | 28 BC and 28 healthy controls | Serum exosome overexpression was associated with larger tumors (p = 0.042), advanced TNM stage (p = 0.0273), and lymph node metastasis (p = 0.0193). | Promotes in vitro proliferation and migration and in vivo tumorigenesis. | FBLN5 and NF-kB signaling | [42] |
| 3 | miR-222 | Oncomir | Lymph node | 38 BC (19 LNM+, 19 LNM−) and 19 healthy controls | Higher expression significantly associated with breast cancer and lymph node metastasis | Promotes in vitro proliferation, migration, and invasion. | PDLIM2 and NF-kB signaling | [43] |
| 4 | miR-148a | Tumor-suppressor miR | Lymph node | 125 BC, 50 benign tumors, and 40 healthy controls | Low expression correlated with lymph node metastases (p = 0.0011), poor tumor differentiation (p = 0.0167), and advanced TNM stage (p = 0.0004). Breast cancer diagnosis biomarker AUC = 0.897 (95% confidence interval = 0.840–0.939, specificity = 80.0%, sensitivity = 84.0%). Independent prognostic factor for breast cancer (HR = 2.460, 95% CI = 1.165–3.620, p = 0.015). Higher expression associated with a longer 5-year OS (p = 0.0232) and DFS (p = 0.0103). | Suppresses in vitro cancer cell migration and invasion and lung metastasis in vivo. | WNT-1 | [43,44] |
| 5 | miR-3662, miR-146a, miR-1290 | Oncomir | Lymph node | 60 BC, 20 healthy controls | Higher expression correlated with lymph node metastasis and later disease stages (II/III). | Promotes proliferation, migration, colony formation in vitro. Sustains tumor growth and metastasis in vivo. | NAT1, TXNIP, HBP1 | [46,47,48] |
| 6 | miR-188-5p | Tumor-suppressor miR | Lymph node | 45 BC, 40 breast fibroadenoma, 50 healthy controls | Exosomal miR-188 downregulated in breast cancer patients. High levels of free-circulating serum miR-188-5p associated with advanced TNM stages and lymph node metastasis. | Inhibits the migration, invasion, and colony formation in vitro. | IL6ST | [51] |
| 7 | miR-21 | Oncomir | Bone metastasis | 51 BC (21 bone metasasis, 21 localized disease, 9 other metastatic sites) | Higher expression correlated with bone metastasis in breast cancer patients. | Promotes osteoclast activity in vitro and the formation of the bone pre-metastatic niche in vivo. | PDCD4 | [56] |
| 8 | miR-218 | Oncomir | Bone metastasis | 47 BC (33 bone metastasis, 14 other metastatic sites) | Higher expression correlated with bone metastasis in breast cancer patients. | Inhibits the deposition of collagen in osteoblasts in vitro and promotes the formation of the bone pre-metastatic niche in vivo. | COL1A1,YY1, INHBB | [57] |
| 9 | mir-576-3p and miR-130a | Tumor-suppressor miR | Brain | 65 BC (15 primary cancer, 18 visceral metastasis, 16 bone metastasis, 16 liver metastasis) and 18 healthy controls | Cerebral metastasis biomarker AUC: 0.699 (p = 0.012, SD: 0.060, 95% CI 0.582–0.816). | Not evaluated in this study. | - | [58] |
| 10 | miR-130a-3p | Tumor-suppressor miR | Lymph node | 40 BC and 40 healthy controls | Lower expression was associated with advanced TNM stage (p = 0.0014) and lymph node metastasis (p = 0.0019). | Inhibits proliferation, migration, and invasion in vitro. | RAB5 | [59] |
| 11 | miR-181c | Oncomir | Brain | 56 BC | Higher serum and serum exosome expression was associated with brain metastasis (p < 0.05). | Promotes blood–brain barrier destruction in vitro and metastatic niche formation in brain in vivo. | PDPK1 | [61] |
| 12 | miR-105 | Oncomir | Not specific | Higher exosome and tissue expression associated with distant metastasis. | Promotes endothelial barrier destruction, trans-endothelial migration, and invasion of cancer cells in vitro and in vivo. | ZO-1 | [62] | |
| 13 | miR-200c, miR-141 | Oncomir | Not specific | 114 BC, 30 benign tumors, 94 healthy controls | Higher plasma expression associated with breast cancer metastasis. AUC: 0.770 for miR-200c and AUC: 0.678 for miR-141. | Promotes tumor metastasis in vivo. | - | [64] |
| 14 | miR-7641 | Oncomir | Not specific | 28 BC (13 metastatic, 15 localized disease) | Higher expression associated with distant metastasis. | Promotes cancer cell proliferation and invasion in vitro and tumor formation in vivo. | - | [65] |
3. Technical Aspects of Biomarker Discovery for the Clinic
3.1. Sample Processing
| Nr crt | Exozomal miRNA | Ser/Plasma | Blood Volume | Serum/Plasma Filtration | Exosomes Isolation/Kit | Exosomes Characterization | RNA Extraction from Exosomes | miRNA System | Normalizer | Year | Refs. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | miR-363-5p | plasma | 5 mL | 0.22 μm filter | Ultracentrifugation at 100,000× g | TEM, NTA, and WB (CD63, TSG101, and calnexin) | miRNeasy Mini kit | miScript SYBR Green | U6 | 2021 | [41] |
| 2 | miR-370-3p | serum | 250 µL serum | No | Evs precipitation (ExoQuick precipitation solution) | not specified | QIAzol LS | miScript SYBR Green | U6 | 2021 | [42] |
| 3 | miR-222 | plasma | not specified | 0.22 μm filter | Evs precipitation (Exoquick Exosome Isolation Kit) | TEM and WB (CD63, TSG101) | Exoquick Exosome Isolation Kit | SYBR Premix ExTaq reagent | U6 | 2018 | [43] |
| 4 | miR-148a | serum | not specified | No | Evs precipitation (Exosome Precipitation Solution) | not specified | miRNeasy Mini kit | miScript SYBR Green PCR Kit | cel-miR-39 | 2020 | [44] |
| 5 | miR-3662, miR-146a, and miR-1290 | serum | 2 mL | No | Evs precipitation (Exosome Isolation Reagent). | TEM, NTA, and WB (TSG101 and CD63) | HiPure Serum miRNA Kit | miDETECT A Track miRNA qRT-PCR Starter Kit | Not specified | 2021 | [47] |
| 6 | miR-188-5p | serum | not specified | 0.22 μm filter | Evs precipitation (ExoQuick exosome precipitation solution) | NTA and WB (CD9 and CD63) | miRNeasy Serum/Plasma Kit | miScript SYBR Green | cel-miR-39 | 2019 | [51] |
| 7 | miR-21 | serum | 1 mL | No | Ultracentrifugation at 120,000× g | TEM, NTA, and WB (TSG101. Alix, and HSP70) | exoRNeasy Serum/Plasma Midi Kit | SYBR Premix Ex Taq reagernt | cel-miR-39 | 2021 | [56] |
| 8 | miR-218 | serum | 500 µL of serum | No | Ultracentrifugation at 110,000× g | NTA | Trizol | miScript SYBR Green | miR-140-3p | 2018 | [57] |
| 9 | mir-576-3p, miR-130a-3p | serum | 0.5–1 mL | No | Evs precipitation (Total Exosome Isolation reagent from serum) | ELISA (CD63 and CD9) | Total Exosome RNA and Protein Isolation Kit | TaqMan miRNA Advanced | miR-320 | 2022 | [58] |
| 10 | miR-130a-3p | circulating blood | not specified | No | Column-based isolation (ExoQuickExosomal Extraction Kit) | not specified | Trizol | TaqMan MicroRNA Assay kits | U6 | 2018 | [59] |
| 11 | miR-181c | serum | not specified | 0.22 μm filter | Evs precipitation (Total Exosome Isolation (from serum)) | WB (CD9, CD63, cytochrome C) | RNeasy Mini Kit | TaqMan Micro-RNA Assays | U6 | 2015 | [61] |
| 12 | miR-105 | serum | not specified | No | Ultracentrifugation at 110,000× g | TEM | TRIZOL LS for serum exosomes | miScript SYBR Green | miR-16 | 2014 | [62] |
| 13 | miR-200c and miR-141 | plasma | 200 µL | No | Ultracentrifugation at 100,000× g | not specified/not performed | miRNeasy Serum/Plasma Kits | TaqMan Micro-RNA Assays | cel-miR-39 | 2017 | [64] |
| 14 | miR-7641 | plasma | not specified | 0.22 μm filter | Ultracentrifugation at 100,000× g | TEM and WB (CD9 and CD63) | miRNeasy Serum/Plasma Kit | miScript SYBR Green | U6 and cel-miR-39 | 2021 | [65] |
3.2. Analytical Platforms
3.3. Data Normalization
4. Perspectives of Exosomal miRNAs from Preclinical Models
| Nr crt | Exozomal miRNA | Regulation | Biological Function in Metastasis | Target | Evaluation | Refs. |
|---|---|---|---|---|---|---|
| 1 | miR-1910-3p | oncomir | Migration and invasion | MTMR3 | in vitro in vivo | [79] |
| 2 | miR-370 | oncomir | Migration and invasion | - | in vitro | [76] |
| 3 | miR-4443 | oncomir | Migration and invasion | TIMP2 | in vitro in vivo | [77] |
| 4 | miR-760 | oncomir | Migration and invasion | ARF6 | in vitro | [78] |
| 5 | miR-9, miR-155 | oncomirs | Migration and invasion | PTEN, DUSP | in vitro | [80] |
| 6 | let-7a | tumor suppressor | Migration and invasion | c-Myc | in vitro in vivo patients tissue | [88] |
| 7 | miR-182-5p | oncomir | Angiogenesis | CMTM7 | in vitro in vivo patients tissue | [82] |
| 8 | miR-210 | oncomir | Angiogenesis | Ephrin-A3 | in vitro in vivo | [81] |
| 9 | miR-939 | oncomir | Extravasation | VE-cadherin | in vitro in vivo patients tissue | [83] |
| 10 | mir-155-5p, miR-205-5p | miR-205 tumor suppressor; miR-155 oncomiR | Immune response | IL-6, IL-17 | in vitro in vivo | [86] |
| 11 | miR-138-5p | oncomir | Immune response | KDM6B | in vitro in vivo patients plasma | [84] |
| 12 | miR-183-5p | oncomir | Immune response | PPP2CA | in vitro in vivo | [85] |
| 13 | let-7a | tumor suppressor | Immune response | - | in vitro in vivo patients tissue | [87] |
| 14 | miR-122 | oncomir | Pre-metastatic niche formation | PKM, GLUT1 | in vitro in vivo | [89] |
| 15 | mir-19a | oncomir | Pre-metastatic niche formation | PTEN | in vitro in vivo patients tissue and plasma | [90] |
| 16 | miR-20a-5p | oncomir | Pre-metastatic niche formation | SRCIN1 | in vitro | [91] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Rivenbark, A.G.; O’Connor, S.M.; Coleman, W.B. Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine. Am. J. Pathol. 2013, 183, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef] [PubMed]
- Gennari, A.; André, F.; Barrios, C.H.; Cortés, J.; de Azambuja, E.; DeMichele, A.; Dent, R.; Fenlon, D.; Gligorov, J.; Hurvitz, S.A. ESMO Clinical Practice Guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 1475–1495. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.; Wang, X. The Metabolic Mechanisms of Breast Cancer Metastasis. Front. Oncol. 2020, 10, 602416. [Google Scholar] [CrossRef]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Seltzer, S.; Corrigan, M.; O’Reilly, S. The clinicomolecular landscape of de novo versus relapsed stage IV metastatic breast cancer. Exp. Mol. Pathol. 2020, 114, 104404. [Google Scholar] [CrossRef]
- Heller, D.R.; Chiu, A.S.; Farrell, K.; Killelea, B.K.; Lannin, D.R. Why Has Breast Cancer Screening Failed to Decrease the Incidence of de Novo Stage IV Disease? Cancers 2019, 11, E500. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Castro, A.C.; Spurr, L.F.; Hughes, M.E.; Li, Y.Y.; Cherniack, A.D.; Kumari, P.; Lloyd, M.R.; Bychkovsky, B.; Barroso-Sousa, R.; Di Lascio, S. Genomic Characterization of de novo Metastatic Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Lobbezoo, D.J.A.; van Kampen, R.J.W.; Voogd, A.C.; Dercksen, M.W.; van den Berkmortel, F.; Smilde, T.J.; Van De Wouw, A.J.; Peters, F.P.J.; Van Riel, J.M.G.H.; Peters, N.A.J.B. Prognosis of metastatic breast cancer: Are there differences between patients with de novo and recurrent metastatic breast cancer? Br. J. Cancer 2015, 112, 1445–1451. [Google Scholar] [CrossRef]
- Lu, W.L.; Jansen, L.; Post, W.J.; Bonnema, J.; Van de Velde, J.C.; De Bock, G.H. Impact on survival of early detection of isolated breast recurrences after the primary treatment for breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2009, 114, 403–412. [Google Scholar] [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Prat, A.; Ellis, M.J.; Perou, C.M. Practical implications of gene-expression-based assays for breast oncologists. Nat. Rev. Clin. Oncol. 2011, 9, 48–57. [Google Scholar] [CrossRef]
- Zeng, C.; Zhang, J. A narrative review of five multigenetic assays in breast cancer. Transl. Cancer Res. 2022, 11, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Venesio, T.; Siravegna, G.; Bardelli, A.; Sapino, A. Liquid Biopsies for Monitoring Temporal Genomic Heterogeneity in Breast and Colon Cancers. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2018, 85, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Fiala, C.; Diamandis, E.P. Circulating tumor DNA (ctDNA) is not a good proxy for liquid biopsies of tumor tissues for early detection. Clin. Chem. Lab. Med. 2020, 58, 1651–1653. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Takatani-Nakase, T. Exosomes: Breast cancer-derived extracellular vesicles; recent key findings and technologies in disease progression, diagnostics, and cancer targeting. Drug Metab. Pharmacokinet. 2022, 42, 100435. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef]
- Tan, Y.; Luo, X.; Lv, W.; Hu, W.; Zhao, C.; Xiong, M.; Yi, Y.; Wang, D.; Wang, Y.; Wang, H. Tumor-derived exosomal components: The multifaceted roles and mechanisms in breast cancer metastasis. Cell Death Dis. 2021, 12, 547. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4. [Google Scholar] [CrossRef]
- Wu, S.-G.; Li, H.; Tang, L.-Y.; Sun, J.-Y.; Zhang, W.-W.; Li, F.-Y.; Chen, Y.X.; He, Z.Y. The effect of distant metastases sites on survival in de novo stage-IV breast cancer: A SEER database analysis. Tumor Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317705082. [Google Scholar] [CrossRef]
- Kast, K.; Link, T.; Friedrich, K.; Petzold, A.; Niedostatek, A.; Schoffer, O.; Werner, C.; Klug, S.J.; Werner, A.; Gatzweiler, A. Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res. Treat. 2015, 150, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-Y.; Gong, Y.; Ye, F.-G.; Ling, H.; Hu, X. Incidence and prognostic factors of patients with synchronous liver metastases upon initial diagnosis of breast cancer: A population-based study. Cancer Manag. Res. 2018, 10, 5937–5950. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, E.M.; Anders, C.K. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann. Transl. Med. 2018, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Han, B.; Siegel, E.; Cui, Y.; Giuliano, A.; Cui, X. Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol. Ther. 2018, 19, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Naxerova, K. Defining the role of lymph node metastasis in systemic breast cancer evolution. EBioMedicine 2020, 57, 102852. [Google Scholar] [CrossRef]
- Zhao, Y.-X.; Liu, Y.-R.; Xie, S.; Jiang, Y.-Z.; Shao, Z.-M. A Nomogram Predicting Lymph Node Metastasis in T1 Breast Cancer based on the Surveillance, Epidemiology, and End Results Program. J. Cancer 2019, 10, 2443–2449. [Google Scholar] [CrossRef]
- Abolghasemi, M.; Tehrani, S.S.; Yousefi, T.; Karimian, A.; Mahmoodpoor, A.; Ghamari, A.; Jadidi-Niaragh, F.; Yousefi, M.; Kafil, H.S.; Bastami, M. MicroRNAs in breast cancer: Roles, functions, and mechanism of actions. J. Cell. Physiol. 2020, 235, 5008–5029. [Google Scholar] [CrossRef]
- Gomarasca, M.; Maroni, P.; Banfi, G.; Lombardi, G. microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics. Int. J. Mol. Sci. 2020, 21, E2805. [Google Scholar] [CrossRef]
- Di Agostino, S.; Vahabi, M.; Turco, C.; Fontemaggi, G. Secreted Non-Coding RNAs: Functional Impact on the Tumor Microenvironment and Clinical Relevance in Triple-Negative Breast Cancer. Non-Coding RNA 2022, 8, 5. [Google Scholar] [CrossRef]
- Carter, C.L.; Allen, C.; Henson, D.E. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989, 63, 181–187. [Google Scholar] [CrossRef]
- Wang, X.; Qian, T.; Bao, S.; Zhao, H.; Chen, H.; Xing, Z.; Li, Y.; Zhang, M.; Meng, X.; Wang, C. Circulating exosomal miR-363-5p inhibits lymph node metastasis by downregulating PDGFB and serves as a potential noninvasive biomarker for breast cancer. Mol. Oncol. 2021, 15, 2466–2479. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Wang, L.; Wu, J.; Wang, Y.; Wen, H.; Zhu, X.; Wang, B.; Yang, H. miR-370-3p as a Novel Biomarker Promotes Breast Cancer Progression by Targeting FBLN5. Stem Cells Int. 2021, 2021, 4649890. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Xu, Z.; Zhang, Y.; Tan, C.; Hu, W.; Wang, M.; Xu, Y.; Tang, J. Exosome-mediated miR-222 transferring: An insight into NF-κB-mediated breast cancer metastasis. Exp. Cell Res. 2018, 369, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, J.; Ma, L.-J.; Yang, H.-B.; Jing, J.-F.; Jia, M.-M.; Zhang, X.J.; Guo, F.; Gao, J.N. Identification of serum exosomal miR-148a as a novel prognostic biomarker for breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7303–7309. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Jasper, J.; Lykken, E.; Alexander, P.B.; Markowitz, G.J.; McDonnell, D.P.; Li, Q.J.; Wang, X.F. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget 2016, 7, 20381–20394. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.; Ma, M.-T.; Wu, H.-Z.; Yu, Z.-J.; Guan, S.; Jiang, L.Y.; Wang, Y.; Zheng, D.D.; Jin, F. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol. Rep. 2016, 35, 1425–1432. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Xu, F.; Wang, Y.; Leng, D. Detection significance of miR-3662, miR-146a, and miR-1290 in serum exosomes of breast cancer patients. J. Cancer Res. Ther. 2021, 17, 749–755. [Google Scholar] [CrossRef]
- Endo, Y.; Yamashita, H.; Takahashi, S.; Sato, S.; Yoshimoto, N.; Asano, T.; Hato, Y.; Dong, Y.; Fujii, Y.; Toyama, T. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer 2014, 14, 990. [Google Scholar] [CrossRef]
- Yang, S.-S.; Ma, S.; Dou, H.; Liu, F.; Zhang, S.-Y.; Jiang, C.; Xiao, M.; Huang, Y.X. Breast cancer-derived exosomes regulate cell invasion and metastasis in breast cancer via miR-146a to activate cancer associated fibroblasts in tumor microenvironment. Exp. Cell Res. 2020, 391, 111983. [Google Scholar] [CrossRef]
- Yi, B.; Wang, S.; Wang, X.; Liu, Z.; Zhang, C.; Li, M.; Gao, S.; Wei, S.; Bae, S.; Stringer-Reasor, E. CRISPR interference and activation of the microRNA-3662-HBP1 axis control progression of triple-negative breast cancer. Oncogene 2022, 41, 268–279. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, H.; Yang, F.; Qiu, R.; Zhao, X.; Gong, Z.; Yu, W.; Zhou, B.; Shen, B.; Zhu, W. miR-188-5p suppresses cellular proliferation and migration via IL6ST: A potential noninvasive diagnostic biomarker for breast cancer. J. Cell. Physiol. 2020, 235, 4890–4901. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-H.; Tsao, C.-J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Mirza, M.; Nandi, K.; Jain, S.K.; Kaza, R.C.M.; Khurana, N.; Ray, P.C.; Saxena, A. Serum microRNA-21 expression as a prognostic and therapeutic biomarker for breast cancer patients. Tumor Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 15275–15282. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, A.; de Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. BCR 2019, 21, 21. [Google Scholar] [CrossRef]
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445. [Google Scholar] [CrossRef]
- Liu, X.; Cao, M.; Palomares, M.; Wu, X.; Li, A.; Yan, W.; Fong, M.Y.; Chan, W.C.; Wang, S.E. Metastatic breast cancer cells overexpress and secrete miR-218 to regulate type I collagen deposition by osteoblasts. Breast Cancer Res. BCR 2018, 20, 127. [Google Scholar] [CrossRef]
- Curtaz, C.J.; Reifschläger, L.; Strähle, L.; Feldheim, J.; Feldheim, J.J.; Schmitt, C.; Kiesel, M.; Herbert, S.L.; Wöckel, A.; Meybohm, P. Analysis of microRNAs in Exosomes of Breast Cancer Patients in Search of Molecular Prognostic Factors in Brain Metastases. Int. J. Mol. Sci. 2022, 23, 3683. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, J.; Li, J.; Shao, J.; Fang, L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem. Biophys. Res. Commun. 2018, 501, 486–493. [Google Scholar] [CrossRef]
- Yang, P.-S.; Yin, P.-H.; Tseng, L.-M.; Yang, C.-H.; Hsu, C.-Y.; Lee, M.-Y.; Horng, C.F.; Chi, C.W. Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 2011, 102, 2172–2178. [Google Scholar] [CrossRef]
- Tominaga, N.; Kosaka, N.; Ono, M.; Katsuda, T.; Yoshioka, Y.; Tamura, K.; Lötvall, J.; Nakagama, H.; Ochiya, T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat. Commun. 2015, 6, 6716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Han, J.; Li, X.; Huang, T.; Gao, Y.; Wang, B.; Zhang, K.; Wang, S.; Zhang, W.; Li, W. FOXP3 Inhibits the Metastasis of Breast Cancer by Downregulating the Expression of MTA1. Front. Oncol. 2021, 11, 656190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, W.; Li, B.; Stringer-Reasor, E.; Chu, C.; Sun, L.; Bae, S.; Chen, D.; Wei, S.; Jiao, K. MicroRNA-200c and microRNA- 141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. BCR 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Song, Y.; Zhao, B.; Xu, Y.; Ren, X.; Zhou, Y.; Sun, Q. Cancer-derived exosomal miR-7641 promotes breast cancer progression and metastasis. Cell Commun. Signal. CCS 2021, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Preethi, K.A.; Selvakumar, S.C.; Ross, K.; Jayaraman, S.; Tusubira, D.; Sekar, D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol. Cancer 2022, 21, 54. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Song, E.J. Considerations and Suggestions for the Reliable Analysis of miRNA in Plasma Using qRT-PCR. Genes 2022, 13, 328. [Google Scholar] [CrossRef] [PubMed]
- Chugh, P.; Dittmer, D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 2012, 3, 601–616. [Google Scholar] [CrossRef] [PubMed]
- D’haene, B.; Mestdagh, P.; Hellemans, J.; Vandesompele, J. miRNA expression profiling: From reference genes to global mean normalization. In Next-Generation MicroRNA Expression Profiling Technology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 822, pp. 261–272. [Google Scholar] [CrossRef]
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of circulating miRNAs in plasma: Effect of preanalytical and analytical parameters on their isolation and stability. J. Mol. Diagn. JMD 2013, 15, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.; Jiang, J.; Yang, Y.; Wang, W.; Jia, Z. CircRNA circFOXK2 facilitates oncogenesis in breast cancer via IGF2BP3/miR-370 axis. Aging 2021, 13, 18978–18992. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Q.; Wang, D.; Yang, S.; Zhou, S.; Xu, H.; Zhang, H.; Zhong, S.; Feng, J. Microenvironment-induced TIMP2 loss by cancer-secreted exosomal miR-4443 promotes liver metastasis of breast cancer. J. Cell. Physiol. 2020, 235, 5722–5735. [Google Scholar] [CrossRef]
- Huang, X.; Lai, S.; Qu, F.; Li, Z.; Fu, X.; Li, Q.; Zhong, X.; Wang, C.; Li, H. CCL18 promotes breast cancer progression by exosomal miR-760 activation of ARF6/Src/PI3K/Akt pathway. Mol. Ther. Oncolytics 2022, 25, 1–15. [Google Scholar] [CrossRef]
- Wang, B.; Mao, J.-H.; Wang, B.-Y.; Wang, L.-X.; Wen, H.-Y.; Xu, L.-J.; Fu, J.X.; Yang, H. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020, 489, 87–99. [Google Scholar] [CrossRef]
- Kia, V.; Mortazavi, Y.; Paryan, M.; Biglari, A.; Mohammadi-Yeganeh, S. Exosomal miRNAs from highly metastatic cells can induce metastasis in non-metastatic cells. Life Sci. 2019, 220, 162–168. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Lu, C.; Zhao, Y.; Wang, J.; Shi, W.; Dong, F.; Xin, Y.; Zhao, X.; Liu, C. Breast cancer cell-derived extracellular vesicles transfer miR-182-5p and promote breast carcinogenesis via the CMTM7/EGFR/AKT axis. Mol. Med. 2021, 27, 78. [Google Scholar] [CrossRef] [PubMed]
- Di Modica, M.; Regondi, V.; Sandri, M.; Iorio, M.V.; Zanetti, A.; Tagliabue, E.; Casalini, P.; Triulzi, T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017, 384, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Xun, J.; Du, L.; Gao, R.; Shen, L.; Wang, D.; Kang, L.; Zhang, Z.; Zhang, Y.; Yue, S.; Feng, S.; et al. Cancer-derived exosomal miR-138-5p modulates polarization of tumor-associated macrophages through inhibition of KDM6B. Theranostics 2021, 11, 6847–6859. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Duan, Z.; Zhang, C.; Wang, W.; He, H.; Liu, Y.; Wu, P.; Wang, S.; Song, M.; Chen, H. Mouse 4T1 Breast Cancer Cell-Derived Exosomes Induce Proinflammatory Cytokine Production in Macrophages via miR-183. J. Immunol. 2020, 205, 2916–2925. [Google Scholar] [CrossRef]
- Gorczynski, R.M.; Zhu, F.; Chen, Z.; Kos, O.; Khatri, I. A comparison of serum miRNAs influencing metastatic growth of EMT6 vs 4THM tumor cells in wild-type and CD200R1KO mice. Breast Cancer Res. Treat. 2017, 162, 255–266. [Google Scholar] [CrossRef]
- Qi, M.; Xia, Y.; Wu, Y.; Zhang, Z.; Wang, X.; Lu, L.; Dai, C.; Song, Y.; Xu, K.; Ji, W. Lin28B-high breast cancer cells promote immune suppression in the lung pre-metastatic niche via exosomes and support cancer progression. Nat. Commun. 2022, 13, 897. [Google Scholar] [CrossRef]
- Du, J.; Fan, J.-J.; Dong, C.; Li, H.-T.; Ma, B.-L. Inhibition effect of exosomes-mediated Let-7a on the development and metastasis of triple negative breast cancer by down-regulating the expression of c-Myc. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5301–5314. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Wu, K.; Feng, J.; Lyu, F.; Xing, F.; Sharma, S.; Liu, Y.; Wu, S.Y.; Zhao, D.; Tyagi, A.; Deshpande, R.P. Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nat. Commun. 2021, 12, 5196. [Google Scholar] [CrossRef]
- Guo, L.; Zhu, Y.; Li, L.; Zhou, S.; Yin, G.; Yu, G.; Cui, H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019, 8, 5687–5701. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldasici, O.; Pileczki, V.; Cruceriu, D.; Gavrilas, L.I.; Tudoran, O.; Balacescu, L.; Vlase, L.; Balacescu, O. Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. Int. J. Mol. Sci. 2022, 23, 9371. https://doi.org/10.3390/ijms23169371
Baldasici O, Pileczki V, Cruceriu D, Gavrilas LI, Tudoran O, Balacescu L, Vlase L, Balacescu O. Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. International Journal of Molecular Sciences. 2022; 23(16):9371. https://doi.org/10.3390/ijms23169371
Chicago/Turabian StyleBaldasici, Oana, Valentina Pileczki, Daniel Cruceriu, Laura Ioana Gavrilas, Oana Tudoran, Loredana Balacescu, Laurian Vlase, and Ovidiu Balacescu. 2022. "Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability" International Journal of Molecular Sciences 23, no. 16: 9371. https://doi.org/10.3390/ijms23169371
APA StyleBaldasici, O., Pileczki, V., Cruceriu, D., Gavrilas, L. I., Tudoran, O., Balacescu, L., Vlase, L., & Balacescu, O. (2022). Breast Cancer-Delivered Exosomal miRNA as Liquid Biopsy Biomarkers for Metastasis Prediction: A Focus on Translational Research with Clinical Applicability. International Journal of Molecular Sciences, 23(16), 9371. https://doi.org/10.3390/ijms23169371








