How Microbiota-Derived Metabolites Link the Gut to the Brain during Neuroinflammation
Abstract
1. Introduction
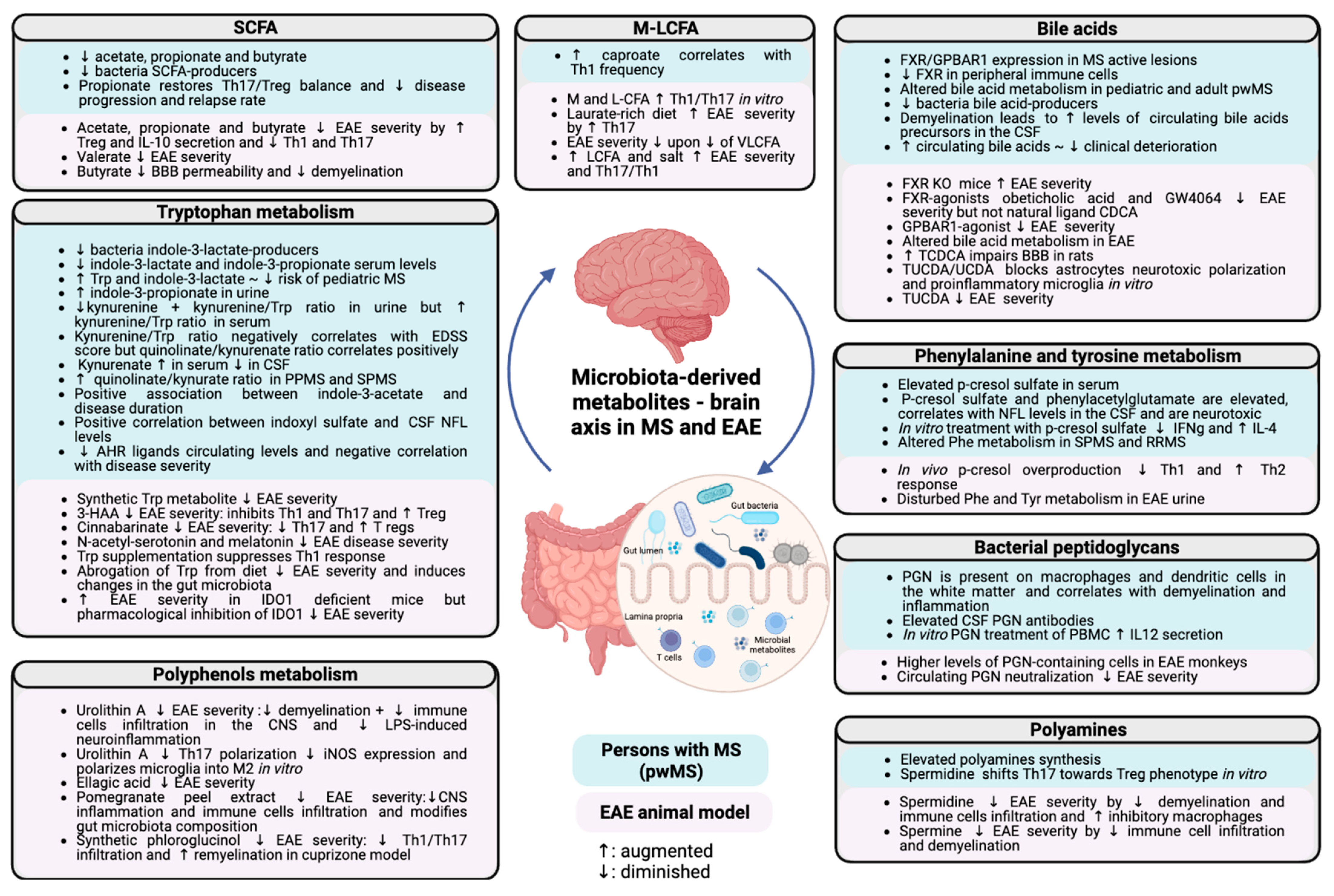
2. Lipid Metabolism
2.1. Short-Chain Fatty Acids
2.2. Bile Acids
3. Amino acid Metabolism
3.1. Tryptophan Metabolism
3.2. Phenylalanine and Tyrosine Metabolism
4. Trimethylamine N-Oxide (TMAO)
5. Polyphenols Metabolism
6. Polyamines
7. Bacterial Peptidoglycan
8. Non-Ribosomal Peptides and Polyketides
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feigin, V.L.; Nichols, E.; Alam, T.; Bannick, M.S.; Beghi, E.; Blake, N.; Culpepper, W.J.; Dorsey, E.R.; Elbaz, A.; Ellenbogen, R.G.; et al. Global, Regional, and National Burden of Neurological Disorders, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of Multiple Sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Nylander, A.; Hafler, D.A. Multiple Sclerosis. J. Clin. Investig. 2012, 122, 1180–1188. [Google Scholar] [CrossRef]
- van den Hoogen, W.J.; Laman, J.D.; ’t Hart, B.A. Modulation of Multiple Sclerosis and Its Animal Model Experimental Autoimmune Encephalomyelitis by Food and Gut Microbiota. Front. Immunol. 2017, 8, 1081. [Google Scholar] [CrossRef] [PubMed]
- Lublin, F.D.; Reingold, S.C. National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis* Defining the Clinical Course of Multiple Sclerosis: Results of an International Survey. Neurology 1996, 46, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Titus, H.E.; Chen, Y.; Podojil, J.R.; Robinson, A.P.; Balabanov, R.; Popko, B.; Miller, S.D. Pre-Clinical and Clinical Implications of “Inside-Out” vs. “Outside-In” Paradigms in Multiple Sclerosis Etiopathogenesis. Front. Cell. Neurosci. 2020, 14, 599717. [Google Scholar] [CrossRef]
- Ruiz, F.; Vigne, S.; Pot, C. Resolution of Inflammation during Multiple Sclerosis. Semin. Immunopathol. 2019, 41, 711–726. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two Types of Murine Helper T Cell Clone. I. Definition According to Profiles of Lymphokine Activities and Secreted Proteins. J. Immunol. 1986, 136, 2348. [Google Scholar]
- Atarashi, K.; Nishimura, J.; Shima, T.; Umesaki, Y.; Yamamoto, M.; Onoue, M.; Yagita, H.; Ishii, N.; Evans, R.; Honda, K.; et al. ATP Drives Lamina Propria TH17 Cell Differentiation. Nature 2008, 455, 808–812. [Google Scholar] [CrossRef]
- Miyake, S.; Kim, S.; Suda, W.; Oshima, K.; Nakamura, M.; Matsuoka, T.; Chihara, N.; Tomita, A.; Sato, W.; Kim, S.-W.; et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS ONE 2015, 10, e0137429. [Google Scholar] [CrossRef]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the Human Gut Microbiome in Multiple Sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chia, N.; Kalari, K.R.; Yao, J.Z.; Novotna, M.; Paz Soldan, M.M.; Luckey, D.H.; Marietta, E.V.; Jeraldo, P.R.; Chen, X.; et al. Multiple Sclerosis Patients Have a Distinct Gut Microbiota Compared to Healthy Controls. Sci. Rep. 2016, 6, 28484. [Google Scholar] [CrossRef] [PubMed]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.E.; et al. Gut Microbiota from Multiple Sclerosis Patients Enables Spontaneous Autoimmune Encephalomyelitis in Mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut Bacteria from Multiple Sclerosis Patients Modulate Human T Cells and Exacerbate Symptoms in Mouse Models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef]
- Correale, J.; Farez, M.F. The Impact of Parasite Infections on the Course of Multiple Sclerosis. J. Neuroimmunol. 2011, 233, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Cosorich, I.; Dalla-Costa, G.; Sorini, C.; Ferrarese, R.; Messina, M.J.; Dolpady, J.; Radice, E.; Mariani, A.; Testoni, P.A.; Canducci, F.; et al. High Frequency of Intestinal T H 17 Cells Correlates with Microbiota Alterations and Disease Activity in Multiple Sclerosis. Sci. Adv. 2017, 3, e1700492. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef]
- Takiishi, T.; Fenero, C.I.M.; Câmara, N.O.S. Intestinal Barrier and Gut Microbiota: Shaping Our Immune Responses throughout Life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef]
- Esplugues, E.; Huber, S.; Gagliani, N.; Hauser, A.E.; Town, T.; Wan, Y.Y.; O’Connor, W.; Rongvaux, A.; Van Rooijen, N.; Haberman, A.M.; et al. Control of TH17 Cells Occurs in the Small Intestine. Nature 2011, 475, 514–518. [Google Scholar] [CrossRef]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-Cell Responses to Gut Microbiota Promote Experimental Autoimmune Encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108, 4615–4622. [Google Scholar] [CrossRef] [PubMed]
- Miyake, S.; Yamamura, T. Gut Environmental Factors and Multiple Sclerosis. J. Neuroimmunol. 2019, 329, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Tremlett, H.; Fadrosh, D.W.; Faruqi, A.A.; Zhu, F.; Hart, J.; Roalstad, S.; Graves, J.; Lynch, S.; Waubant, E. The US Network of Pediatric MS Centers Gut Microbiota in Early Pediatric Multiple Sclerosis: A Case−control Study. Eur. J. Neurol. 2016, 23, 1308–1321. [Google Scholar] [CrossRef] [PubMed]
- Duc, D.; Vigne, S.; Bernier-Latmani, J.; Yersin, Y.; Ruiz, F.; Gaïa, N.; Leo, S.; Lazarevic, V.; Schrenzel, J.; Petrova, T.V.; et al. Disrupting Myelin-Specific Th17 Cell Gut Homing Confers Protection in an Adoptive Transfer Experimental Autoimmune Encephalomyelitis. Cell Rep. 2019, 29, 378–390.e4. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Soergel, K.H. Colonic Fermentation: Metabolic and Clinical Implications. Clin. Investig. 1994, 72, 742–748. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy Contributions of Volatile Fatty Acids from the Gastrointestinal Tract in Various Species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Høverstad, T.; Midtvedt, T. Short-Chain Fatty Acids in Germfree Mice and Rats. J. Nutr. 1986, 116, 1772–1776. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Pathways of Acetate, Propionate, and Butyrate Formation by the Human Fecal Microbial Flora. Appl. Environ. Microbiol. 1996, 62, 1589–1592. [Google Scholar] [CrossRef]
- Koh, A.; de Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Luu, M.; Pautz, S.; Kohl, V.; Singh, R.; Romero, R.; Lucas, S.; Hofmann, J.; Raifer, H.; Vachharajani, N.; Carrascosa, L.C.; et al. The Short-Chain Fatty Acid Pentanoate Suppresses Autoimmunity by Modulating the Metabolic-Epigenetic Crosstalk in Lymphocytes. Nat. Commun. 2019, 10, 760. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G Protein-Coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef]
- Oldendorf, W. Carrier-Mediated Blood-Brain Barrier Transport of Short-Chain Monocarboxylic Organic Acids. Am. J. Physiol. -Leg. Content 1973, 224, 1450–1453. [Google Scholar] [CrossRef] [PubMed]
- Roediger, W.E. Role of Anaerobic Bacteria in the Metabolic Welfare of the Colonic Mucosa in Man. Gut 1980, 21, 793–798. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Erratum: Commensal Microbe-Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells. Nature 2014, 506, 254. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kang, S.G.; Jannasch, A.H.; Cooper, B.; Patterson, J.; Kim, C.H. Short-Chain Fatty Acids Induce Both Effector and Regulatory T Cells by Suppression of Histone Deacetylases and Regulation of the MTOR–S6K Pathway. Mucosal Immunol. 2015, 8, 80–93. [Google Scholar] [CrossRef]
- Duscha, A.; Gisevius, B.; Hirschberg, S.; Yissachar, N.; Stangl, G.I.; Eilers, E.; Bader, V.; Haase, S.; Kaisler, J.; David, C.; et al. Propionic Acid Shapes the Multiple Sclerosis Disease Course by an Immunomodulatory Mechanism. Cell 2020, 180, 1067–1080.e16. [Google Scholar] [CrossRef]
- Saresella, M.; Marventano, I.; Barone, M.; La Rosa, F.; Piancone, F.; Mendozzi, L.; d’Arma, A.; Rossi, V.; Pugnetti, L.; Roda, G.; et al. Alterations in Circulating Fatty Acid Are Associated With Gut Microbiota Dysbiosis and Inflammation in Multiple Sclerosis. Front. Immunol. 2020, 11, 1390. [Google Scholar] [CrossRef]
- Olsson, A.; Gustavsen, S.; Nguyen, T.D.; Nyman, M.; Langkilde, A.R.; Hansen, T.H.; Sellebjerg, F.; Oturai, A.B.; Bach Søndergaard, H. Serum Short-Chain Fatty Acids and Associations With Inflammation in Newly Diagnosed Patients With Multiple Sclerosis and Healthy Controls. Front. Immunol. 2021, 12, 661493. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, C.; Lin, Q.; Dorsett, Y.; Ghezzi, L.; Liu, Z.; Pan, Y.; Chen, K.; Han, Y.; Li, Z.; Xiao, H.; et al. Alterations of Host-Gut Microbiome Interactions in Multiple Sclerosis. eBioMedicine 2022, 76, 103798. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Abuazab, M.; Schwiertz, A.; Walter, S.; Faßbender, K.C.; Fousse, M.; Unger, M.M. Short-Chain Fatty Acids and Intestinal Inflammation in Multiple Sclerosis: Modulation of Female Susceptibility by Microbial Products? Autoimmun. Highlights 2021, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Cuello, J.P.; Martínez Ginés, M.L.; García Domínguez, J.M.; Tejeda-Velarde, A.; Lozano Ros, A.; Higueras, Y.; Meldaña Rivera, A.; Goicochea Briceño, H.; Garcia-Tizon, S.; de León-Luis, J.; et al. Short-chain Fatty Acids during Pregnancy in Multiple Sclerosis: A Prospective Cohort Study. Eur. J. Neurol. 2022, 29, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Levi, I.; Gurevich, M.; Perlman, G.; Magalashvili, D.; Menascu, S.; Bar, N.; Godneva, A.; Zahavi, L.; Chermon, D.; Kosower, N.; et al. Potential Role of Indolelactate and Butyrate in Multiple Sclerosis Revealed by Integrated Microbiome-Metabolome Analysis. Cell Rep. Med. 2021, 2, 100246. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Waubant, E.; Chehoud, C.; Kuczynski, J.; DeSantis, T.Z.; Warrington, J.; Venkatesan, A.; Fraser, C.M.; Mowry, E.M. Gut Microbiota in Multiple Sclerosis: Possible Influence of Immunomodulators. J. Investig. Med. 2015, 63, 729–734. [Google Scholar] [CrossRef]
- Haghikia, A.; Jörg, S.; Duscha, A.; Berg, J.; Manzel, A.; Waschbisch, A.; Hammer, A.; Lee, D.-H.; May, C.; Wilck, N.; et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 2015, 43, 817–829. [Google Scholar] [CrossRef]
- Park, J.; Wang, Q.; Wu, Q.; Mao-Draayer, Y.; Kim, C.H. Bidirectional Regulatory Potentials of Short-Chain Fatty Acids and Their G-Protein-Coupled Receptors in Autoimmune Neuroinflammation. Sci. Rep. 2019, 9, 8837. [Google Scholar] [CrossRef]
- Mizuno, M.; Noto, D.; Kaga, N.; Chiba, A.; Miyake, S. The Dual Role of Short Fatty Acid Chains in the Pathogenesis of Autoimmune Disease Models. PLoS ONE 2017, 12, e0173032. [Google Scholar] [CrossRef]
- Chevalier, A.C.; Rosenberger, T.A. Increasing Acetyl-CoA Metabolism Attenuates Injury and Alters Spinal Cord Lipid Content in Mice Subjected to Experimental Autoimmune Encephalomyelitis. J. Neurochem. 2017, 141, 721–737. [Google Scholar] [CrossRef]
- Calvo-Barreiro, L.; Eixarch, H.; Cornejo, T.; Costa, C.; Castillo, M.; Mestre, L.; Guaza, C.; del Carmen Martínez-Cuesta, M.; Tanoue, T.; Honda, K.; et al. Selected Clostridia Strains from The Human Microbiota and Their Metabolite, Butyrate, Improve Experimental Autoimmune Encephalomyelitis. Neurotherapeutics 2021, 18, 920–937. [Google Scholar] [CrossRef]
- Haase, S.; Mäurer, J.; Duscha, A.; Lee, D.-H.; Balogh, A.; Gold, R.; Müller, D.N.; Haghikia, A.; Linker, R.A. Propionic Acid Rescues High-Fat Diet Enhanced Immunopathology in Autoimmunity via Effects on Th17 Responses. Front. Immunol. 2021, 12, 701626. [Google Scholar] [CrossRef] [PubMed]
- Rohrbeck, L.; Adori, M.; Wang, S.; He, C.; Tibbitt, C.A.; Chernyshev, M.; Sirel, M.; Ribacke, U.; Murrell, B.; Bohlooly-Y, M.; et al. GPR43 Regulates Marginal Zone B-cell Responses to Foreign and Endogenous Antigens. Immunol. Cell Biol. 2021, 99, 234–243. [Google Scholar] [CrossRef]
- Vinolo, M.A.R.; Rodrigues, H.G.; Hatanaka, E.; Sato, F.T.; Sampaio, S.C.; Curi, R. Suppressive Effect of Short-Chain Fatty Acids on Production of Proinflammatory Mediators by Neutrophils. J. Nutr. Biochem. 2011, 22, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Shu, D.; Zheng, M.; Wang, J.; Luo, C.; Wang, Y.; Guo, F.; Zou, X.; Lv, X.; Li, Y.; et al. Microbial Metabolite Butyrate Facilitates M2 Macrophage Polarization and Function. Sci. Rep. 2016, 6, 24838. [Google Scholar] [CrossRef]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Tóth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The Gut Microbiota Influences Blood-Brain Barrier Permeability in Mice. Sci. Transl. Med. 2014, 6, 3009759. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Noto, D.; Hoshino, Y.; Mizuno, M.; Miyake, S. Butyrate Suppresses Demyelination and Enhances Remyelination. J. Neuroinflamm. 2019, 16, 165. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L. Bile Acids: Regulation of Synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Hofmann, A.F. The Continuing Importance of Bile Acids in Liver and Intestinal Disease. Arch. Intern. Med. 1999, 159, 2647. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.-U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-Beta-Muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, P.; Smith, M.D.; Mische, L.; Harrington, E.; Fitzgerald, K.C.; Martin, K.; Kim, S.; Reyes, A.A.; Gonzalez-Cardona, J.; Volsko, C.; et al. Bile Acid Metabolism Is Altered in Multiple Sclerosis and Supplementation Ameliorates Neuroinflammation. J. Clin. Investig. 2020, 130, 3467–3482. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.P.; Steinman, L. Obeticholic Acid, a Synthetic Bile Acid Agonist of the Farnesoid X Receptor, Attenuates Experimental Autoimmune Encephalomyelitis. Proc. Natl. Acad. Sci. USA 2016, 113, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Hucke, S.; Herold, M.; Liebmann, M.; Freise, N.; Lindner, M.; Fleck, A.-K.; Zenker, S.; Thiebes, S.; Fernandez-Orth, J.; Buck, D.; et al. The Farnesoid-X-Receptor in Myeloid Cells Controls CNS Autoimmunity in an IL-10-Dependent Fashion. Acta Neuropathol. 2016, 132, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.D.; Patnaude, L.A.; Pelletier, J.; Souza, D.J.; Lukas, S.M.; King, F.J.; Hill, J.D.; Stefanopoulos, D.E.; Ryan, K.; Desai, S.; et al. A GPBAR1 (TGR5) Small Molecule Agonist Shows Specific Inhibitory Effects on Myeloid Cell Activation In Vitro and Reduces Experimental Autoimmune Encephalitis (EAE) In Vivo. PLoS ONE 2014, 9, e100883. [Google Scholar] [CrossRef]
- Ak, M. Profile of Circulatory Metabolites in an Animal Model of Multiple Sclerosis Using Global Metabolomics. J. Clin. Cell. Immunol. 2013, 4, 1000150. [Google Scholar] [CrossRef]
- Greenwood, J.; Adu, J.; Davey, A.J.; Abbott, N.J.; Bradbury, M.W.B. The Effect of Bile Salts on the Permeability and Ultrastructure of the Perfused, Energy-Depleted, Rat Blood-Brain Barrier. J. Cereb. Blood Flow Metab. 1991, 11, 644–654. [Google Scholar] [CrossRef]
- Joo, S.S.; Won, T.J.; Lee, D.I. Potential Role of Ursodeoxycholic Acid in Suppression of Nuclear Factor Kappa B in Microglial Cell Line (BV-2). Arch. Pharm. Res. 2004, 27, 954–960. [Google Scholar] [CrossRef]
- Yanguas-Casás, N.; Barreda-Manso, M.A.; Nieto-Sampedro, M.; Romero-Ramírez, L. TUDCA: An Agonist of the Bile Acid Receptor GPBAR1/TGR5 With Anti-Inflammatory Effects in Microglial Cells: Anti-Inflammatory Effect of Tudca in Microglia. J. Cell. Physiol. 2017, 232, 2231–2245. [Google Scholar] [CrossRef]
- Crick, P.J.; Griffiths, W.J.; Zhang, J.; Beibel, M.; Abdel-Khalik, J.; Kuhle, J.; Sailer, A.W.; Wang, Y. Reduced Plasma Levels of 25-Hydroxycholesterol and Increased Cerebrospinal Fluid Levels of Bile Acid Precursors in Multiple Sclerosis Patients. Mol. Neurobiol. 2017, 54, 8009–8020. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.; Bjornevik, K.; Clish, C.B.; Edan, G.; Freedman, M.; Hartung, H.-P.; Montalban, X.; Sandbrink, R.; Radue, E.-W.; Barkhof, F.; et al. Bile Acids Metabolites as Predictors of Long-Term Multiple Sclerosis Progression (S5.004). Neurology 2022, 98, 2787. [Google Scholar]
- Le Floc’h, N.; Otten, W.; Merlot, E. Tryptophan Metabolism, from Nutrition to Potential Therapeutic Applications. Amino Acids 2011, 41, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Imanishi, J.; Oku, T.; Kishida, T.; Hayaishi, O. Induction of Pulmonary Indoleamine 2,3-Dioxygenase by Interferon. Proc. Natl. Acad. Sci. USA 1981, 78, 129–132. [Google Scholar] [CrossRef]
- Laurans, L.; Venteclef, N.; Haddad, Y.; Chajadine, M.; Alzaid, F.; Metghalchi, S.; Sovran, B.; Denis, R.G.P.; Dairou, J.; Cardellini, M.; et al. Genetic Deficiency of Indoleamine 2,3-Dioxygenase Promotes Gut Microbiota-Mediated Metabolic Health. Nat. Med. 2018, 24, 1113–1120. [Google Scholar] [CrossRef]
- Funatake, C.J.; Marshall, N.B.; Steppan, L.B.; Mourich, D.V.; Kerkvliet, N.I. Cutting Edge: Activation of the Aryl Hydrocarbon Receptor by 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Generates a Population of CD4+ CD25+ Cells with Characteristics of Regulatory T Cells. J. Immunol. 2005, 175, 4184–4188. [Google Scholar] [CrossRef]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The Aryl Hydrocarbon Receptor Interacts with C-Maf to Promote the Differentiation of Type 1 Regulatory T Cells Induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- DeMoss, R.D.; Moser, K. Tryptophanase in Diverse Bacterial Species. J. Bacteriol. 1969, 98, 167–171. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Chyan, Y.-J.; Poeggeler, B.; Omar, R.A.; Chain, D.G.; Frangione, B.; Ghiso, J.; Pappolla, M.A. Potent Neuroprotective Properties against the Alzheimer β-Amyloid by an Endogenous Melatonin-Related Indole Structure, Indole-3-Propionic Acid. J. Biol. Chem. 1999, 274, 21937–21942. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, B.; Bhargava, P.; Tremlett, H.; Hart, J.; Graves, J.; Waubant, E. Altered Tryptophan Metabolism Is Associated with Pediatric Multiple Sclerosis Risk and Course. Ann. Clin. Transl. Neurol. 2018, 5, 1211–1221. [Google Scholar] [CrossRef]
- Gaetani, L.; Boscaro, F.; Pieraccini, G.; Calabresi, P.; Romani, L.; Di Filippo, M.; Zelante, T. Host and Microbial Tryptophan Metabolic Profiling in Multiple Sclerosis. Front. Immunol. 2020, 11, 157. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.J.; Guillemin, G.J. Kynurenine Pathway Metabolomics Predicts and Provides Mechanistic Insight into Multiple Sclerosis Progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef]
- Ntranos, A.; Park, H.-J.; Wentling, M.; Tolstikov, V.; Amatruda, M.; Inbar, B.; Kim-Schulze, S.; Frazier, C.; Button, J.; Kiebish, M.A.; et al. Bacterial Neurotoxic Metabolites in Multiple Sclerosis Cerebrospinal Fluid and Plasma. Brain 2022, 145, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Tömösi, F.; Kecskeméti, G.; Cseh, E.K.; Szabó, E.; Rajda, C.; Kormány, R.; Szabó, Z.; Vécsei, L.; Janáky, T. A Validated UHPLC-MS Method for Tryptophan Metabolites: Application in the Diagnosis of Multiple Sclerosis. J. Pharm. Biomed. Anal. 2020, 185, 113246. [Google Scholar] [CrossRef]
- Stone, T. Neuropharmacology of Quinolinic and Kynurenic Acids. Pharmacol. Rev. 1993, 45, 309–379. [Google Scholar]
- Foster, A.C.; Vezzani, A.; French, E.D.; Schwarcz, R. Kynurenic Acid Blocks Neurotoxicity and Seizures Induced in Rats by the Related Brain Metabolite Quinolinic Acid. Neurosci. Lett. 1984, 48, 273–278. [Google Scholar] [CrossRef]
- Herman, S.; Åkerfeldt, T.; Spjuth, O.; Burman, J.; Kultima, K. Biochemical Differences in Cerebrospinal Fluid between Secondary Progressive and Relapsing–Remitting Multiple Sclerosis. Cells 2019, 8, 84. [Google Scholar] [CrossRef]
- Platten, M.; Ho, P.P.; Youssef, S.; Fontoura, P.; Garren, H.; Hur, E.M.; Gupta, R.; Lee, L.Y.; Kidd, B.A.; Robinson, W.H.; et al. Treatment of Autoimmune Neuroinflammation with a Synthetic Tryptophan Metabolite. Science 2005, 310, 850–855. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, G.-X.; Gran, B.; Fallarino, F.; Yu, S.; Li, H.; Cullimore, M.L.; Rostami, A.; Xu, H. IDO Upregulates Regulatory T Cells via Tryptophan Catabolite and Suppresses Encephalitogenic T Cell Responses in Experimental Autoimmune Encephalomyelitis. J. Immunol. 2010, 185, 5953–5961. [Google Scholar] [CrossRef] [PubMed]
- Fazio, F.; Zappulla, C.; Notartomaso, S.; Busceti, C.; Bessede, A.; Scarselli, P.; Vacca, C.; Gargaro, M.; Volpi, C.; Allegrucci, M.; et al. Cinnabarinic Acid, an Endogenous Agonist of Type-4 Metabotropic Glutamate Receptor, Suppresses Experimental Autoimmune Encephalomyelitis in Mice. Neuropharmacology 2014, 81, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Ariyannur, P.S.; Ribeiro, R.; Tanaka, M.; Moffett, J.R.; Kirmani, B.F.; Namboodiri, A.M.A.; Zhang, Y. Efficacy of N-Acetylserotonin and Melatonin in the EAE Model of Multiple Sclerosis. J. Neuroimmune Pharmacol. 2016, 11, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Dopkins, N.; Becker, W.; Miranda, K.; Walla, M.; Nagarkatti, P.; Nagarkatti, M. Tryptamine Attenuates Experimental Multiple Sclerosis Through Activation of Aryl Hydrocarbon Receptor. Front. Pharmacol. 2021, 11, 619265. [Google Scholar] [CrossRef] [PubMed]
- Lanz, T.V.; Becker, S.; Mohapatra, S.R.; Opitz, C.A.; Wick, W.; Platten, M. Suppression of Th1 Differentiation by Tryptophan Supplementation in Vivo. Amino Acids 2017, 49, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Sonner, J.K.; Keil, M.; Falk-Paulsen, M.; Mishra, N.; Rehman, A.; Kramer, M.; Deumelandt, K.; Röwe, J.; Sanghvi, K.; Wolf, L.; et al. Dietary Tryptophan Links Encephalogenicity of Autoreactive T Cells with Gut Microbial Ecology. Nat. Commun. 2019, 10, 4877. [Google Scholar] [CrossRef]
- Rothhammer, V.; Borucki, D.M.; Garcia Sanchez, M.I.; Mazzola, M.A.; Hemond, C.C.; Regev, K.; Paul, A.; Kivisäkk, P.; Bakshi, R.; Izquierdo, G.; et al. Dynamic Regulation of Serum Aryl Hydrocarbon Receptor Agonists in MS. Neurol. Neuroimmunol. Neuroinflamm 2017, 4, e359. [Google Scholar] [CrossRef]
- Quintana, F.J.; Murugaiyan, G.; Farez, M.F.; Mitsdoerffer, M.; Tukpah, A.-M.; Burns, E.J.; Weiner, H.L. An Endogenous Aryl Hydrocarbon Receptor Ligand Acts on Dendritic Cells and T Cells to Suppress Experimental Autoimmune Encephalomyelitis. Proc. Natl. Acad. Sci. USA 2010, 107, 20768–20773. [Google Scholar] [CrossRef]
- Hwang, S.-J.; Hwang, Y.-J.; Yun, M.-O.; Kim, J.-H.; Oh, G.-S.; Park, J.-H. Indoxyl 3-Sulfate Stimulates Th17 Differentiation Enhancing Phosphorylation of c-Src and STAT3 to Worsen Experimental Autoimmune Encephalomyelitis. Toxicol. Lett. 2013, 220, 109–117. [Google Scholar] [CrossRef]
- Wetzel, L.A.; Hurtado, M.; MacDowell Kaswan, Z.A.; McCusker, R.H.; Steelman, A.J. Deletion of Indoleamine 2,3 Dioxygenase (Ido)1 but Not Ido2 Exacerbates Disease Symptoms of MOG35-55-Induced Experimental Autoimmune Encephalomyelitis. Brain Behav. Immun. Health 2020, 7, 100116. [Google Scholar] [CrossRef]
- Zarzecki, M.S.; Cattelan Souza, L.; Giacomeli, R.; Silva, M.R.P.; Prigol, M.; Boeira, S.P.; Jesse, C.R. Involvement of Indoleamine-2,3-Dioxygenase and Kynurenine Pathway in Experimental Autoimmune Encephalomyelitis in Mice. Neurochem. Res. 2020, 45, 2959–2977. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Macfarlane, G.T. Enumeration of Human Colonic Bacteria Producing Phenolic and Indolic Compounds: Effects of PH, Carbohydrate Availability and Retention Time on Dissimilatory Aromatic Amino Acid Metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.D.; Mills, R.G.; Coleman, D.J. Improved Gas-Liquid Chromatography Method for the Identification of Clostridium Difficile. J. Clin. Pathol. 1985, 38, 108–110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ward, L.A.; Johnson, K.A.; Robinson, I.M.; Yokoyama, M.T. Isolation from Swine Feces of a Bacterium Which Decarboxylates P-Hydroxyphenylacetic Acid to 4-Methylphenol (p-Cresol). Appl. Environ. Microbiol. 1987, 53, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Kawakami, K.; Sasaki, T.; Makino, I.; Kato, I.; Kobayashi, T.; Uchida, K.; Kaneko, K. Effects of Intestinal Bacteria-Derived p-Cresyl Sulfate on Th1-Type Immune Response in Vivo and in Vitro. Toxicol. Appl. Pharmacol. 2014, 274, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Lutz, N.W.; Viola, A.; Malikova, I.; Confort-Gouny, S.; Audoin, B.; Ranjeva, J.-P.; Pelletier, J.; Cozzone, P.J. Inflammatory Multiple-Sclerosis Plaques Generate Characteristic Metabolic Profiles in Cerebrospinal Fluid. PLoS ONE 2007, 2, e595. [Google Scholar] [CrossRef]
- Nicoli, F. Cerebrospinal Fluid Metabolic Profiles in Multiple Sclerosis and Degenerative Dementias Obtained by High Resolution Proton Magnetic Resonance Spectroscopy. C. R. Acad. Sci. Ser. III 1996, 319, 623–631. [Google Scholar]
- Singh, J.; Cerghet, M.; Poisson, L.M.; Datta, I.; Labuzek, K.; Suhail, H.; Rattan, R.; Giri, S. Urinary and Plasma Metabolomics Identify the Distinct Metabolic Profile of Disease State in Chronic Mouse Model of Multiple Sclerosis. J. Neuroimmune Pharmacol. 2019, 14, 241–250. [Google Scholar] [CrossRef]
- Fennema, D.; Phillips, I.R.; Shephard, E.A. Trimethylamine and Trimethylamine N-Oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-Mediated Host-Microbiome Metabolic Axis Implicated in Health and Disease. Drug Metab. Dispos. 2016, 44, 1839–1850. [Google Scholar] [CrossRef]
- Al-waiz, M.; Mitchell, S.C.; Idle, J.R.; Smith, R.L. The Metabolism of 14 C-Labelled Trimethylamine and Its N-Oxide in Man. Xenobiotica 1987, 17, 551–558. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Kira, Y.; Nishikawa, M.; Ochi, A.; Sato, E.; Inoue, M. L-Carnitine Suppresses the Onset of Neuromuscular Degeneration and Increases the Life Span of Mice with Familial Amyotrophic Lateral Sclerosis. Brain Res. 2006, 1070, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, Y.; Zhao, M.; Zheng, L.; Fan, D. Changes in the Concentrations of Trimethylamine N-Oxide (TMAO) and Its Precursors in Patients with Amyotrophic Lateral Sclerosis. Sci. Rep. 2020, 10, 15198. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Shen, T.; Lou, H. Dietary Polyphenols and Their Biological Significance. Int. J. Mol. Sci. 2007, 8, 950–988. [Google Scholar] [CrossRef] [PubMed Central]
- Hervert-Hernández, D.; Goñi, I. Dietary Polyphenols and Human Gut Microbiota: A Review. Food Rev. Int. 2011, 27, 154–169. [Google Scholar] [CrossRef]
- Espín, J.C.; Larrosa, M.; García-Conesa, M.T.; Tomás-Barberán, F. Biological Significance of Urolithins, the Gut Microbial Ellagic Acid-Derived Metabolites: The Evidence So Far. Evid. -Based Complement. Altern. Med. 2013, 2013, 1–15. [Google Scholar] [CrossRef]
- Romo-Vaquero, M.; García-Villalba, R.; González-Sarrías, A.; Beltrán, D.; Tomás-Barberán, F.A.; Espín, J.C.; Selma, M.V. Interindividual Variability in the Human Metabolism of Ellagic Acid: Contribution of Gordonibacter to Urolithin Production. J. Funct. Foods 2015, 17, 785–791. [Google Scholar] [CrossRef]
- Zhang, M.; Cui, S.; Mao, B.; Zhang, Q.; Zhao, J.; Zhang, H.; Tang, X.; Chen, W. Ellagic Acid and Intestinal Microflora Metabolite Urolithin A: A Review on Its Sources, Metabolic Distribution, Health Benefits, and Biotransformation. Crit. Rev. Food Sci. Nutr. 2022, 1–23. [Google Scholar] [CrossRef]
- Shen, P.-X.; Li, X.; Deng, S.-Y.; Zhao, L.; Zhang, Y.-Y.; Deng, X.; Han, B.; Yu, J.; Li, Y.; Wang, Z.-Z.; et al. Urolithin A Ameliorates Experimental Autoimmune Encephalomyelitis by Targeting Aryl Hydrocarbon Receptor. eBioMedicine 2021, 64, 103227. [Google Scholar] [CrossRef]
- Busto, R.; Serna, J.; Perianes-Cachero, A.; Quintana-Portillo, R.; García-Seisdedos, D.; Canfrán-Duque, A.; Paino, C.L.; Lerma, M.; Casado, M.E.; Martín-Hidalgo, A.; et al. Ellagic Acid Protects from Myelin-Associated Sphingolipid Loss in Experimental Autoimmune Encephalomyelitis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 958–967. [Google Scholar] [CrossRef]
- Kiasalari, Z.; Afshin-Majd, S.; Baluchnejadmojarad, T.; Azadi-Ahmadabadi, E.; Esmaeil-Jamaat, E.; Fahanik-Babaei, J.; Fakour, M.; Fereidouni, F.; Ghasemi-Tarie, R.; Jalalzade-Ogvar, S.; et al. Ellagic Acid Ameliorates Neuroinflammation and Demyelination in Experimental Autoimmune Encephalomyelitis: Involvement of NLRP3 and Pyroptosis. J. Chem. Neuroanat. 2021, 111, 101891. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-Y.; Han, B.; Deng, X.; Deng, S.-Y.; Zhang, Y.-Y.; Shen, P.-X.; Hui, T.; Chen, R.-H.; Li, X.; Zhang, Y. Pomegranate Peel Extract Ameliorates the Severity of Experimental Autoimmune Encephalomyelitis via Modulation of Gut Microbiota. Gut Microbes 2020, 12, 1857515. [Google Scholar] [CrossRef] [PubMed]
- Vallarino, G.; Salis, A.; Lucarini, E.; Turrini, F.; Olivero, G.; Roggeri, A.; Damonte, G.; Boggia, R.; Di Cesare Mannelli, L.; Ghelardini, C.; et al. Healthy Properties of a New Formulation of Pomegranate-Peel Extract in Mice Suffering from Experimental Autoimmune Encephalomyelitis. Molecules 2022, 27, 914. [Google Scholar] [CrossRef]
- Toney, A.M.; Albusharif, M.; Works, D.; Polenz, L.; Schlange, S.; Chaidez, V.; Ramer-Tait, A.E.; Chung, S. Differential Effects of Whole Red Raspberry Polyphenols and Their Gut Metabolite Urolithin A on Neuroinflammation in BV-2 Microglia. IJERPH 2020, 18, 68. [Google Scholar] [CrossRef]
- Zhao, Z.; Bao, X.; Zhang, Z.; Liu, H.; Zhang, D. Phloroglucinol Derivative Compound 21 Attenuates Cuprizone-Induced Multiple Sclerosis Mice through Promoting Remyelination and Inhibiting Neuroinflammation. Sci. China Life Sci. 2020, 63, 905–914. [Google Scholar] [CrossRef]
- Xie, L.; Li, X.-K.; Funeshima-Fuji, N.; Kimura, H.; Matsumoto, Y.; Isaka, Y.; Takahara, S. Amelioration of Experimental Autoimmune Encephalomyelitis by Curcumin Treatment through Inhibition of IL-17 Production. Int. Immunopharmacol. 2009, 9, 575–581. [Google Scholar] [CrossRef]
- Ciftci, O.; Ozcan, C.; Kamisli, O.; Cetin, A.; Basak, N.; Aytac, B. Hesperidin, a Citrus Flavonoid, Has the Ameliorative Effects Against Experimental Autoimmune Encephalomyelitis (EAE) in a C57BL/J6 Mouse Model. Neurochem. Res. 2015, 40, 1111–1120. [Google Scholar] [CrossRef]
- Imler, T.J.; Petro, T.M. Decreased Severity of Experimental Autoimmune Encephalomyelitis during Resveratrol Administration Is Associated with Increased IL-17+IL-10+ T Cells, CD4− IFN-Γ+ Cells, and Decreased Macrophage IL-6 Expression. Int. Immunopharmacol. 2009, 9, 134–143. [Google Scholar] [CrossRef]
- Ginwala, R.; McTish, E.; Raman, C.; Singh, N.; Nagarkatti, M.; Nagarkatti, P.; Sagar, D.; Jain, P.; Khan, Z.K. Apigenin, a Natural Flavonoid, Attenuates EAE Severity Through the Modulation of Dendritic Cell and Other Immune Cell Functions. J. Neuroimmune Pharmacol. 2016, 11, 36–47. [Google Scholar] [CrossRef]
- Razeghi, J. Alleviation of Experimental Allergic Encephalomyelitis in C57BL/6 Mice by Soy Daidzein. Iran. J. Allergy Asthma Immunol. 2014, 13. [Google Scholar]
- Jensen, S.N.; Cady, N.M.; Shahi, S.K.; Peterson, S.R.; Gupta, A.; Gibson-Corley, K.N.; Mangalam, A.K. Isoflavone Diet Ameliorates Experimental Autoimmune Encephalomyelitis through Modulation of Gut Bacteria Depleted in Patients with Multiple Sclerosis. Sci. Adv. 2021, 7, eabd4595. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. The Functional Role of Polyamines in Eukaryotic Cells. Int. J. Biochem. Cell Biol. 2019, 107, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Pugin, B.; Barcik, W.; Westermann, P.; Heider, A.; Wawrzyniak, M.; Hellings, P.; Akdis, C.A.; O’Mahony, L. A Wide Diversity of Bacteria from the Human Gut Produces and Degrades Biogenic Amines. Microb. Ecol. Health Dis. 2017, 28, 1353881. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.M.S.; Doty, D.J.; DePaula-Silva, A.B.; Brown, D.G.; Bell, R.; Klag, K.A.; Truong, A.; Libbey, J.E.; Round, J.L.; Fujinami, R.S. Molecular Patterns from a Human Gut-Derived Lactobacillus Strain Suppress Pathogenic Infiltration of Leukocytes into the Central Nervous System. J. Neuroinflamm. 2020, 17, 291. [Google Scholar] [CrossRef]
- Puleston, D.J.; Baixauli, F.; Sanin, D.E.; Edwards-Hicks, J.; Villa, M.; Kabat, A.M.; Kamiński, M.M.; Stanckzak, M.; Weiss, H.J.; Grzes, K.M.; et al. Polyamine Metabolism Is a Central Determinant of Helper T Cell Lineage Fidelity. Cell 2021, 184, 4186–4202.e20. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Wang, C.; Fessler, J.; DeTomaso, D.; Avila-Pacheco, J.; Kaminski, J.; Zaghouani, S.; Christian, E.; Thakore, P.; Schellhaass, B.; et al. Metabolic Modeling of Single Th17 Cells Reveals Regulators of Autoimmunity. Cell 2021, 184, 4168–4185.e21. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, H.; Abujrais, S.; Herman, S.; Khoonsari, P.E.; Åkerfeldt, T.; Svenningsson, A.; Burman, J.; Kultima, K. Targeted Metabolomics of CSF in Healthy Individuals and Patients with Secondary Progressive Multiple Sclerosis Using High-Resolution Mass Spectrometry. Metabolomics 2020, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Bolayir, A.; Celik, V.K.; Bolayir, H.A.; Kapancik, S.; Kilicgun, H.; Gokce, S.F. The Possible Effects Of Polyamines In Multiple Sclerosis Patients On New Lesion Development And Disability. Int. J. Res. Granthaalayah 2018, 6, 536–543. [Google Scholar] [CrossRef]
- Guo, X.; Harada, C.; Namekata, K.; Kimura, A.; Mitamura, Y.; Yoshida, H.; Matsumoto, Y.; Harada, T. Spermidine Alleviates Severity of Murine Experimental Autoimmune Encephalomyelitis. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2696. [Google Scholar] [CrossRef]
- Yang, Q.; Zheng, C.; Cao, J.; Cao, G.; Shou, P.; Lin, L.; Velletri, T.; Jiang, M.; Chen, Q.; Han, Y.; et al. Spermidine Alleviates Experimental Autoimmune Encephalomyelitis through Inducing Inhibitory Macrophages. Cell Death Differ. 2016, 23, 1850–1861. [Google Scholar] [CrossRef]
- Carriche, G.M.; Almeida, L.; Stüve, P.; Velasquez, L.; Dhillon-LaBrooy, A.; Roy, U.; Lindenberg, M.; Strowig, T.; Plaza-Sirvent, C.; Schmitz, I.; et al. Regulating T-Cell Differentiation through the Polyamine Spermidine. J. Allergy Clin. Immunol. 2021, 147, 335–348.e11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Kong, M.; Wang, S.; He, B.; Xie, X. Spermine Alleviates Experimental Autoimmune Encephalomyelitis via Regulating T Cell Activation and Differentiation. Int. Immunopharmacol. 2022, 107, 108702. [Google Scholar] [CrossRef] [PubMed]
- Royet, J.; Dziarski, R. Peptidoglycan Recognition Proteins: Pleiotropic Sensors and Effectors of Antimicrobial Defences. Nat. Rev. Microbiol. 2007, 5, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Santana, A.; Diaz Heijtz, R. Bacterial Peptidoglycans from Microbiota in Neurodevelopment and Behavior. Trends Mol. Med. 2020, 26, 729–743. [Google Scholar] [CrossRef]
- Schrijver, I.A. Bacterial Peptidoglycan and Immune Reactivity in the Central Nervous System in Multiple Sclerosis. Brain 2001, 124, 1544–1554. [Google Scholar] [CrossRef]
- Kriesel, J.D.; Bhetariya, P.; Wang, Z.-M.; Renner, D.; Palmer, C.; Fischer, K.F. Spectrum of Microbial Sequences and a Bacterial Cell Wall Antigen in Primary Demyelination Brain Specimens Obtained from Living Patients. Sci. Rep. 2019, 9, 1387. [Google Scholar] [CrossRef]
- Branton, W.G.; Lu, J.Q.; Surette, M.G.; Holt, R.A.; Lind, J.; Laman, J.D.; Power, C. Brain Microbiota Disruption within Inflammatory Demyelinating Lesions in Multiple Sclerosis. Sci. Rep. 2016, 6, 37344. [Google Scholar] [CrossRef]
- Visser, L.; Melief, M.-J.; van Riel, D.; van Meurs, M.; Sick, E.A.; Inamura, S.; Bajramovic, J.J.; Amor, S.; Hintzen, R.Q.; Boven, L.A.; et al. Phagocytes Containing a Disease-Promoting Toll-Like Receptor/Nod Ligand Are Present in the Brain during Demyelinating Disease in Primates. Am. J. Pathol. 2006, 169, 1671–1685. [Google Scholar] [CrossRef]
- Visser, L.; Jan de Heer, H.; Boven, L.A.; van Riel, D.; van Meurs, M.; Melief, M.-J.; Zähringer, U.; van Strijp, J.; Lambrecht, B.N.; Nieuwenhuis, E.E.; et al. Proinflammatory Bacterial Peptidoglycan as a Cofactor for the Development of Central Nervous System Autoimmune Disease. J. Immunol. 2005, 174, 808–816. [Google Scholar] [CrossRef]
- Shaw, P.J.; Barr, M.J.; Lukens, J.R.; McGargill, M.A.; Chi, H.; Mak, T.W.; Kanneganti, T.-D. Signaling via the RIP2 Adaptor Protein in Central Nervous System-Infiltrating Dendritic Cells Promotes Inflammation and Autoimmunity. Immunity 2011, 34, 75–84. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, J.; Xu, X.; Wang, H.; Qiao, Y.; Chu, W.C.; Xu, S.; Chai, L.; Cottier, F.; Pavelka, N.; et al. Antibody Neutralization of Microbiota-Derived Circulating Peptidoglycan Dampens Inflammation and Ameliorates Autoimmunity. Nat. Microbiol. 2019, 4, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Balskus, E.P. Colibactin: Understanding an Elusive Gut Bacterial Genotoxin. Nat. Prod. Rep. 2015, 32, 1534–1540. [Google Scholar] [CrossRef] [PubMed]
- Dornisch, E.; Pletz, J.; Glabonjat, R.A.; Martin, F.; Lembacher-Fadum, C.; Neger, M.; Högenauer, C.; Francesconi, K.; Kroutil, W.; Zangger, K.; et al. Biosynthesis of the Enterotoxic Pyrrolobenzodiazepine Natural Product Tilivalline. Angew. Chem. Int. Ed. 2017, 56, 14753–14757. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-J.; Chang, F.-Y.; Wyche, T.P.; Backus, K.M.; Acker, T.M.; Funabashi, M.; Taketani, M.; Donia, M.S.; Nayfach, S.; Pollard, K.S.; et al. Discovery of Reactive Microbiota-Derived Metabolites That Inhibit Host Proteases. Cell 2017, 168, 517–526.e18. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ravichandran, V.; Yin, Y.; Yin, J.; Zhang, Y. Natural Products from Mammalian Gut Microbiota. Trends Biotechnol. 2019, 37, 492–504. [Google Scholar] [CrossRef]
- Milshteyn, A.; Colosimo, D.A.; Brady, S.F. Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe 2018, 23, 725–736. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rebeaud, J.; Peter, B.; Pot, C. How Microbiota-Derived Metabolites Link the Gut to the Brain during Neuroinflammation. Int. J. Mol. Sci. 2022, 23, 10128. https://doi.org/10.3390/ijms231710128
Rebeaud J, Peter B, Pot C. How Microbiota-Derived Metabolites Link the Gut to the Brain during Neuroinflammation. International Journal of Molecular Sciences. 2022; 23(17):10128. https://doi.org/10.3390/ijms231710128
Chicago/Turabian StyleRebeaud, Jessica, Benjamin Peter, and Caroline Pot. 2022. "How Microbiota-Derived Metabolites Link the Gut to the Brain during Neuroinflammation" International Journal of Molecular Sciences 23, no. 17: 10128. https://doi.org/10.3390/ijms231710128
APA StyleRebeaud, J., Peter, B., & Pot, C. (2022). How Microbiota-Derived Metabolites Link the Gut to the Brain during Neuroinflammation. International Journal of Molecular Sciences, 23(17), 10128. https://doi.org/10.3390/ijms231710128







