BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways
Abstract
:1. Introduction
2. BES1/BZR1 Family in Plants
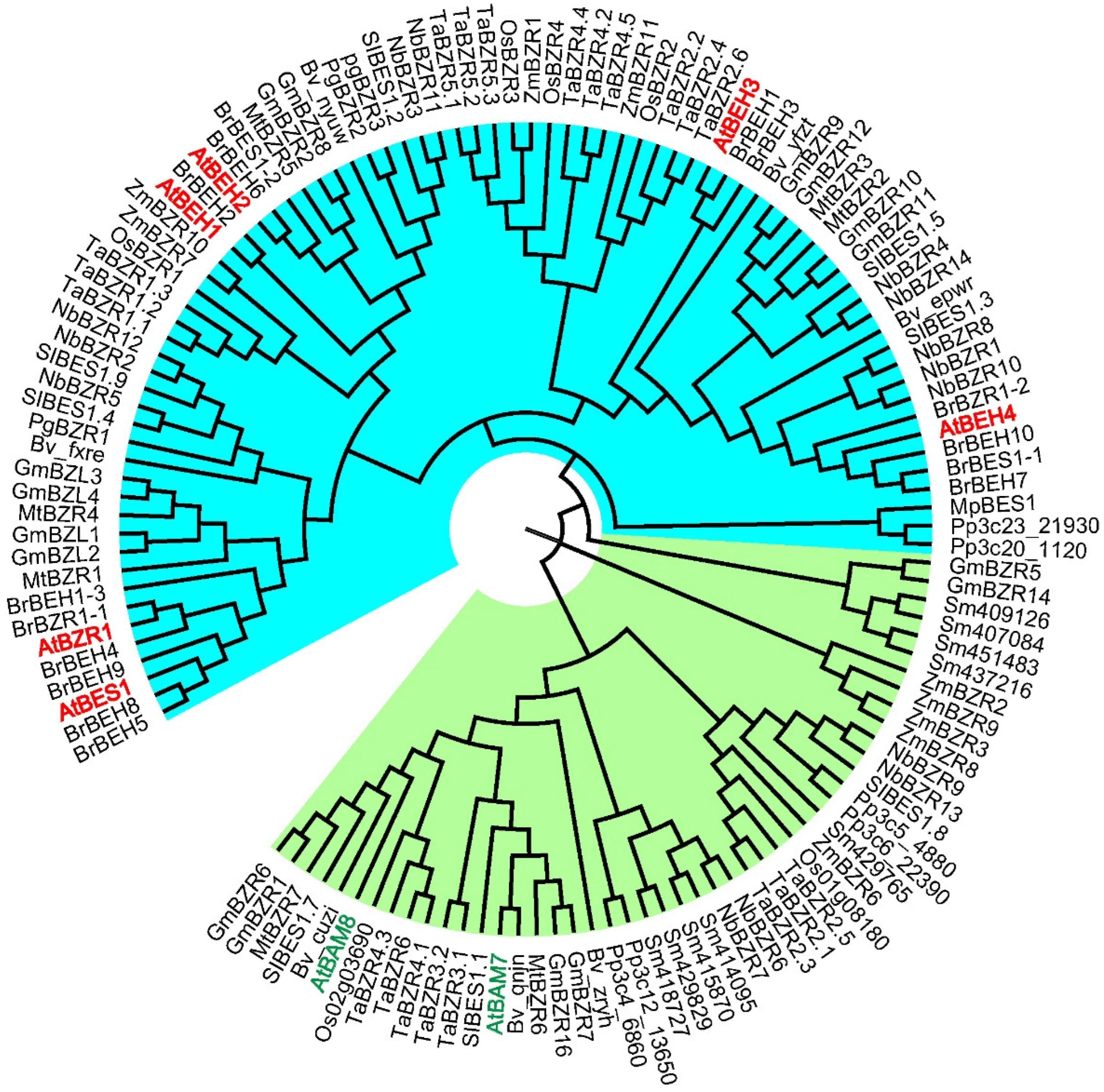
3. Activation of BES1 and BZR1 via Post-Translational Modifications
4. DNA-Binding Motifs of the BES1/BZR1 Family
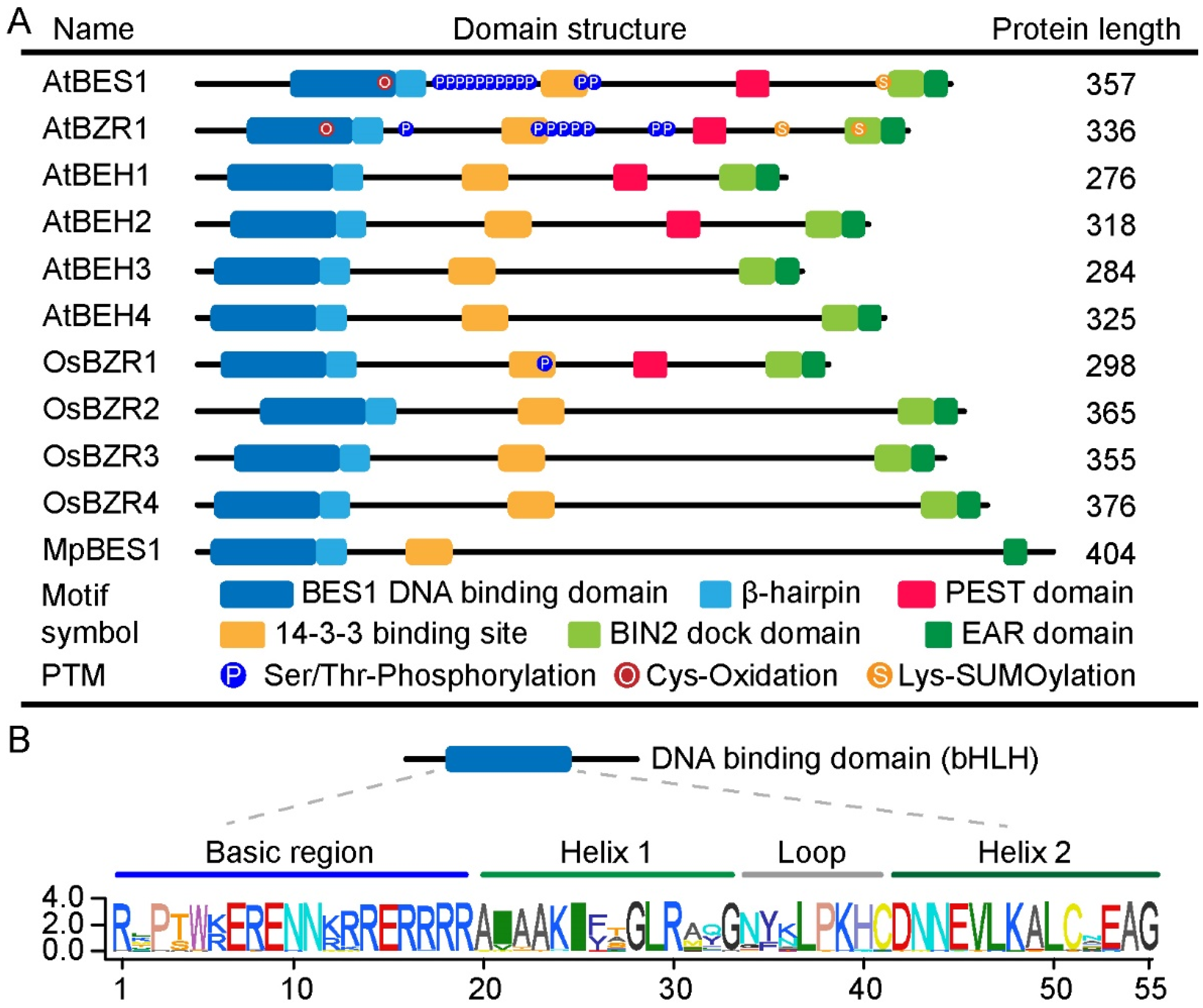
5. Other Conserved Motifs in the BES1/BZR1 Family
6. Target Genes of the BES1/BZR1 Family
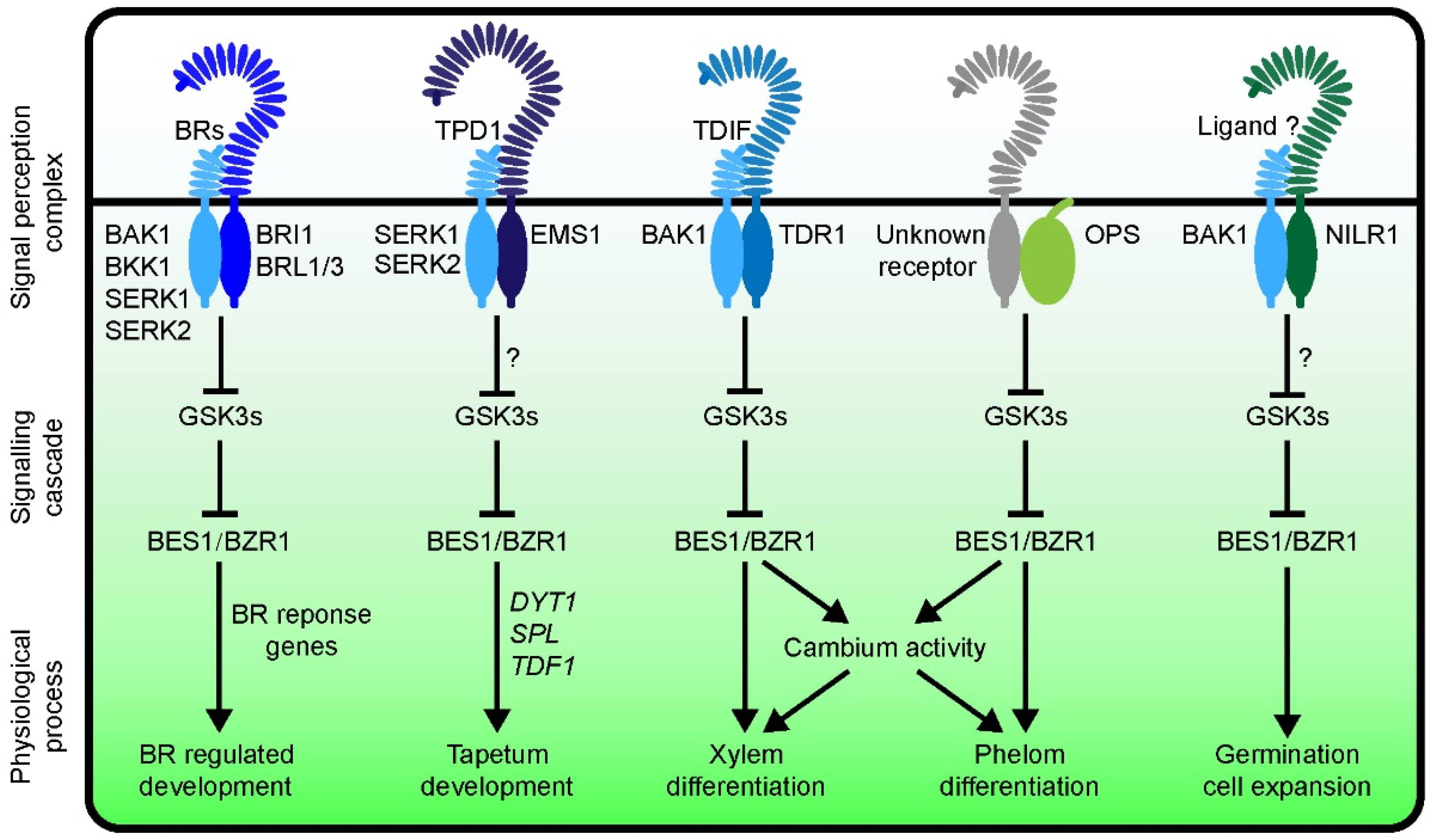
7. Functions of BES1/BZR1 in Regulating Plant Growth and Development through a Canonical BRI1-BR-BAK1 Pathway
7.1. BES1 Regulates the Development of Female Gametophyte and Ovuled through a BR-Dependent Pathway
7.2. BES1 Regulates Root Development through a BR-Dependent Pathway
8. Functions of the BES1/BZR1 Family in Regulating Plant Growth and Development through a BR-Independent Pathway
8.1. BES1/BZR1 Family Regulates Tapetum Development
8.2. BES1/BZR1 Family Functions in Vascular Development
8.3. BES1 Function in Other Signaling Pathways
9. Conclusions and Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.Q.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- He, K.; Xu, S.; Li, J. BAK1 directly regulates brassinosteroid perception and BRI1 activation. J. Integr. Plant Biol. 2013, 55, 1264–1270. [Google Scholar] [CrossRef]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.D.; Li, J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012, 8, e1002452. [Google Scholar] [CrossRef]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.-C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef]
- Zhou, A.F.; Wang, H.C.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef]
- Sun, Y.; Han, Z.; Tang, J.; Hu, Z.; Chai, C.; Zhou, B.; Chai, J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013, 23, 1326. [Google Scholar] [CrossRef]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.X.; Ma, H.; Wang, X. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 2011, 21, 825–834. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, J.J.; Wang, J.; Chen, L.; Fan, S.-L.; Wu, J.-W.; Wang, X.; Wang, Z.-X. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014, 24, 1328–1341. [Google Scholar] [CrossRef]
- Li, J.M.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.-X.; Sun, Y.; Burlingame, A.L.; Wang, Z.-Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Peng, P.; Schmitz, R.J.; Decker, A.D.; Tax, F.E.; Li, J.M. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002, 130, 1221–1229. [Google Scholar] [CrossRef]
- Vert, G.; Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef]
- Kim, T.-W.; Wang, Z.-Y. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef]
- Wang, R.J.; Wang, R.X.; Liu, M.M.; Yuan, W.W.; Zhao, Z.Y.; Liu, X.Q.; Peng, Y.M.; Yang, X.R.; Sun, Y.; Tang, W.Q. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.-Y.; Cao, D.-M.; Tang, W.; He, K.; Zhu, J.-Y.; He, J.-X.; Bai, M.-Y.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef]
- Yu, X.; Li, L.; Zola, J.; Aluru, M.; Ye, H.; Foudree, A.; Guo, H.Q.; Anderson, S.; Aluru, S.; Liu, P.; et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BES1 target genes in Arabidopsis thaliana. Plant J. 2011, 65, 634–646. [Google Scholar] [CrossRef]
- Nolan, T.; Vukasinovic, N.; Liu, D.R.; Russinova, E.; Yin, Y.H. Brassinosteroids: Multi-dimensional regulators of plant growth, development, and stress responses. Plant Cell 2019, 32, 295–318. [Google Scholar] [CrossRef]
- Lv, M.H.; Li, J. Molecular mechanisms of brassinosteroid-mediated responses to changing environments in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2737. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Lv, M.H.; Wang, Y.Z.; Wang, P.A.; Cui, Y.W.; Li, M.Z.; Wang, R.S.; Gou, X.P.; Li, J. BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat. Commun. 2019, 10, 4164. [Google Scholar] [CrossRef]
- Zheng, B.; Bai, Q.W.; Wu, L.; Liu, H.; Liu, Y.P.; Xu, W.J.; Li, G.S.; Ren, H.Y.; She, X.P.; Wu, G. EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation. Nat. Commun. 2019, 10, 4165. [Google Scholar] [CrossRef]
- Bai, Q.; Li, C.; Wu, L.; Liu, H.; Ren, H.; Li, G.; Wang, Q.; Wu, G.; Zheng, B. Engineering chimeras by fusing plant receptor-like kinase EMS1 and BRI1 reveals the two receptors’ structural specificity and molecular mechanisms. Int. J. Mol. Sci. 2022, 23, 2155. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF–TDR signalling. Nat. Commun. 2014, 5, 3504. [Google Scholar] [CrossRef]
- Albertos, P.; Dündar, G.; Schenk, P.; Carrera, S.; Cavelius, P.; Sieberer, T.; Poppenberger, B. Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J. 2022, 41, e108664. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Bai, Q.; Li, C.; Wang, L.; Wei, Q.; Ali, K.; Li, W.; Huang, S.; Xu, H.; Li, G.; et al. Pan-brassinosteroid signaling revealed by functional analysis of NILR1 in land plants. New Phytol. 2022, 235. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Soyk, S.; Šimková, K.; Zürcher, E.; Luginbühl, L.; Brand, L.H.; Vaughan, C.K.; Wanke, D.; Zeeman, S.C. The enzyme-like domain of arabidopsis nuclear β-Amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 2014, 26, 1746–1763. [Google Scholar] [CrossRef]
- Monroe, J.D.; Storm, A.R. Review: The Arabidopsis β-amylase (BAM) gene family: Diversity of form and function. Plant Sci. 2018, 276, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, M.; Coiro, M.; Meier, T.; Wicker, T.; Zeeman, S.C.; Santelia, D. The evolution of functional complexity within the β-amylase gene family in land plants. BMC Evol. Biol. 2019, 19, 1–18. [Google Scholar] [CrossRef]
- Vriet, C.; Lemmens, K.; Vandepoele, K.; Reuzeau, C.; Russinova, E. Evolutionary trails of plant steroid genes. Trends Plant Sci. 2015, 20, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Mecchia, M.A.; García-Hourquet, M.; Lozano-Elena, F.; Planas-Riverola, A.; Blasco-Escamez, D.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 2021, 31, 4860–4869. [Google Scholar] [CrossRef]
- Hwang, H.; Lee, H.-Y.; Ryu, H.; Cho, H. Functional characterization of BRASSINAZOLE-RESISTANT 1 in Panax ginseng (PgBZR1) and brassinosteroid response during storage root formation. Int. J. Mol. Sci. 2020, 21, 9666. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-Wide identification and characterization of the brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Feng, W.Q.; Liu, Y.; Cao, Y.; Zhao, Y.R.; Zhang, H.W.J.; Sun, F.; Yang, Q.Q.; Li, W.C.; Lu, Y.L.; Zhang, X.C.; et al. Maize ZmBES1/BZR1-3 and -9 transcription factors negatively regulate drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2022, 23, 6025. [Google Scholar] [CrossRef]
- Bradai, M.; Amorim-Silva, V.; Belgaroui, N.; Esteban del Valle, A.; Chabouté, M.E.; Schmit, A.C.; Lozano-Duran, R.; Botella, M.A.; Hanin, M.; Ebel, C. Wheat type one protein phosphatase participates in the brassinosteroid control of root growth via activation of BES1. Int. J. Mol. Sci. 2021, 22, 10424. [Google Scholar] [CrossRef]
- Chen, X.; Wu, X.; Qiu, S.; Zheng, H.; Lu, Y.; Peng, J.; Wu, G.; Chen, J.; Rao, S.; Yan, F. Genome-Wide identification and expression profiling of the BZR transcription factor gene family in Nicotiana benthamiana. Int. J. Mol. Sci. 2021, 22, 10379. [Google Scholar] [CrossRef]
- Cui, C.; Wang, H.; Hong, L.; Xu, Y.; Zhao, Y.; Zhou, C. MtBZR1 plays an important role in nodule development in Medicago truncatula. Int. J. Mol. Sci. 2019, 20, 2941. [Google Scholar] [CrossRef]
- Yu, H.Q.; Feng, W.Q.; Sun, F.; Zhang, Y.Y.; Qu, J.T.; Liu, B.L.; Lu, F.Z.; Yang, L.; Fu, F.L.; Li, W.C. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Bai, M.-Y.; Zhang, L.-Y.; Gampala, S.S.; Zhu, S.-W.; Song, W.-Y.; Chong, K.; Wang, Z.-Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.-I.; Jung, H.-J.; Ahmed, N.U.; Kayum, A.; Kang, J.-G.; Nou, I.-S. Molecular characterization of BZR transcription factor family and abiotic stress induced expression profiling in Brassica rapa. Plant Physiol. Biochem. 2015, 92, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, L.; Li, X. GmBEHL1, a BES1/BZR1 family protein, negatively regulates soybean nodulation. Sci. Rep. 2018, 8, 7614. [Google Scholar] [CrossRef]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef]
- Ma, S.; Ji, T.; Liang, M.; Li, S.; Tian, Y.; Gao, L. Genome-Wide identification, structural, and gene expression analysis of BRI1-EMS-Suppressor 1 transcription factor family in Cucumis sativus. Front. Genet. 2020, 11, 583996. [Google Scholar] [CrossRef]
- Wang, W.; Sun, Y.-Q.; Li, G.-L.; Zhang, S.-Y. Genome-wide identification, characterization, and expression patterns of the BZR transcription factor family in sugar beet (Beta vulgaris L.). BMC Plant Biol. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Ji, Y.; Qu, Y.; Jiang, Z.; Yan, J.; Chu, J.; Xu, M.; Su, X.; Yuan, H.; Wang, A. The mechanism for brassinosteroids suppressing climacteric fruit ripening. Plant Physiol. 2021, 185, 1875–1893. [Google Scholar] [CrossRef]
- Shi, Z.M.; Chen, X.; Xue, H.D.; Jia, T.T.; Meng, F.N.; Liu, Y.F.; Luo, X.M.; Xiao, G.H.; Zhu, S.W. GhBZR3 suppresses cotton fiber elongation by inhibiting very-long-chain fatty acid biosynthesis. Plant J. 2022, 111, 785–799. [Google Scholar] [CrossRef]
- Gao, X.Y.; Zhang, J.Q.; Zhang, X.J.; Zhou, J.; Jiang, Z.S.; Huang, P.; Tang, Z.B.; Bao, Y.M.; Cheng, J.P.; Tang, H.J.; et al. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 2019, 31, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Y.-J.; Yang, B.-J.; Yu, X.-X.; Wang, D.; Zu, S.-H.; Xue, H.-W.; Lin, W.-H. Functional characterization of GmBZL2 (AtBZR1 like gene) reveals the conserved BR signaling regulation in Glycine max. Sci. Rep. 2016, 6, 31134. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Chen, W.; Wang, B.; Yao, Q.-M.; Valliyodan, B.; Bai, M.-Y.; Zhao, M.-Z.; Ye, H.; Wang, Z.-Y.; Nguyen, H.T. GmBZL3 acts as a major BR signaling regulator through crosstalk with multiple pathways in Glycine max. BMC Plant Biol. 2019, 19, 86. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Chen, L.-G.; Gao, Z.; Zhao, Z.; Liu, X.; Li, Y.; Zhang, Y.; Liu, X.; Sun, Y.; Tang, W. BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol. Plant 2019, 12, 1408–1415. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, J.; Tang, L.; Zhao, Y.; Gu, X.; Gao, G.; Luo, J. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2010, 39 (Suppl. 1), D1114–D1117. [Google Scholar] [CrossRef]
- Nosaki, S.; Miyakawa, T.; Xu, Y.Q.; Nakamura, A.; Hirabayashi, K.; Asami, T.; Nakano, T.; Tanokura, M. Structural basis for brassinosteroid response by BIL1/BZR1. Nat. Plants 2018, 4, 771–776. [Google Scholar] [CrossRef]
- Yan, Z.; Zhao, J.; Peng, P.; Chihara, R.K.; Li, J. BIN2 functions redundantly with other arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009, 150, 710–721. [Google Scholar] [CrossRef]
- Gampala, S.S.; Kim, T.-W.; He, J.-X.; Tang, W.; Deng, Z.; Bai, M.-Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef]
- Ryu, H.; Kim, K.; Cho, H.; Park, J.; Choe, S.; Hwang, I. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 2007, 19, 2749–2762. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.-W.; Zhou, H.-W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Min, H.J.; Cui, L.H.; Oh, T.R.; Kim, J.H.; Kim, T.W.; Kim, W.T. OsBZR1 turnover mediated by OsSK22-regulated U-box E3 ligase OsPUB24 in rice BR response. Plant J. 2019, 99, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Ji, Y.; Hu, J.; Guo, R.; Sun, S.; Wang, X. Strigolactones and brassinosteroids antagonistically regulate the stability of the D53–OsBZR1 complex to determine FC1 expression in rice tillering. Mol. Plant 2020, 13, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.-Q.; et al. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.; Cai, Z.; Hu, Y.; Nolan, T.; Yu, F.; Yin, Y.; Xie, Q.; Tang, G.; Wang, X. SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 2017, 41, 47–58. [Google Scholar] [CrossRef]
- Feng, Z.; Shi, H.; Lv, M.; Ma, Y.; Li, J. Protein farnesylation negatively regulates brassinosteroid signaling via reducing BES1 stability in Arabidopsis thaliana. J. Integr. Plant Biol. 2021, 63, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.R.; Wang, X. Multiple ways of BES1/BZR1 degradation to decode distinct developmental and environmental cues in plants. Mol. Plant. 2017, 10, 915–917. [Google Scholar] [CrossRef]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46. [Google Scholar] [CrossRef]
- Zhang, L.; Han, Q.; Xiong, J.; Zheng, T.; Han, J.; Zhou, H.; Lin, H.; Yin, Y.; Zhang, D. Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 2282–2292. [Google Scholar] [CrossRef]
- Kono, A.; Yin, Y. Updates on BES1/BZR1 regulatory networks coordinating plant growth and stress responses. Front. Plant Sci. 2020, 11, 617162. [Google Scholar] [CrossRef]
- Srivastava, M.; Srivastava, A.K.; Orosa-Puente, B.; Campanaro, A.; Zhang, C.; Sadanandom, A. SUMO conjugation to BZR1 enables brassinosteroid signaling to integrate environmental cues to shape plant growth. Curr. Biol. 2020, 30, 1410–1423. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Rozhon, W.; Unterholzner, S.J.; Chen, T.; Eremina, M.; Wurzinger, B.; Bachmair, A.; Teige, M.; Sieberer, T.; Isono, E.; et al. Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 2014, 5, 4687. [Google Scholar] [CrossRef] [PubMed]
- Morrell, R.; Sadanandom, A. Dealing with stress: A review of plant SUMO proteases. Front. Plant Sci. 2019, 10, 1122. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, N.; Wang, M.; Zhou, W.; Guo, J.; Han, C.; Zhou, C.; Wang, W.; Wu, S.; Tang, W.; et al. Integrated regulation of periclinal cell division by transcriptional module of BZR1-SHR in Arabidopsis roots. New Phytol. 2021, 233, 795–808. [Google Scholar] [CrossRef]
- Nishida, K.; Frith, M.; Nakai, K. Pseudocounts for transcription factor binding sites. Nucleic Acids Res. 2008, 37, 939–944. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; López-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- He, J.-X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.L.; Gendron, N.; Sun, C.Q.; Wang, Z.-Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef]
- Furuya, T.; Saito, M.; Uchimura, H.; Satake, A.; Nosaki, S.; Miyakawa, T.; Shimadzu, S.; Yamori, W.; Tanokura, M.; Fukuda, H.; et al. Gene co-expression network analysis identifies BEH3 as a stabilizer of secondary vascular development in Arabidopsis. Plant Cell 2021, 33, 2618–2636. [Google Scholar] [CrossRef]
- Nosaki, S.; Terada, T.; Nakamura, A.; Hirabayashi, K.; Xu, Y.; Bui, T.B.C.; Nakano, T.; Tanokura, M.; Miyakawa, T. Highlighting the potential utility of MBP crystallization chaperone for Arabidopsis BIL1/BZR1 transcription factor-DNA complex. Sci. Rep. 2021, 11, 3879. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Zhao, J.; Zhu, Y.; Asami, T.; Li, J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J. Biol. Chem. 2010, 285, 24646–24653. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.-Y.; Ryu, H.; Hwang, I.; Wang, Z.-Y. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 2014, 5, 4140. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, H.; Kim, K.; Hwang, I. Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 2010, 29, 283–290. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef]
- Vilarrasa-Blasi, J.; González-García, M.P.; Frigola, D.; Fàbregas, N.; Alexiou Konstantinos, G.; López-Bigas, N.; Rivas, S.; Jauneau, A.; Lohmann Jan, U.; Benfey Philip, N.; et al. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell 2014, 30, 36–47. [Google Scholar] [CrossRef]
- Galstyan, A.; Nemhauser, J.L. Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4. Plant Direct 2019, 3, e00166. [Google Scholar] [CrossRef]
- Van Nguyen, T.; Park, C.R.; Lee, K.H.; Lee, S.; Kim, C.S. BES1/BZR1 homolog 3 cooperates with E3 ligase AtRZF1 to regulate osmotic stress and brassinosteroid responses in Arabidopsis. J. Exp. Bot. 2020, 72, 636–653. [Google Scholar] [CrossRef]
- Otani, Y.; Kawanishi, M.; Kamimura, M.; Sasaki, A.; Nakamura, Y.; Nakamura, T.; Okamoto, S. Behavior and possible function of Arabidopsis BES1/BZR1 homolog 2 in brassinosteroid signaling. Plant Signal. Behav. 2022, 17, 2084277. [Google Scholar] [CrossRef] [PubMed]
- Lachowiec, J.; Mason, G.A.; Schultz, K.; Queitsch, C. Redundancy, feedback, and robustness in the Arabidopsis thaliana BZR/BEH gene family. Front. Genet. 2018, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 2013, 27, 681–688. [Google Scholar] [CrossRef]
- Clouse, S.D.; Sasse, J.M. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef]
- Hacham, Y.; Holland, N.; Butterfield, C.; Tomas, S.U.; Bennett, M.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.-Y.; Bai, M.-Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Bai, M.-Y.; Oh, E.; Zhu, J.-Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef]
- Wei, Z.; Li, J. Brassinosteroids regulate root growth, development, and symbiosis. Mol. Plant 2016, 9, 86–100. [Google Scholar] [CrossRef]
- Yamamoto, R.; Fujioka, S.; Demura, T.; Takatsuto, S.; Yoshida, S.; Fukuda, H. Brassinosteroid levels increase drastically prior to morphogenesis of tracheary elements. Plant Physiol. 2001, 125, 556–563. [Google Scholar] [CrossRef]
- Cai, H.; Liu, L.; Huang, Y.; Zhu, W.; Qi, J.; Xi, X.; Aslam, M.; Dresselhaus, T.; Qin, Y. Brassinosteroid signaling regulates female germline specification in Arabidopsis. Curr. Biol. 2022, 32, 1102–1114. [Google Scholar] [CrossRef]
- Jia, D.; Chen, L.; Yin, G.; Yang, X.; Gao, Z.; Guo, Y.; Sun, Y.; Tang, W. Brassinosteroids regulate outer ovule integument growth in part via the control of INNER NO OUTER by BRASSINOZOLE-RESISTANT family transcription factors. J. Integr. Plant Biol. 2020, 63, 1353–1366. [Google Scholar]
- Graeff, M.; Rana, S.; Wendrich, J.R.; Dorier, J.; Eekhout, T.; Aliaga Fandino, A.C.; Guex, N.; Bassel, G.W.; De Rybel, B.; Hardtke, C.S. A single-cell morpho-transcriptomic map of brassinosteroid action in the Arabidopsis root. Mol. Plant 2021, 14, 1985–1999. [Google Scholar] [CrossRef]
- Vukašinović, N.; Wang, Y.; Vanhoutte, I.; Fendrych, M.; Guo, B.; Kvasnica, M.; Jiroutová, P.; Oklestkova, J.; Strnad, M.; Russinova, E. Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 2021, 7, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Kui, H.; Li, J. Receptor-like kinases in root development: Current progress and future directions. Mol. Plant 2020, 14, 166–185. [Google Scholar] [CrossRef]
- Yang, S.-L.; Xie, L.-F.; Mao, H.-Z.; Puah, C.S.; Yang, W.-C.; Jiang, L.; Sundaresan, V.; Ye, D. TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 2003, 15, 2792–2804. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; Jiang, L.X.; Puah, C.S.; Xie, L.F.; Zhang, X.Q.; Chen, L.Q.; Yang, W.-C.; Ye, D. Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS. Plant Physiol. 2005, 139, 186–191. [Google Scholar] [CrossRef]
- Colcombet, J.; Boisson-Dernier, A.; Ros-Palau, R.; Vera, C.E.; Schroeder, J.I. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 2005, 17, 3350–3361. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, T.; Linstroth, L.; Tillman, Z.; Otegui, M.S.; Owen, H.A.; Zhao, D. Control of anther cell differentiation by the small protein ligand TPD1 and its receptor EMS1 in Arabidopsis. PLoS Genet. 2016, 12, e1006147. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Huang, J.; Ahsan, N.; Biener, G.; Paprocki, J.; Thelen, J.J.; Raicu, V.; Zhao, D. Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol. 2016, 173, 326–337. [Google Scholar] [CrossRef]
- Fisher, K.; Turner, S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007, 17, 1061–1066. [Google Scholar] [CrossRef]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Fujita, T.; Sugiyama, M.; Fukuda, H. A novel system for xylem cell differentiation in Arabidopsis thaliana. Mol. Plant. 2015, 8, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 redundantly promote phloem and xylem differentiation. Plant Cell Physiol. 2018, 59, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Villalon, A.; Gujas, B.; Kang, Y.H.; Breda, A.S.; Cattaneo, P.; Depuydt, S.; Hardtke, C.S. Molecular genetic framework for protophloem formation. Proc. Natl. Acad. Sci. USA 2014, 111, 11551–11556. [Google Scholar] [CrossRef] [PubMed]
- Anne, P.; Azzopardi, M.; Gissot, L.; Beaubiat, S.; Hématy, K.; Palauqui, J.-C. OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol. 2015, 25, 2584–2590. [Google Scholar] [CrossRef]
- Hu, J.; Hu, X.; Yang, Y.; He, C.; Hu, J.; Wang, X. Strigolactone signaling regulates cambial activity through repression of WOX4 by transcription factor BES1. Plant Physiol. 2021, 188, 255–267. [Google Scholar] [CrossRef]
- Tamaki, T.; Oya, S.; Naito, M.; Ozawa, Y.; Furuya, T.; Saito, M.; Sato, M.; Wakazaki, M.; Toyooka, K.; Fukuda, H.; et al. VISUAL-CC system uncovers the role of GSK3 as an orchestrator of vascular cell type ratio in plants. Commun. Biol. 2020, 3, 184. [Google Scholar] [CrossRef]
- Truernit, E.; Bauby, H.; Belcram, K.; Barthélémy, J.; Palauqui, J.C. OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 2012, 139, 1306–1315. [Google Scholar] [CrossRef]
- Ruiz Sola, M.A.; Coiro, M.; Crivelli, S.; Zeeman, S.C.; Schmidt Kjølner Hansen, S.; Truernit, E. OCTOPUS-LIKE 2, a novel player in Arabidopsis root and vascular development, reveals a key role for OCTOPUS family genes in root metaphloem sieve tube differentiation. New Phytol. 2017, 216, 1191–1204. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, S.; Song, W.; Lin, G.; Wang, W.; Zhang, H.; Shan, L.; Chai, J. Functional and structural characterization of a receptor-like kinase involved in germination and cell expansion in Arabidopsis. Front. Plant Sci. 2017, 8, 1999. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.; Jiang, H.; Tang, B.; Zhang, M.; Li, Z.; Yin, Y. The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 2019, 31, 1788–1806. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Guerra, M.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. Delving into the evolutionary origin of steroid sensing in plants. Curr. Opin. Plant Biol. 2020, 57, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Furumizu, C.; Sawa, S. Insight into early diversification of leucine-rich repeat receptor-like kinases provided by the sequenced moss and hornwort genomes. Plant Mol. Biol. 2021, 107, 337–353. [Google Scholar] [CrossRef]
- Liang, T.; Shi, C.; Peng, Y.; Tan, H.; Xin, P.; Yang, Y.; Wang, F.; Li, X.; Chu, J.; Huang, J.; et al. Brassinosteroid-activated BRI1-EMS-SUPPRESSOR 1 inhibits flavonoid biosynthesis and coordinates growth and UV-B Stress responses in plants. Plant Cell 2020, 32, 3224–3239. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lu, X.; Li, L.; Lian, H.; Mao, Z.; Xu, P.; Guo, T.; Xu, F.; Du, S.; Cao, X.; et al. Photoexcited CRYPTOCHROME1 signaling and photomorphogenesis in Arabidopsis. Plant Cell 2018, 30, 1989–2005. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, W.; Xu, P.; Pan, J.; Zhang, T.; Li, Y.; Li, G.; Yang, H.; Lian, H. phyB interacts with BES1 to regulate brassinosteroid signaling in Arabidopsis. Plant Cell Physiol. 2018, 60, 353–366. [Google Scholar] [CrossRef]
- Chen, K.; Łyskowski, A.; Jaremko, Ł.; Jaremko, M. Genetic and molecular factors determining grain weight in rice. Front Plant Sci. 2021, 12, 605799. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Li, X.; Lv, M.; Li, J. BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways. Int. J. Mol. Sci. 2022, 23, 10149. https://doi.org/10.3390/ijms231710149
Shi H, Li X, Lv M, Li J. BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways. International Journal of Molecular Sciences. 2022; 23(17):10149. https://doi.org/10.3390/ijms231710149
Chicago/Turabian StyleShi, Hongyong, Xiaopeng Li, Minghui Lv, and Jia Li. 2022. "BES1/BZR1 Family Transcription Factors Regulate Plant Development via Brassinosteroid-Dependent and Independent Pathways" International Journal of Molecular Sciences 23, no. 17: 10149. https://doi.org/10.3390/ijms231710149




