Comparative Physiological and Transcriptomic Analyses of Two Contrasting Pepper Genotypes under Salt Stress Reveal Complex Salt Tolerance Mechanisms in Seedlings
Abstract
:1. Introduction
2. Results
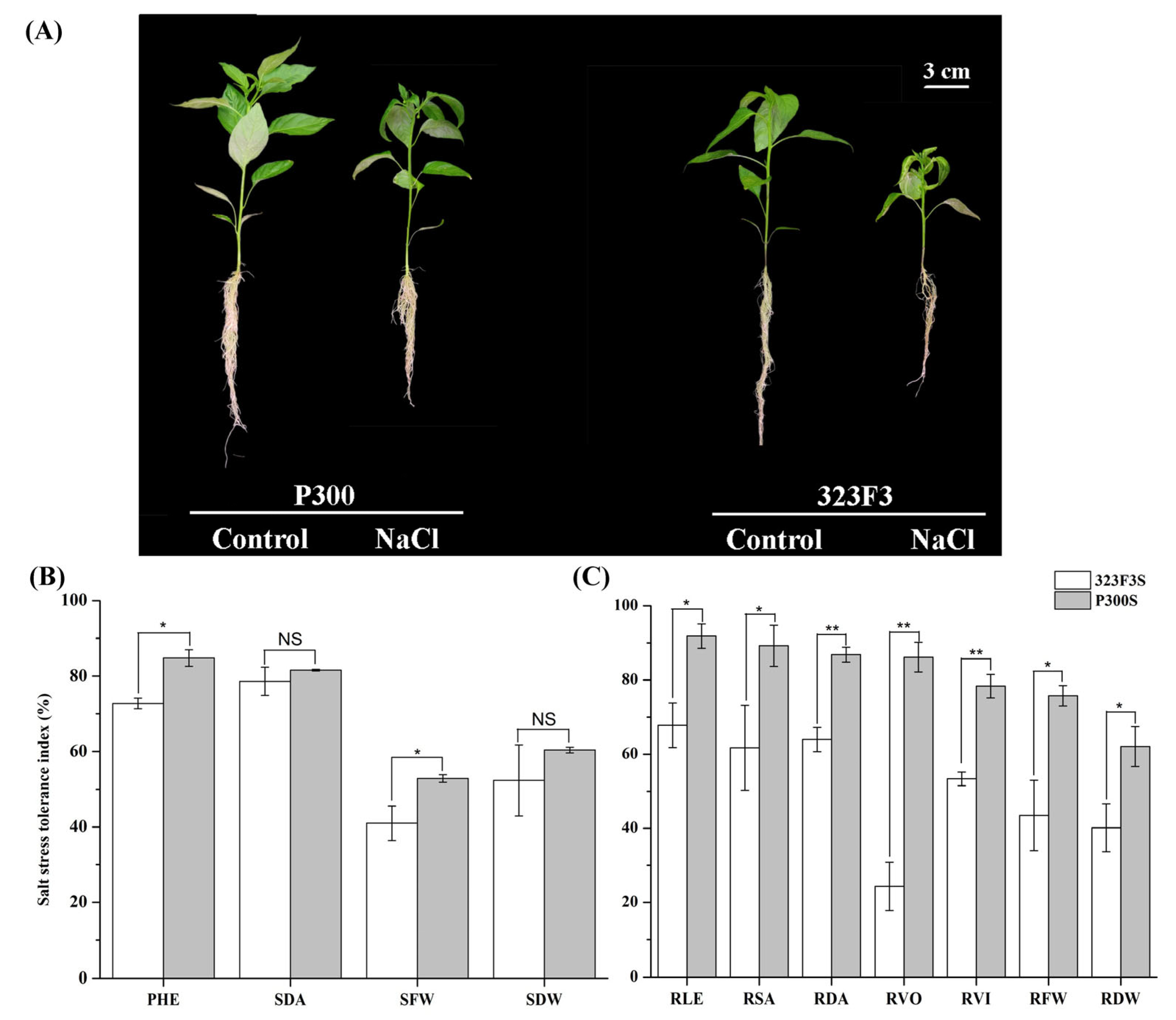
2.1. Performance of 323F3 and P300 under Salt Stress
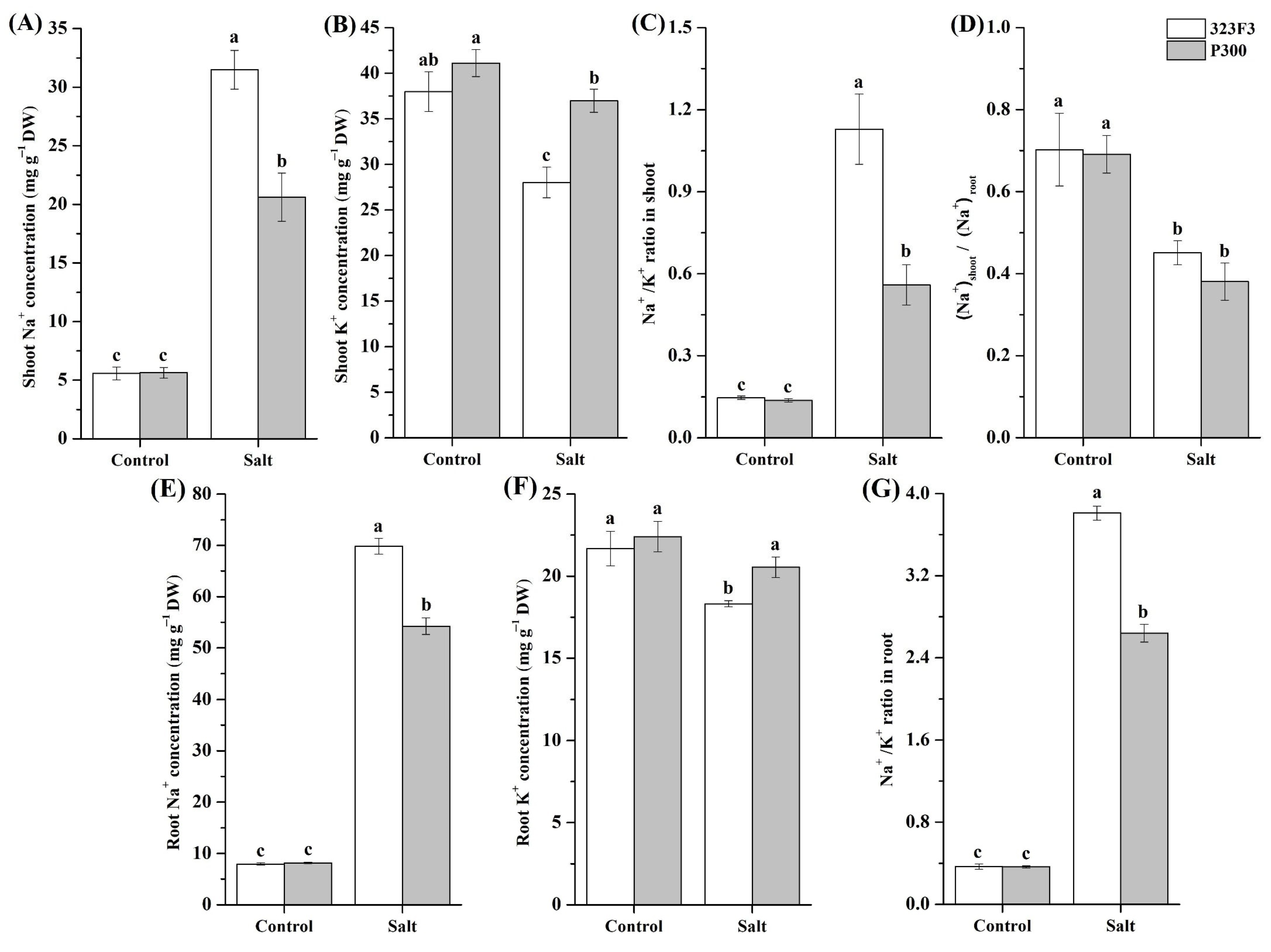
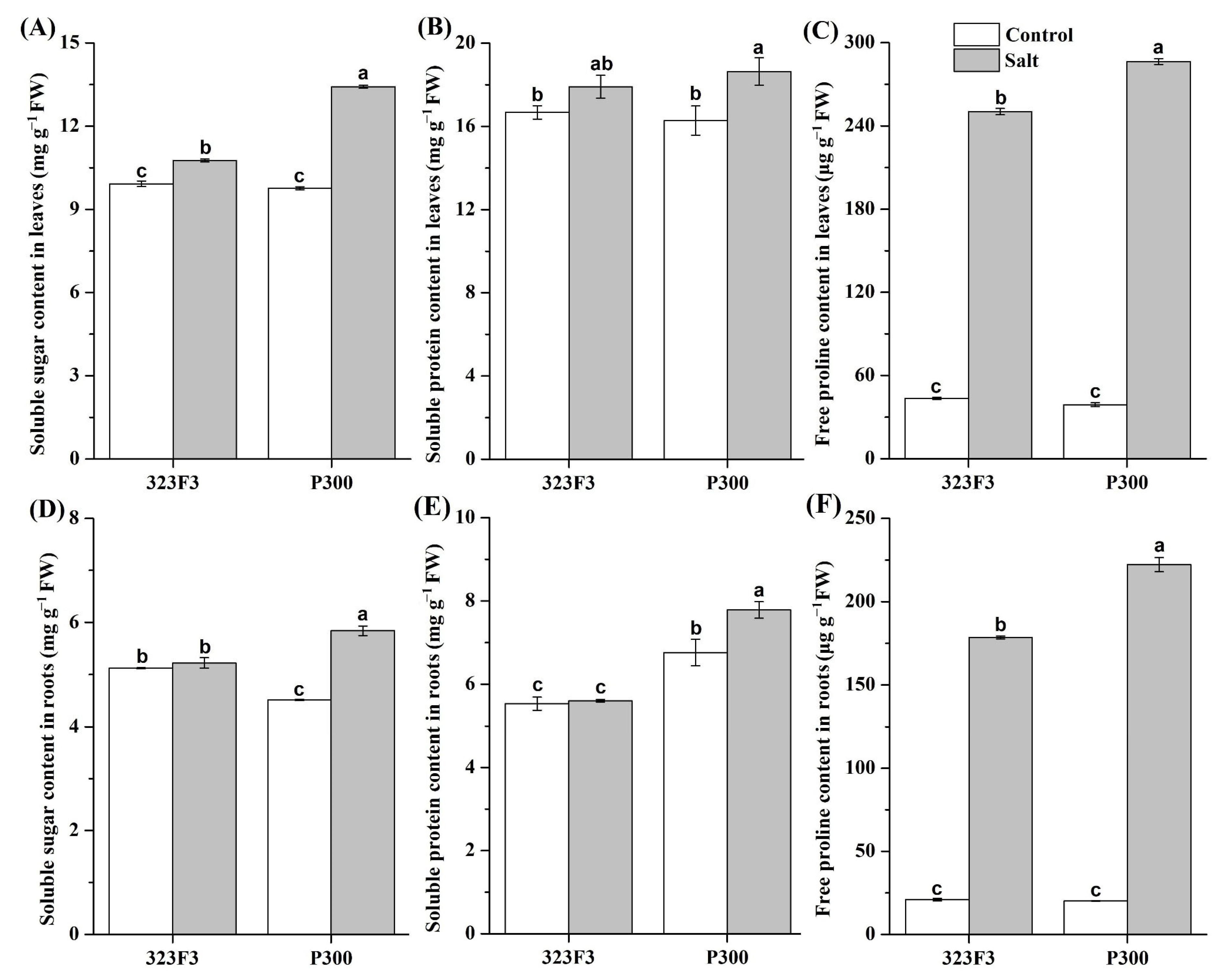
2.2. Physiological Responses to NaCl Stress in 323F3 and P300
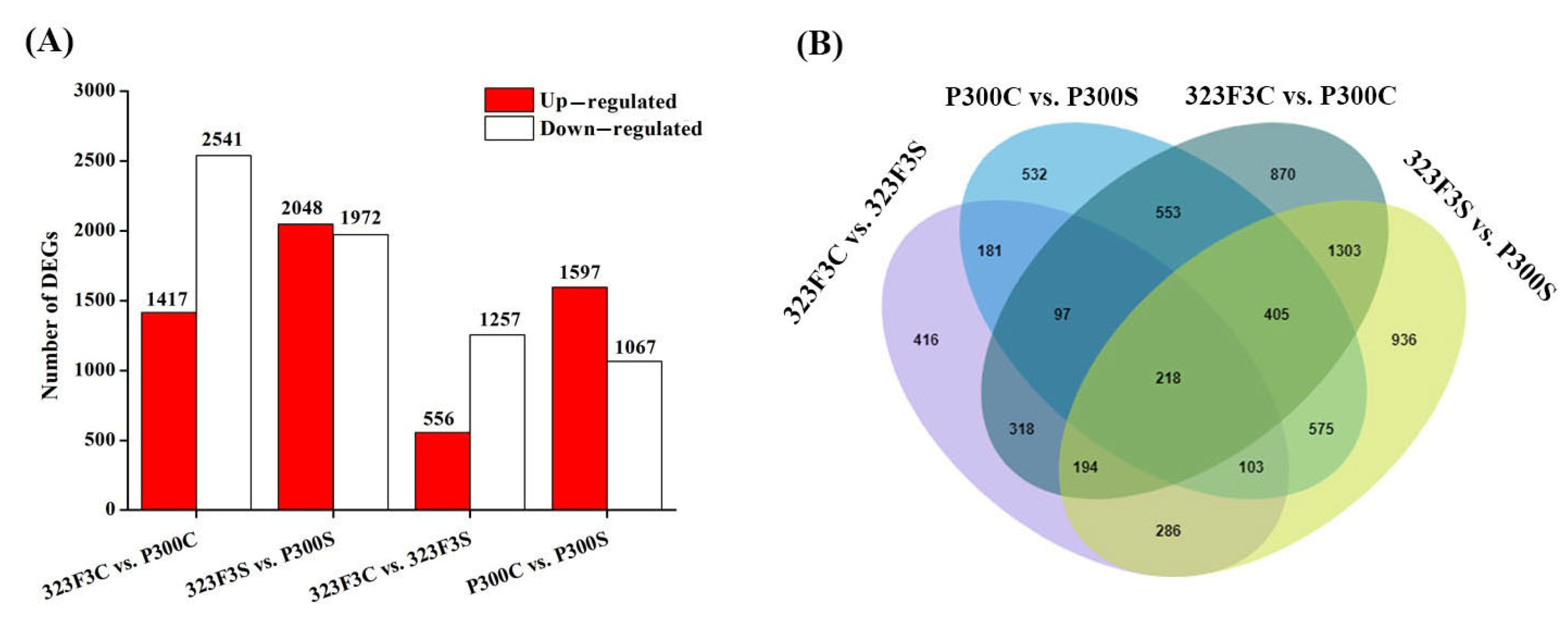
2.3. RNA-seq Analysis and Identification of DEGs
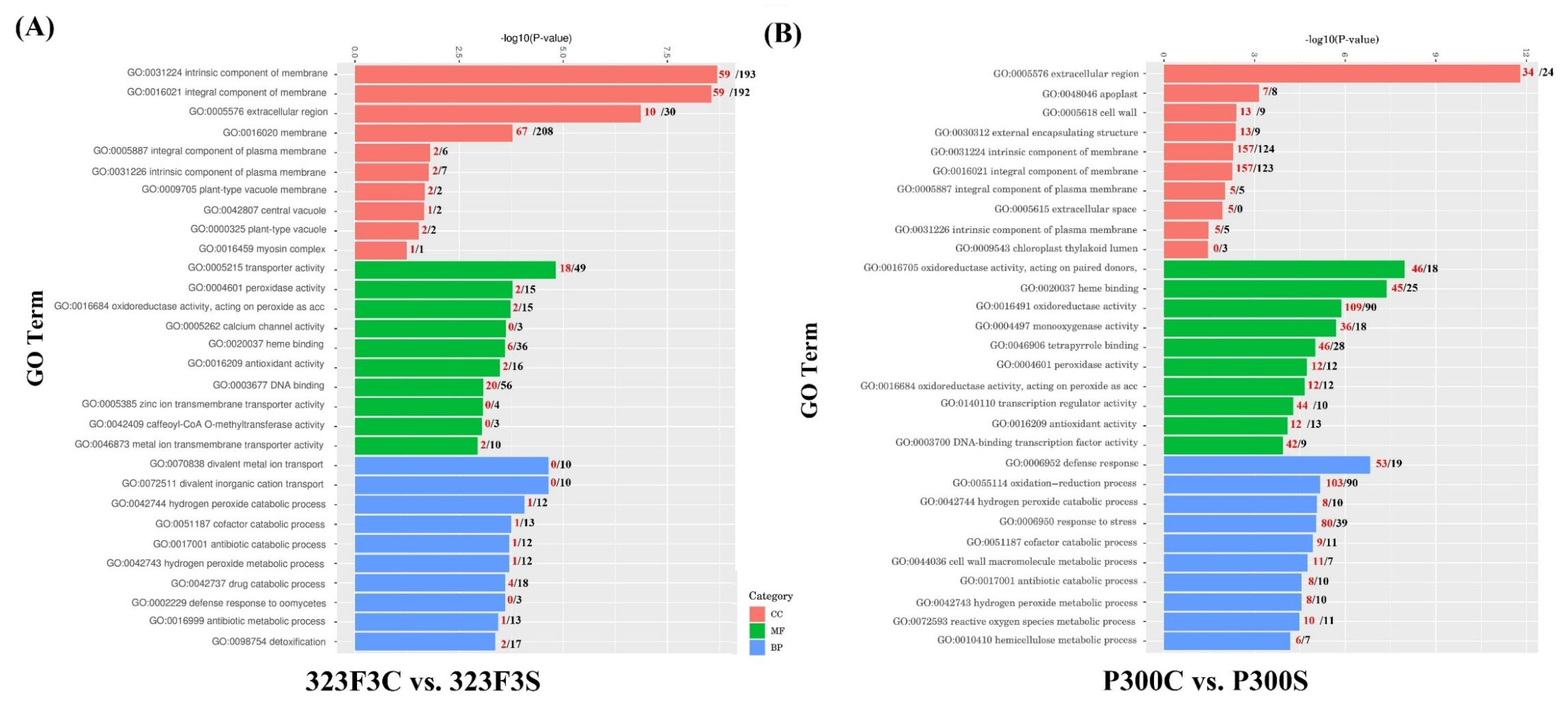
2.4. Gene Ontology Enrichment Analysis of DEGs
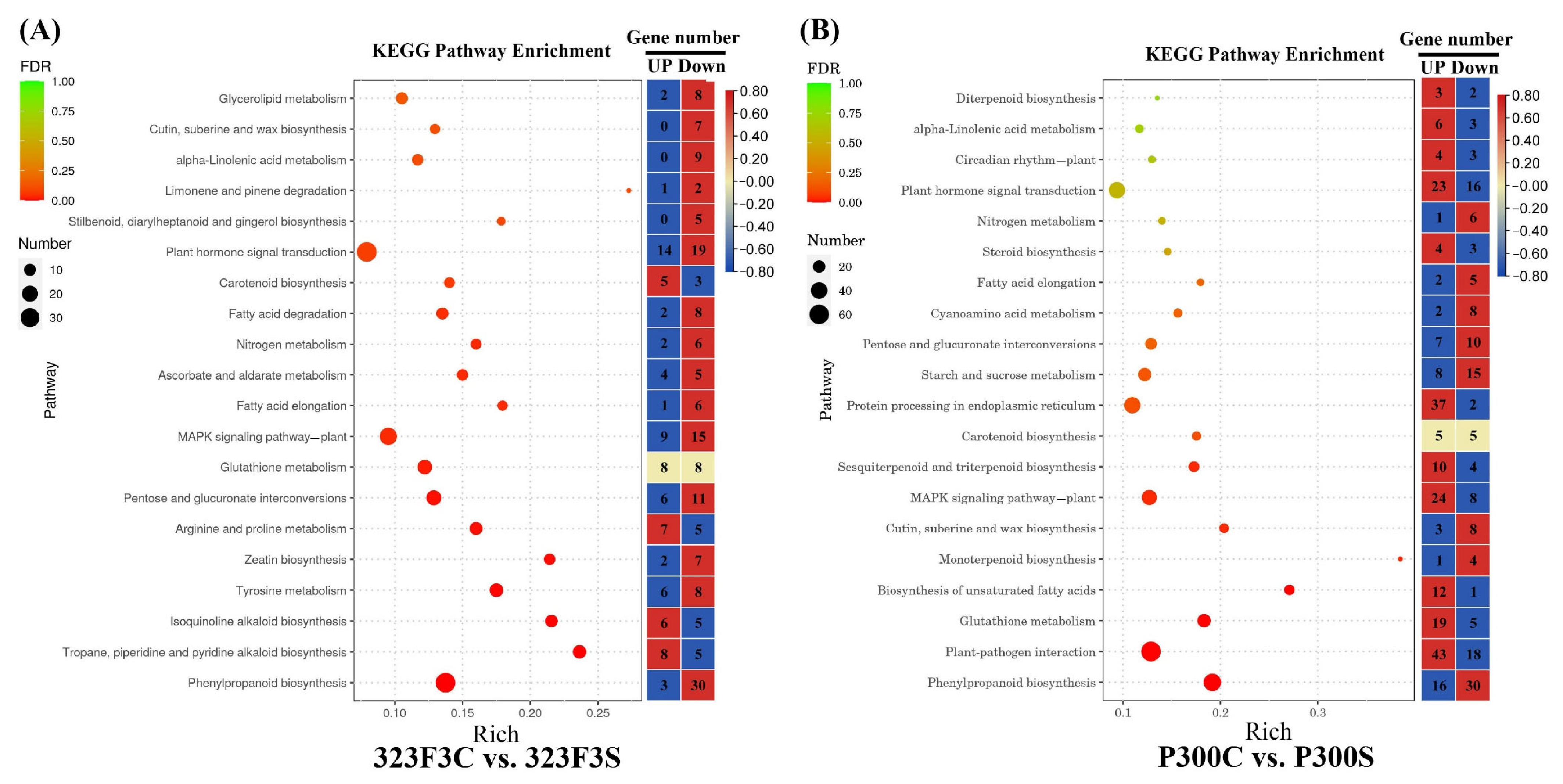
2.5. Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis of DEGs
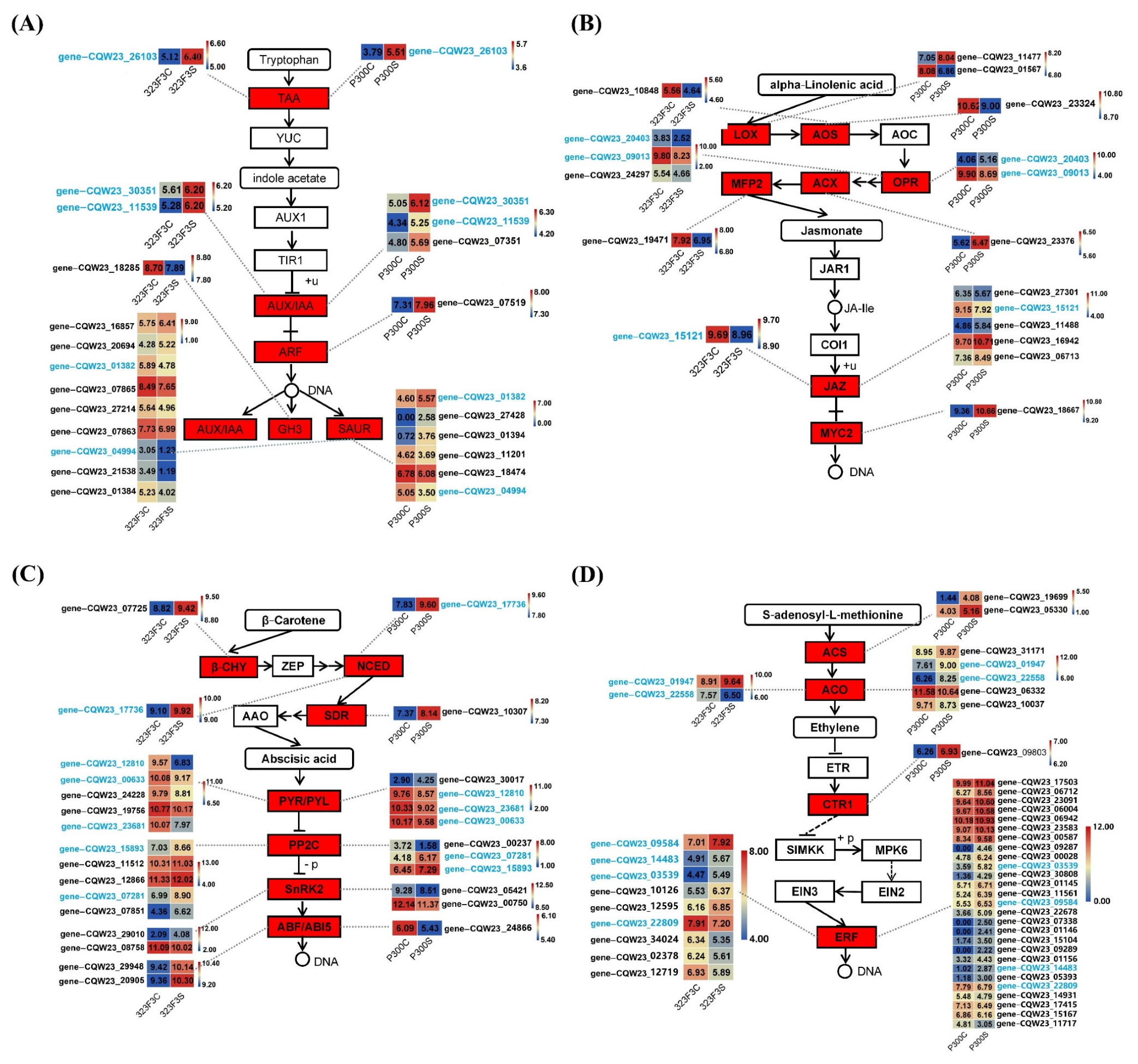
2.6. Analysis of DEGs Associated with Plant Hormones
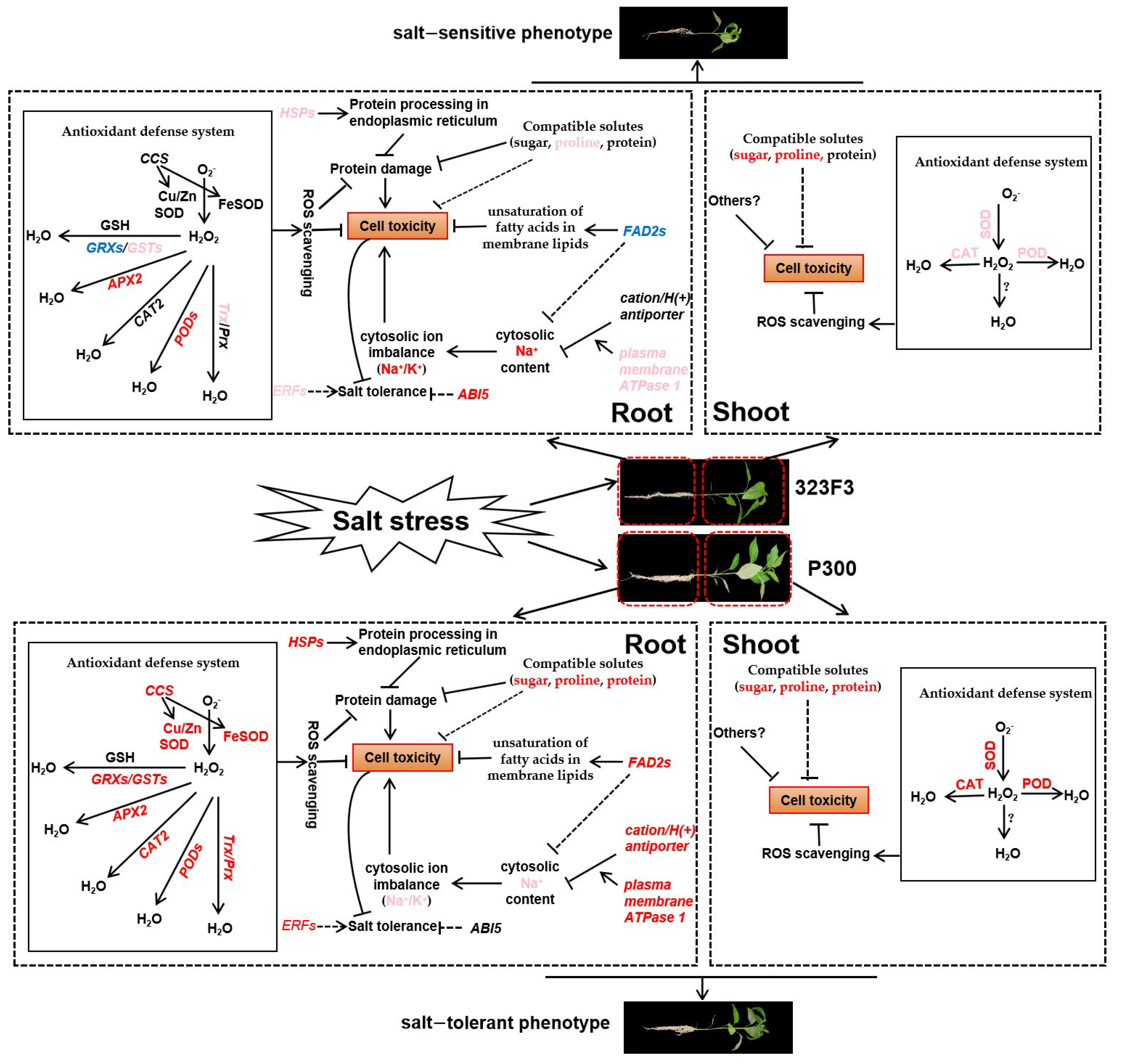
2.7. Analysis of DEGs Associated with ROS Scavenging
2.8. Analysis of Fatty Acid Desaturase and Heat-Shock-Protein-Related DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Salt Stress Treatment
4.2. Physiological Analysis
4.3. RNA Extraction and Transcriptome Analysis
4.4. Quantitative Real-Time PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUX/IAA | Auxin/indole-3-acetic acid |
| JA | Jasmonic acid |
| ABA | Abscisic acid |
| NCED | 9-cis-epoxycarotenoid dioxygenase |
| ABI5 | Abscisic acid insensitive 5 |
| ETH | Ethylene |
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| ACO | 1-aminocyclopropane-1-carboxylate oxidase |
| ERF | Ethylene response factor |
| DEG | Differential expressed gene |
| ROS | Reactive oxygen species |
| POD | Peroxidase |
| SOD | superoxide dismutase |
| CAT | catalase |
| APX | Ascorbate peroxidase |
| GST | Glutathione S-transferase |
| GSH | Glutathione |
| GRX | Glutaredoxin |
| Trx | Thioredoxin |
| Prx | Peroxiredoxin |
| HSP | Heat-shock protein |
| FAD | Fatty acid desaturase |
| CCS | Copper chaperone for superoxide dismutase |
| CPA | Cation/H+ antiporter |
| PM-ATPase | Plasma membrane ATPase |
| MDA | Malondialdehyde |
| REC | Relative electric conductivity |
| RNA-seq | RNA sequencing |
| SOS | Salt overly sensitive |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| qRT-PCR | Quantitative real-time PCR |
| 323F3C | Control group of 323F3 |
| 323F3S | Salt treatment group of cmh15 |
| P300C | Control group of P300 |
| P300S | Salt treatment group of P300 |
References
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Huang, B.R. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef]
- Cushman, J.C.; Bohnert, H.J. Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 2000, 3, 117–124. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.; Xiang, X.; Chen, J.; Rong, J. Genetic diversity of crop wild relatives under threat in Yangtze River Basin: Call for enhanced in situ conservation and utilization. Mol. Plant. 2019, 12, 1535–1538. [Google Scholar] [CrossRef]
- Perrino, E.V.; Wagensommer, R.P. Crop wild relatives (CWRs) threatened and endemic to Italy: Urgent actions for protection and use. Biology 2022, 11, 193. [Google Scholar] [CrossRef]
- Abenavoli, L.; Milanovic, M.; Procopio, A.C.; Spampinato, G.; Maruca, G.; Perrino, E.V.; Mannino, G.C.; Fagoonee, S.; Luzza, F.; Musarella, C.M. Ancient wheats: Beneficial effects on insulin resistance. Minerva Med. 2021, 112, 641–650. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Goudarzi, M.; Pakniyat, H. Salinity causes increase in proline and protein contents and peroxidase activity in wheat Cultivars. J. Appl. Sci. 2009, 9, 348–353. [Google Scholar] [CrossRef]
- Sakr, M.T.; El-Sarkassy, N.M.; Fuller, M.P. Osmoregulators proline and glycine betaine counteract salinity stress in canola. Agron. Sustain. Dev. 2012, 32, 747–754. [Google Scholar] [CrossRef]
- Zhang, M.; Fang, Y.; Ji, Y.; Jiang, Z.; Wang, L. Effects of salt stress on ion content, antioxidant enzymes and protein profile in different tissues of Broussonetia papyrifera. S. Afr. J. Bot. 2013, 85, 1–9. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta Gene Struct. Expr. 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Allakhverdiev, S.I.; Nishiyama, Y.; Suzuki, I.; Tasaka, Y.; Murata, N. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc. Natl. Acad. Sci. USA 1999, 96, 5862–5867. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 516. [Google Scholar] [CrossRef]
- Cao, Y.R.; Chen, S.Y.; Zhang, J.S. Ethylene signaling regulates salt stress response: An overview. Plant Signal. Behav. 2008, 3, 761–763. [Google Scholar] [CrossRef] [Green Version]
- Freitas, V.S.; Miranda, R.D.; Costa, J.H.; de Oliveira, D.F.; Paula, S.D.; Miguel, E.D.; Freire, R.S.; Prisco, J.T.; Gomes-Filho, E. Ethylene triggers salt tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ. Exp. Bot. 2018, 145, 75–86. [Google Scholar] [CrossRef]
- Zhai, Y.; Wang, Y.; Li, Y.J.; Lei, T.T.; Yan, F.; Su, L.T.; Li, X.; Zhao, Y.; Sun, X.; Li, J.; et al. Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 2013, 513, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Guo, J.; Zhu, A.; Zhang, L.; Zhang, M. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014, 104, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C.F. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Carmody, M.; Waszczak, C.; Idänheimo, N.; Saarinen, T.; Kangasjärvi, J. ROS signalling in a destabilised world: A molecular understanding of climate change. J. Plant Physiol. 2016, 203, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Kibria, M.G.; Hossain, M.; Murata, Y.; Hoque, M.A. Antioxidant defense mechanisms of salinity tolerance in rice genotypes. Rice Sci. 2017, 24, 155–162. [Google Scholar] [CrossRef]
- Hassen, A.; Maher, S.; Cherif, H. Effect of salt stress (NaCl) on germination and early seedling parameters of three pepper cultivars (Capsicum annuum L.). J. Stress Physiol. Biochem. 2014, 10, 14–25. [Google Scholar]
- Kaouther, Z.; Nina, H.; Ahmad, R.; Cherif, H. Evaluation of salt tolerance (NaCl) in Tunisian chili pepper (Capsicum frutescens L.) on growth, mineral analysis and solutes synthesis. J. Stress Physiol. Biochem. 2013, 9, 209–228. [Google Scholar]
- Giorio, P.; Cirillo, V.; Caramante, M.; Oliva, M.; Guida, G.; Venezia, A.; Grillo, S.; Maggio, A.; Albrizio, R. Physiological basis of salt stress tolerance in a landrace and a commercial variety of sweet pepper (Capsicum annuum L.). Plants 2020, 9, 795. [Google Scholar] [CrossRef] [PubMed]
- Genzel, F.; Dicke, M.D.; Junker-Frohn, L.V.; Neuwohner, A.; Thiele, B.; Putz, A.; Usadel, B.; Wormit, A.; Wiese-Klinkenberg, A. Impact of moderate cold and salt stress on the accumulation of antioxidant flavonoids in the leaves of two capsicum cultivars. J. Agric. Food Chem. 2021, 69, 6431–6443. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, L.; Zhang, X.; Wu, S.; Han, X.; Li, X.; Xu, J. Comparative transcriptomics analysis of roots and leaves under Cd stress in Calotropis gigantea L. Int. J. Mol. Sci. 2022, 23, 3329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, J.; Huang, Y.; Wu, H.; Hu, X.; Cheng, B.; Ma, Q.; Zhao, Y. Comparative transcriptomics Reveals the molecular mechanism of the parental lines of maize hybrid An’nong876 in response to salt stress. Int. J. Mol. Sci. 2022, 23, 5231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in Plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef]
- Shi, H.; Zhu, J.K. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol. Biol. 2002, 50, 543–550. [Google Scholar] [CrossRef]
- Møller, J.V.; Juul, B.; Le Maire, M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta. 1996, 1286, 1–51. [Google Scholar] [CrossRef]
- Li, J.; Guo, Y.; Yang, Y. The molecular mechanism of plasma membrane H+-ATPases in plant responses to abiotic stress. J. Genet. Genom. 2022, 49, 35654346. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar] [CrossRef]
- Cho, S.C.; Chao, Y.Y.; Hong, C.Y.; Kao, C.H. The role of hydrogen peroxide in cadmium-inhibited root growth of rice seedlings. Plant Growth Regul. 2012, 66, 27–35. [Google Scholar] [CrossRef]
- Kim, S.; Lim, J.H.; Park, M.; Kim, Y.; Park, T.; Seo, Y.; Choi, K.; Yun, S. Enhanced antioxidant enzymes are associated with reduced hydrogen peroxide in barley roots under saline stress. J. Biochem. Mol. Biol. 2005, 38, 218–224. [Google Scholar] [CrossRef]
- Chu, C.C.; Lee, W.C.; Guo, W.Y.; Pan, S.M.; Chen, L.J.; Li, H.M.; Jinn, T.L. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef]
- Cohu, C.M.; Abdel-Ghany, S.E.; Gogolin Reynolds, K.A.; Onofrio, A.M.; Bodecker, J.R.; Kimbrel, J.A.; Niyogi, K.K.; Pilon, M. Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: Regulation and unexpected phenotypes in an Arabidopsis mutant. Mol. Plant 2009, 2, 1336–1350. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, C.; Xue, X.; Xu, M.; Li, J.; Wu, J.X. Overexpression of a cytosolic ascorbate peroxidase gene, OsAPX2, increases salt tolerance in transgenic alfalfa. J. Integr. Agric. 2014, 13, 2500–2507. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.I.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Yang, G.; Wang, Y.; Xia, D.; Gao, C.; Wang, C.; Yang, C. Overexpression of a GST gene (ThGSTZ1) from Tamarix hispida improves drought and salinity tolerance by enhancing the ability to scavenge reactive oxygen species. Plant Cell Tiss. Organ Cult. 2014, 117, 99–112. [Google Scholar] [CrossRef]
- Sevilla, F.; Camejo, D.; Ortiz-Espín, A.; Calderón, A.; Lázaro, J.J.; Jiménez, A. The thioredoxin/peroxiredoxin/sulfiredoxin system: Current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015, 66, 2945–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, P.; Yan, J.; Li, W.; Wang, L.; Zhao, J.; Ma, X.; Li, A.; Wang, H.; Kong, L. A member of wheat class III peroxidase gene family, TaPRX-2A, enhanced the tolerance of salt stress. BMC Plant Biol. 2020, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]
- Siddikee, M.A.; Chauhan, P.S.; Sa, T. Regulation of ethylene biosynthesis under salt stress in red pepper (Capsicum annuum L.) by 1-aminocyclopropane-1-carboxylic Acid (ACC) deaminase-producing halotolerant bacteria. J. Plant Growth Regul. 2012, 31, 265–272. [Google Scholar] [CrossRef]
- Naing, A.H.; Campol, J.R.; Kang, H.; Xu, J.; Chung, M.Y.; Kim, C.K. Role of ethylene biosynthesis genes in the regulation of salt stress and drought stress tolerance in petunia. Front. Plant Sci. 2022, 13, 844449. [Google Scholar] [CrossRef]
- Yang, R.; Liu, J.; Lin, Z.; Sun, W.; Wu, Z.; Hu, H.; Zhang, Y. ERF transcription factors involved in salt response in tomato. Plant Growth Regul. 2018, 84, 573–582. [Google Scholar] [CrossRef]
- Lu, C.W.; Shao, Y.; Li, L.; Chen, A.J.; Xu, W.Q.; Wu, K.J.; Luo, Y.B.; Zhu, B.Z. Overexpression of SlERF1 tomato gene encoding an ERF-type transcription activator enhances salt tolerance. Russ. J. Plant Physiol. 2011, 58, 118–125. [Google Scholar] [CrossRef]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Martínez-Pérez, A.; Albacete, A.; Martínez-Melgarejo, P.A.; Dodd, I.C.; Thompson, A.J.; Mohareb, F.; Estelles-Lopez, L.; Kevei, Z.; Ferrández-Ayela, A.; et al. Overproduction of ABA in rootstocks alleviates salinity stress in tomato shoots. Plant Cell Environ. 2021, 44, 2966–2986. [Google Scholar] [CrossRef] [PubMed]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Sun, J.; Li, B.; Zhu, Q.; Chen, S.; Zhang, H. Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS ONE 2012, 7, e30355. [Google Scholar] [CrossRef]
- Zhang, J.T.; Zhu, J.Q.; Zhu, Q.; Liu, H.; Gao, X.S.; Zhang, H.X. Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2009, 390, 469–474. [Google Scholar] [CrossRef]
- Mishra, D.; Shekhar, S.; Singh, D.; Chakraborty, S.; Chakraborty, N. Heat shock proteins and abiotic stress tolerance in plants. In Regulation of Heat Shock Protein Responses; Asea, A.A.A., Kaur, P., Eds.; Springer International Publishing: Cham, Switzerland, 2018; Volume 13, pp. 41–69. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Han, Y.; Chang, X.; Song, D.; Yao, Q. Study on salt tolerance screening of 120 Capsicum annuum L. germplasm resources at seedling stage. J. Henan Agric. Univ. 2020, 54, 793–802. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Han, Y.; Chang, X.; Yao, Q. Comprehensive evaluation on salt tolerance of 100 pepper germplasm resources and screening salt tolerant tarieties. Shandong Agric. Sci. 2020, 52, 7–15. [Google Scholar] [CrossRef]
- Clemensson-Lindell, A. Triphenyltetrazolium chloride as an indicator of fine-root vitality and environmental stress in coniferous forest stands: Applications and limitations. Plant Soil 1994, 159, 297–300. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Yemm, E.W.; Willis, A.J. The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 1954, 57, 508–514. [Google Scholar] [CrossRef]
- Read, S.M.; Northcote, D.H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal. Biochem. 1981, 116, 53–64. [Google Scholar] [CrossRef]
- Song, S.Y.; Chen, Y.; Chen, J.; Dai, X.Y.; Zhang, W.H. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Protein Type | Gene ID | Gene Description | Gene Name | 323F3C vs. 323F3S | P300C vs. P300S | ||
|---|---|---|---|---|---|---|---|
| Fold | p-Value | Fold | p-Value | ||||
| CAT | gene–CQW23_04867 | Catalase isozyme 2 | CAT2 | 1.24 | 3.99 × 10−3 | 1.88↑ | 8.86 × 10−25 |
| APX | gene–CQW23_21948 | L-ascorbate peroxidase 2, cytosolic | APX2 | 2.00↑ | 1.95 × 10−29 | 1.54↑ | 5.71 × 10−28 |
| Prx | gene–CQW23_27780 | Peroxiredoxin-2C | Prx-2C | 1.02 | 0.8449117 | 21.79↑ | 4.86 × 10−208 |
| Trx | gene–CQW23_08596 | Thioredoxin-like protein 4A | Trx-4A | 2.03↑ | 3.18 × 10−8 | 2.07↑ | 1.22 × 10−18 |
| gene–CQW23_23211 | TPR repeat-containing thioredoxin TTL1 | TTL1 | 1.62↑ | 3.72 × 10−3 | 1.01 | 0.96941233 | |
| gene–CQW23_34207 | TPR repeat-containing thioredoxin TDX | TDX | 0.78 | 0.6865073 | 11.13↑ | 1.86 × 10−4 | |
| gene–CQW23_18479 | TPR repeat-containing thioredoxin TDX | TDX | 0.72 | 0.2916553 | 7.81↑ | 8.08 × 10−3 | |
| POD | gene–CQW23_09144 | Suberization-associated anionic peroxidase 1 | POD1 | 0.59↓ | 6.22 × 10−10 | 2.34↑ | 1.65 × 10−60 |
| gene–CQW23_06899 | Peroxidase 28 | POD28 | 1.51↑ | 8.34 × 10−9 | 0.71 | 1.54 × 10−5 | |
| gene–CQW23_32462 | Peroxidase 4 | POD4 | 1.79↑ | 4.96 × 10−10 | 6.69↑ | 5.17 × 10−14 | |
| gene–CQW23_05116 | Peroxidase 15 | POD15 | 1.01 | 0.8866078 | 2.04↑ | 1.0371 × 10−52 | |
| gene–CQW23_08677 | Peroxidase 5 | POD5 | 1.41 | 1.44 × 10−5 | 2.16↑ | 4.59 × 10−44 | |
| gene–CQW23_07657 | Peroxidase 4 | POD4 | 1.19 | 0.7675621 | 3.34↑ | 0.02781563 | |
| gene–CQW23_18616 | Peroxidase 6 | POD6 | 1.10 | 0.858594 | 1.95↑ | 0.0165733 | |
| gene–CQW23_01286 | Lignin-forming anionic peroxidase | POD | 0.81 | 0.0972802 | 2.44↑ | 1.20 × 10−3 | |
| gene–CQW23_07523 | Peroxidase 66 | POD66 | 1.18 | 0.1376819 | 2.10↑ | 5.91 × 10−12 | |
| gene–CQW23_10318 | Peroxidase 12 | POD12 | 1.12 | 2.42 × 10−3 | 1.63↑ | 2.77 × 10−28 | |
| gene–CQW23_16671 | Peroxidase 72 | POD72 | 1.13 | 0.5396232 | Inf a↑ | 3.52 × 10−23 | |
| GRX | gene–CQW23_02281 | Glutaredoxin-C1 | GRX-C1 | 0.45↓ | 6.30 × 10−3 | 4.22↑ | 3.23 × 10−7 |
| gene–CQW23_22057 | Monothiol glutaredoxin-S2 | GRX-S2 | 0.59 | 0.3190946 | 2.78↑ | 1.93 × 10−3 | |
| gene–CQW23_17543 | Glutaredoxin-C9 | GRX-C9 | 0.68 | 3.47 × 10−3 | 1.62↑ | 0.01676165 | |
| GST | gene–CQW23_03952 | Putative glutathione S-transferase | GST | 1.57↑ | 3.96× 10−22 | 3.98↑ | 2.55 × 10−224 |
| gene–CQW23_21928 | Putative glutathione S-transferase | GST | 1.68↑ | 2.34 × 10−6 | 3.04↑ | 5.79 × 10−16 | |
| gene–CQW23_21929 | Putative glutathione S-transferase | GST | 1.64↑ | 0.0198286 | 2.63↑ | 1.56 × 10−6 | |
| gene–CQW23_22418 | Glutathione S-transferase U9 | GST-U9 | 1.93↑ | 6.24 × 10−30 | 4.57↑ | 5.97 × 10−145 | |
| gene–CQW23_25828 | Glutathione S-transferase U18 | GST-U18 | 1.71↑ | 1.46 × 10−74 | 1.09 | 1.99 × 10−3 | |
| gene–CQW23_24335 | Putative glutathione S-transferase | GST | 0.81 | 3.94 × 10−4 | 2.54↑ | 5.87 × 10−45 | |
| gene–CQW23_25825 | Glutathione S-transferase U18 | GST-U18 | 0.78 | 3.26 × 10−4 | 2.01↑ | 6.59 × 10−23 | |
| gene–CQW23_21930 | Putative glutathione S-transferase | GST | 1.45 | 1.55 × 10−13 | 1.86↑ | 2.62 × 10−15 | |
| gene–CQW23_04818 | Putative glutathione S-transferase | GST | 0.28 | 0.6107545 | 19.02↑ | 1.38 × 10−10 | |
| gene–CQW23_25838 | Glutathione S-transferase U18 | GST-U18 | 0.39 | 0.4708276 | 12.18↑ | 2.22 × 10−9 | |
| gene–CQW23_22421 | Glutathione S-transferase U9 | GST-U9 | 1.28 | 4.42 × 10−3 | 1.81↑ | 5.99 × 10−9 | |
| gene–CQW23_17729 | Putative glutathione S-transferase | GST | 0.48 | 1 | 13.81↑ | 1.63 × 10−7 | |
| gene–CQW23_02556 | Putative glutathione S-transferase | GST | 1.23 | 0.1406508 | 2.01↑ | 3.56 × 10−7 | |
| gene–CQW23_21533 | Putative glutathione S-transferase | GST | 1.61 | 0.0656395 | 3.64↑ | 5.19 × 10−5 | |
| gene–CQW23_21187 | Putative glutathione S-transferase | GST | 1.03 | 0.9176443 | 1.99↑ | 0.03055018 | |
| Protein Type | Gene ID | Gene Description | Gene Name | 323F3C vs. 323F3S | P300C vs. P300S | ||
|---|---|---|---|---|---|---|---|
| Fold | p-Value | Fold | p-Value | ||||
| HSP | gene–CQW23_08818 | Heat-shock cognate 70 kDa protein | HSP70 | 3.43↑ | 0 | 8.27↑ | 0 |
| gene–CQW23_14285 | Heat-shock protein 90-1 | HSP90-1 | 7.73↑ | 0 | 12.91↑ | 0 | |
| gene–CQW23_15919 | Heat-shock cognate 70 kDa protein | HSP70 | 2.37↑ | 0 | 2.74↑ | 0 | |
| gene–CQW23_08264 | 17.9 kDa class II heat-shock protein | HSP17.9 | 3.79↑ | 1.09 × 10−59 | 7.09↑ | 1.91 × 10−112 | |
| gene–CQW23_00534 | Heat-shock 22 kDa protein, mitochondrial | HSP22 | 2.60↑ | 1.04 × 10−50 | 6.99↑ | 6.98 × 10−126 | |
| gene–CQW23_08265 | 17.9 kDa class II heat-shock protein | HSP17.9 | 5.09↑ | 2.16 × 10−21 | 10.94↑ | 6.73 × 10−123 | |
| gene–CQW23_08902 | 15.7 kDa heat-shock protein, peroxisomal | HSP15.7 | 3.02↑ | 4.62 × 10−21 | 3.19↑ | 3.39 × 10−45 | |
| gene–CQW23_31624 | 18.5 kDa class I heat-hock protein | HSP18.5 | 2.02↑ | 1.17 × 10−19 | 4.86↑ | 2.08 × 10−100 | |
| gene–CQW23_11457 | 17.4 kDa class III heat-shock protein | HSP17.4 | 4.90↑ | 9.92 × 10−17 | 5.01↑ | 6.07 × 10−17 | |
| gene–CQW23_00535 | Heat-shock 22 kDa protein, mitochondrial | HSP22 | 2.64↑ | 7.74 × 10−14 | 5.95↑ | 1.085 × 10−87 | |
| gene–CQW23_08718 | 22.0 kDa class IV heat-shock protein | HSP22 | 2.01↑ | 5.70 × 10−4 | 1.69↑ | 2.88 × 10−5 | |
| gene–CQW23_31623 | 18.5 kDa class I heat-shock protein | HSP18.5 | 2.68↑ | 1.03 × 10−3 | 2.31↑ | 2.09 × 10−4 | |
| gene–CQW23_11673 | Heat-shock 70 kDa protein | HSP70 | 1.99↑ | 0.015001 | 2.44↑ | 0.02448657 | |
| gene–CQW23_23188 | Heat-shock protein 90-2 | HSP90-1 | 1.37 | 4.04 × 10−76 | 1.58↑ | 6.33 × 10−106 | |
| gene–CQW23_24043 | Heat-shock cognate 70 kDa protein | HSP70 | 1.29 | 4.27 × 10−45 | 1.51↑ | 6.56 × 10−84 | |
| gene–CQW23_31620 | 18.5 kDa class I heat-shock protein | HSP18.5 | 1.02 | 0.9619644 | 2.14↑ | 1.43 × 10−3 | |
| gene–CQW23_01345 | 26.5 kDa heat-shock protein, mitochondrial | HSP26.5 | 1.54 | 0.4367286 | 3.02↑ | 0.01976457 | |
| FAD | gene–CQW23_29307 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.16↓ | 0 | 1.62↑ | 0.02217258 |
| gene–CQW23_29533 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.60↓ | 3.46 × 10−8 | 1.62↑ | 7.31 × 10−5 | |
| gene–CQW23_29306 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.18↓ | 6.23 × 10−3 | 29.17↑ | 1.12 × 10−10 | |
| gene–CQW23_29537 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.77 | 7.26 × 10−31 | 2.06↑ | 5.75 × 10−136 | |
| gene–CQW23_21403 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 1.08 | 0.3271504 | 3.77↑ | 9.26 × 10−67 | |
| gene–CQW23_29304 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.84 | 0.2026156 | 4.04↑ | 1.28 × 10−36 | |
| gene–CQW23_35257 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.79 | 3.40 × 10−14 | 1.55↑ | 1.01 × 10−20 | |
| gene–CQW23_29539 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.74 | 1.30 × 10−6 | 1.73↑ | 6.26 × 10−15 | |
| gene–CQW23_29540 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.89 | 0.0626088 | 1.75↑ | 6.35 × 10−9 | |
| gene–CQW23_29534 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.94 | 0.6231729 | 1.69↑ | 3.66 × 10−8 | |
| gene–CQW23_29305 | Omega-6 fatty acid desaturase, endoplasmic reticulum | FAD2 | 0.70 | 0.6677045 | 11.45↑ | 2.39 × 10−5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Sun, K.; Chang, X.; Ouyang, Z.; Meng, G.; Han, Y.; Shen, S.; Yao, Q.; Piao, F.; Wang, Y. Comparative Physiological and Transcriptomic Analyses of Two Contrasting Pepper Genotypes under Salt Stress Reveal Complex Salt Tolerance Mechanisms in Seedlings. Int. J. Mol. Sci. 2022, 23, 9701. https://doi.org/10.3390/ijms23179701
Zhang T, Sun K, Chang X, Ouyang Z, Meng G, Han Y, Shen S, Yao Q, Piao F, Wang Y. Comparative Physiological and Transcriptomic Analyses of Two Contrasting Pepper Genotypes under Salt Stress Reveal Complex Salt Tolerance Mechanisms in Seedlings. International Journal of Molecular Sciences. 2022; 23(17):9701. https://doi.org/10.3390/ijms23179701
Chicago/Turabian StyleZhang, Tao, Kaile Sun, Xiaoke Chang, Zhaopeng Ouyang, Geng Meng, Yanan Han, Shunshan Shen, Qiuju Yao, Fengzhi Piao, and Yong Wang. 2022. "Comparative Physiological and Transcriptomic Analyses of Two Contrasting Pepper Genotypes under Salt Stress Reveal Complex Salt Tolerance Mechanisms in Seedlings" International Journal of Molecular Sciences 23, no. 17: 9701. https://doi.org/10.3390/ijms23179701
APA StyleZhang, T., Sun, K., Chang, X., Ouyang, Z., Meng, G., Han, Y., Shen, S., Yao, Q., Piao, F., & Wang, Y. (2022). Comparative Physiological and Transcriptomic Analyses of Two Contrasting Pepper Genotypes under Salt Stress Reveal Complex Salt Tolerance Mechanisms in Seedlings. International Journal of Molecular Sciences, 23(17), 9701. https://doi.org/10.3390/ijms23179701






