Evaluation of the Possible Contribution of Various Regulatory Genes to Determination of Carpel Number as a Potential Mechanism for Optimal Agricultural Yield
Abstract
:1. Introduction
2. Natural Variation in Gynoecium Structure
3. The Structural Organization in Floral Meristem
4. Genes Defining Identity of Floral Organs and Their Possible Contribution to Multicarpelly
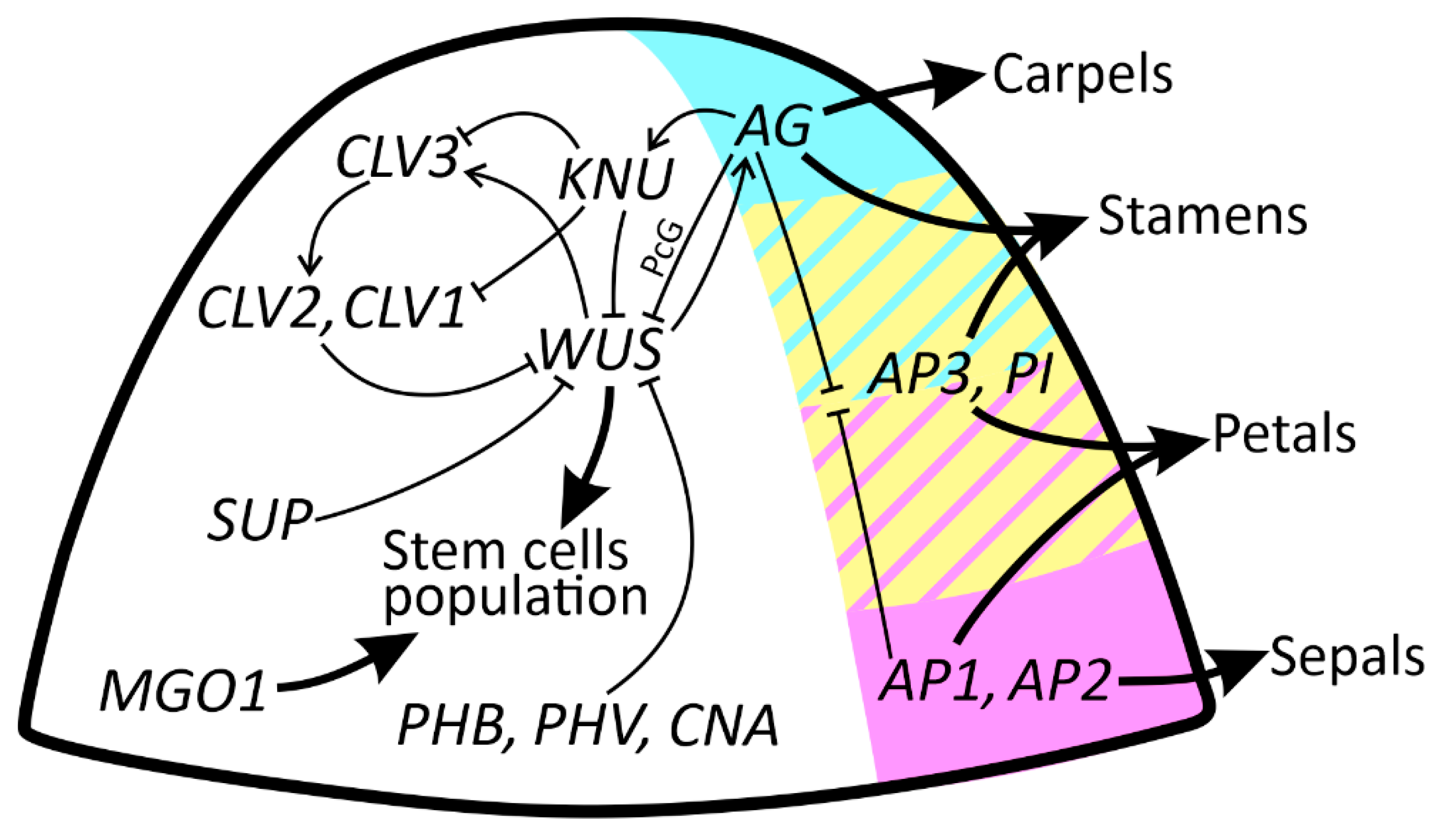
5. Key Players in Regulation of FM Size: CLV/WUS Negative Feedback Loop
6. FM Should Not Proliferate for Too Long: AG/WUS Interaction
7. Further Regulatory Genes Potentially Contributing to Multicarpelly
8. Potential Use of Certain Mutations in Agriculture
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Allaby, R.G. Domestication Syndrome in Plants. In Encyclopedia of Global Archaeology; Smith, C., Ed.; Springer: New York, NY, USA, 2014; pp. 2182–2184. [Google Scholar]
- Stetter, M.G. Limits and constraints to crop domestication. Am. J. Bot. 2020, 107, 1617–1621. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.C.; Wu, Y.D.; Yang, Q.Y.; Yang, Y.; Meng, Q.W.; Zhang, K.Q.; Li, J.G.; Wang, J.F.; Zhou, Y.M. A novel single-nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa L. Mol. Plant 2014, 7, 1788–1792. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Liu, F.; Zhao, Z.; Xu, L.; Chen, C.P.; Wang, Y.H.; Shang, G.X.; Du, D.Z. Mutations in the CDS and promoter of BjuA07.CLV1 cause a multilocular trait in Brassica juncea. Sci. Rep. 2018, 8, 5339. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, K.Y.; Li, H.L.; Han, S.Q.; Meng, Q.W.; Khan, S.U.; Fan, C.C.; Xie, K.; Zhou, Y.M. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018, 16, 1322–1335. [Google Scholar] [CrossRef] [PubMed]
- Sokoloff, D.D.; Nuraliev, M.S.; Oskolski, A.A.; Remizowa, M.V. Gynoecium evolution in angiosperms: Monomery, pseudomonomery, and mixomery. Mosc. Univ. Biol. Sci. Bull. 2017, 72, 97–108. [Google Scholar] [CrossRef]
- Cucinotta, M.; Di Marzo, M.; Guazzotti, A.; de Folter, S.; Kater, M.M.; Colombo, L. Gynoecium size and ovule number are interconnected traits that impact seed yield. J. Exp. Bot. 2020, 71, 2479–2489. [Google Scholar] [CrossRef]
- Ellis, T.H.N.; Hofer, J.M.I.; Vikeli, E.; Ambrose, M.J.; Higuera-Poveda, P.; Wingen, L.U.; Chayut, N. Diversity of pod shape in Pisum. Diversity 2021, 13, 203. [Google Scholar] [CrossRef]
- Gonzalez, A.M. Floral structure, development of the gynoecium, and embryology in Schinopsis balansae Engler (Anacardiaceae) with particular reference to aporogamy. Int. J. Plant Sci. 2016, 177, 326–338. [Google Scholar] [CrossRef]
- Endress, P.K. Multicarpellate gynoecia in angiosperms: Occurrence, development, organization and architectural constraints. Bot. J. Linn. Soc. 2014, 174, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Endress, P.K. Diversity and Evolutionary Biology of Tropical Flowers, 1st ed.; Cambridge University Press: New York, NY, USA, 1994; pp. xiv, 511p: Ill. [Google Scholar]
- Schoute, J.C. On pleiomery and meiomery in the flower. In Recueil des Travaux Botaniques Néerlandais; Société Botanique Néerlandaise: Nimègue, The Netherlands, 1932; Volume 29. [Google Scholar]
- Paulino, J.V.; Prenner, G.; Mansano, V.F.; Teixeira, S.P. Comparative development of rare cases of a polycarpellate gynoecium in an otherwise monocarpellate family, Leguminosae. Am. J. Bot. 2014, 101, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, C.; Dural, H.; Ertugrul, K.; Kucukoduk, M.; Baran, P.; Sanda, M.A. Morphology and anatomy of endemic Thermopsis turcica Kit Tan, Vural & Kucukoduk. Bangl. J. Bot. 2008, 37, 105–114. [Google Scholar]
- Davis, P.; Miller, R.; Tan, K. Flora of Turkey; Edinburgh University Press: Edinburgh, Scotland, 1988; Volume 10. [Google Scholar]
- Tan, K.; Vural, M.; Küçüködük, M. An unusual new Thermopsis from Turkey. Notes R. Bot. Gard Edinb. 1983, 40, 515–518. [Google Scholar]
- Sinjushin, A.A.; Tekdal, D.; Ciftci, C.; Cetiner, S. Floral development in Thermopsis turcica, an unusual multicarpellate papilionoid legume. Plant Syst. Evol. 2018, 304, 461–471. [Google Scholar] [CrossRef]
- Sinjushin, A.A. Origin and variation of polymerous gynoecia in Fabaceae: Evidence from floral mutants of pea (Pisum sativum L.). Plant Syst. Evol. 2014, 300, 717–727. [Google Scholar] [CrossRef]
- Nagar, P.; Albert, S. An unusual multicarpellary condition in Crotalaria verrucosa L. (Fabaceae) from Gujarat (India). Webbia 2013, 68, 187–190. [Google Scholar] [CrossRef]
- Ramana, P.V.; Swamy, J.; Ahmedullah, M. A striking new unifoliolate and polycarpellate species of Senna (Fabaceae: Caesalpinioideae) from Andhra Pradesh, India. Nord J. Bot. 2019, 37, e02148. [Google Scholar] [CrossRef]
- Li, R.; Wen, J. Phylogeny and diversification of Chinese Araliaceae based on nuclear and plastid DNA sequence data. J. Syst. Evol. 2016, 54, 453–467. [Google Scholar] [CrossRef]
- Nuraliev, M.S.; Oskolsvki, A.A.; Sokoloff, D.D.; Remizowa, M.V. Flowers of Araliaceae: Structural diversity, developmental and evolutionary aspects. Plant Divers. Evol. 2010, 128, 247–268. [Google Scholar] [CrossRef]
- Hufford, L. Ontogeny and morphology of the fertile flowers of Hydrangea and allied genera of tribe Hydrangeeae (Hydrangeaceae). Bot. J. Linn. Soc. 2001, 137, 139–187. [Google Scholar] [CrossRef]
- Sokoloff, D.D.; Oskolski, A.A.; Remizowa, M.V.; Nuraliev, M.S. Flower structure and development in Tupidanthus calyptratus (Araliaceae): An extreme case of polymery among asterids. Plant Syst. Evol. 2007, 268, 209–234. [Google Scholar] [CrossRef]
- Choob, V.V.; Sinyushin, A.A. Flower and shoot fasciation: From phenomenology to the construction of models of apical meristem transformations. Russ. J. Plant Physiol. 2012, 59, 530–545. [Google Scholar] [CrossRef]
- Clark, S.E.; Running, M.P.; Meyerowitz, E.M. Clavata1, a regulator of meristem and flower development in Arabidopsis. Development 1993, 119, 397–418. [Google Scholar] [CrossRef] [PubMed]
- Decraene, L.P.R.; Smets, E.F. Dedoublement revisited—towards a renewed interpretation of the androecium of the Magnoliophytina. Bot. J. Linn. Soc. 1993, 113, 103–124. [Google Scholar]
- Shi, W.; Liu, P.L.; Duan, L.; Pan, B.R.; Su, Z.H. Evolutionary response to the Qinghai-Tibetan Plateau uplift: Phylogeny and biogeography of Ammopiptanthus and tribe Thermopsideae (Fabaceae). PeerJ 2017, 5, e3607. [Google Scholar] [CrossRef]
- De Smet, Y.; Mendoza, C.G.; Wanke, S.; Goetghebeur, P.; Samain, M.S. Molecular phylogenetics and new (infra)generic classification to alleviate polyphyly in tribe Hydrangeeae (Cornales: Hydrangeaceae). Taxon 2015, 64, 741–753. [Google Scholar] [CrossRef]
- Han, H.; Liu, X.; Zhou, Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 2020, 53, 50–56. [Google Scholar] [CrossRef]
- Shang, E.L.; Ito, T.; Sun, B. Control of floral stem cell activity in Arabidopsis. Plant Signal. Behav. 2019, 14, 1659706. [Google Scholar] [CrossRef]
- Xu, Y.F.; Yamaguchi, N.; Gan, E.S.; Ito, T. When to stop: An update on molecular mechanisms of floral meristem termination. J. Exp. Bot. 2019, 70, 1711–1718. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Ferrandiz, C.; Madueno, F.; Parcy, F. How floral meristems are built. Plant Mol. Biol. 2006, 60, 855–870. [Google Scholar] [CrossRef]
- Remizowa, M.V. One upward, two steps down: Order of floral organ initiation. Russ. J. Dev. Biol. 2019, 50, 325–340. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls—genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Li, G.S.; Meng, Z.; Kong, H.Z.; Chen, Z.D.; Lu, A.M. ABC model and floral evolution. Chin. Sci. Bull. 2003, 48, 2651–2657. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The homeotic gene Apetala3 of Arabidopsis thaliana encodes a mads box and is expressed in petals and stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef]
- Krizek, B.A.; Meyerowitz, E.M. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 1996, 122, 11–22. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene Agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef]
- Kanno, A.; Nakada, M.; Akita, Y.; Hirai, M. Class B gene expression and the modified ABC model in nongrass monocots. Sci. World J. 2007, 7, 268–279. [Google Scholar] [CrossRef]
- Hintz, M.; Bartholmes, C.; Nutt, P.; Ziermann, J.; Hameister, S.; Neuffer, B.; Theissen, G. Catching a ‘hopeful monster’: Shepherd’s purse (Capsella bursa-pastoris) as a model system to study the evolution of flower development. J. Exp. Bot. 2006, 57, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.X.; Schoof, H.; Haecker, A.; Lenhard, M.; Jurgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Kayes, J.M.; Clark, S.E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 1998, 125, 3843–3851. [Google Scholar] [CrossRef]
- Leyser, H.M.O.; Furner, I.J. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development 1992, 116, 397–403. [Google Scholar] [CrossRef]
- Liu, X.G.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.Y.; Pan, Y.Y.; Cao, X.F.; Goodrich, J.; Chena, X.M. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Sun, B.; Xu, Y.F.; Ng, K.H.; Ito, T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Gene Dev. 2009, 23, 1791–1804. [Google Scholar] [CrossRef]
- Payne, T.; Johnson, S.D.; Koltunow, A.M. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 2004, 131, 3737–3749. [Google Scholar] [CrossRef]
- Han, G.L.; Lu, C.X.; Guo, J.R.; Qiao, Z.Q.; Sui, N.; Qiu, N.W.; Wang, B.S. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Lyu, T.; Cao, J. Cys2/His2 Zinc-finger proteins in transcriptional regulation of flower development. Int. J. Mol. Sci. 2018, 19, 2589. [Google Scholar] [CrossRef]
- Liang, J.J.; Guan, P.Y.; Liu, Z.H.; Wang, Y.; Xing, J.Y.; Hu, J.F. The VvSUPERMAN-like gene is differentially expressed between bicarpellate and tricarpellate florets of Vitis vinifera L. Cv. ‘Xiangfei’ and its heterologous expression reduces carpel number in tomato. Plant Cell Physiol. 2020, 61, 1760–1774. [Google Scholar] [CrossRef] [PubMed]
- Shang, E.L.; Wang, X.; Li, T.H.; Guo, F.F.; Ito, T.; Sun, B. Robust control of floral meristem determinacy by position-specific multifunctions of KNUCKLES. Proc. Natl. Acad. Sci. USA 2021, 118, e2102826118. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jurgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Qi, M.F.; Sun, M.H.; Liu, Y.; Liu, Y.D.; Xu, T.; Li, Y.B.; Li, T.L. tomato transcription factor SlWUS plays an important role in tomato flower and locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef] [PubMed]
- Bollier, N.; Sicard, A.; Leblond, J.; Latrasse, D.; Gonzalez, N.; Gevaudant, F.; Benhamed, M.; Raynaud, C.; Lenhard, M.; Chevalier, C.; et al. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in arabidopsis and tomato. Plant Cell 2018, 30, 83–100. [Google Scholar] [CrossRef]
- Sakai, H.; Medrano, L.J.; Meyerowitz, E.M. Role of Superman in maintaining Arabidopsis floral whorl boundaries. Nature 1995, 378, 199–203. [Google Scholar] [CrossRef]
- Breuil-Broyer, S.; Trehin, C.; Morel, P.; Boltz, V.; Sun, B.; Chambrier, P.; Ito, T.; Negrutiu, I. Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male-female boundary, flower meristem termination and carpel compartmentalization. Ann. Bot. 2016, 117, 905–923. [Google Scholar] [CrossRef]
- Xu, Y.F.; Prunet, N.; Gan, E.S.; Wang, Y.B.; Stewart, D.; Wellmer, F.; Huang, J.B.; Yamaguchi, N.; Tatsumi, Y.; Kojima, M.; et al. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 2018, 37, e97499. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Weigel, D.; Yanofsky, M.F. NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 2006, 133, 1645–1655. [Google Scholar] [CrossRef]
- Xiao, H.; Tang, J.F.; Li, Y.F.; Wang, W.M.; Li, X.B.; Jin, L.; Xie, R.; Luo, H.F.; Zhao, X.F.; Meng, Z.; et al. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J. 2009, 59, 789–801. [Google Scholar] [CrossRef]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Laufs, P.; Dockx, J.; Kronenberger, J.; Traas, J. MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 1998, 125, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Graf, P.; Dolzblasz, A.; Wurschum, T.; Lenhard, M.; Pfreundt, U.; Laux, T. MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell 2010, 22, 716–728. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J.; Bowman, J.L. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008, 13, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Wall, K.; Leebens-Mack, J.; Wang, Y.J.; Carlson, J.E.; de Pamphilis, C.W. Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. Plant J. 2007, 51, 991–1003. [Google Scholar] [CrossRef]
- Floyd, S.K.; Bowman, J.L. Gene regulation: Ancient microRNA target sequences in plants. Nature 2004, 428, 485–486. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Gene Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Zhang, B.H.; Pan, X.P.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L.H. The critical role of miRNAs in regulation of flowering time and flower development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Zhang, Y.; Rahmani, R.S.; Yang, X.Y.; Chen, J.M.; Shi, T. Integrative expression network analysis of microRNA and gene isoforms in sacred lotus. BMC Genom. 2020, 21, 429. [Google Scholar] [CrossRef]
- Silva, G.F.F.E.; Silva, E.M.; Azevedo, M.D.; Guivin, M.A.C.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.P.; Nogueira, F.T.S. microRNA156-targeted SPL/ SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.L.G.; Tian, Y.Y.; Tan, C.; Bai, S.B.E.; Hao, J.F.; Hasi, A. Genome-wide identification of microRNAs involved in the regulation of fruit ripening and climacteric stages in melon (Cucumis melo). Hortic. Res. 2020, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.B.; Zhang, Z.L.; Liu, D.M.; Zhang, K.; Li, A.L.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Yang, Z.F.; Wang, X.F.; Gu, S.L.; Hu, Z.Q.; Xu, H.; Xu, C.W. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Smith, M.R.; Willmann, M.R.; Wu, G.; Berardini, T.Z.; Moller, B.; Weijers, D.; Poethig, R.S. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5424–5429. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Zhao, H.Y.; Zhao, Z.; Du, D.Z.; Xu, L.; Yao, Y.M.; Zhao, Z.G.; Xing, X.R.; Shang, G.X.; Zhao, H.C. Genetic and physical fine mapping of a multilocular gene Bjln1 in Brassica juncea to a 208-kb region. Mol. Breed. 2013, 32, 373–383. [Google Scholar] [CrossRef]
- Xu, P.; Cao, S.Q.; Hu, K.N.; Wang, X.H.; Huang, W.; Wang, G.; Lv, Z.W.; Liu, Z.S.; Wen, J.; Yi, B.; et al. Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 2017, 7, 3498. [Google Scholar] [CrossRef]
- Chai, L.; Zhang, J.F.; Lu, K.; Li, H.J.; Wu, L.T.; Wan, H.S.; Zheng, B.C.; Cui, C.; Jiang, J.; Jiang, L.C. Identification of genomic regions associated with multi-silique trait in Brassica napus. BMC Genom. 2019, 20, 304. [Google Scholar] [CrossRef]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 2004, 16, S181–S189. [Google Scholar] [CrossRef]
- Nair, R.M.; Peck, D.M.; Dundas, I.S.; Samac, D.A.; Moore, A.; Randles, J.W. Morphological characterisation and genetic analysis of a bi-pistil mutant (bip) in Medicago truncatula Gaertn. Sex. Plant Reprod. 2008, 21, 133–141. [Google Scholar] [CrossRef]
- Liu, J.X.; Wang, J.Y.; She, W.J.; Wang, L.; Luo, M.; Chen, Y.J.; Li, Y.T.; Wang, S.P.; Zhang, C.X. MADS-box genes are involved in cultivar- and temperature-dependent formation of multi-pistil and polycarpy in Prunus avium L. J. Plant Growth Regul. 2019, 38, 1017–1027. [Google Scholar] [CrossRef]
- Watson, M.; Gould, K.S. Development of flat and fan-shaped fruit in Actinidia chinensis var. chinensis and Actinidia deliciosa. Ann. Bot. 1994, 74, 59–68. [Google Scholar] [CrossRef]
- Cam, N.T.; Sunagawa, N.; Sesumi, M.; Kitamura, Y.; Tanaka, Y.; Goto, T.; Yasuba, K.I.; Yoshida, Y. Fasciation in strawberry floral organs and possible implications for floral transition. Hortic. J. 2022, 91, 58–67. [Google Scholar] [CrossRef]
- Hanano, S.; Goto, K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 2011, 23, 3172–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abiri, N.; Sinjushin, A.; Tekdal, D.; Cetiner, S. Evaluation of the Possible Contribution of Various Regulatory Genes to Determination of Carpel Number as a Potential Mechanism for Optimal Agricultural Yield. Int. J. Mol. Sci. 2022, 23, 9723. https://doi.org/10.3390/ijms23179723
Abiri N, Sinjushin A, Tekdal D, Cetiner S. Evaluation of the Possible Contribution of Various Regulatory Genes to Determination of Carpel Number as a Potential Mechanism for Optimal Agricultural Yield. International Journal of Molecular Sciences. 2022; 23(17):9723. https://doi.org/10.3390/ijms23179723
Chicago/Turabian StyleAbiri, Naghmeh, Andrey Sinjushin, Dilek Tekdal, and Selim Cetiner. 2022. "Evaluation of the Possible Contribution of Various Regulatory Genes to Determination of Carpel Number as a Potential Mechanism for Optimal Agricultural Yield" International Journal of Molecular Sciences 23, no. 17: 9723. https://doi.org/10.3390/ijms23179723





