Genome Analyses of Ten New Ape Adenoviruses with Similarity to Human Mastadenovirus C
Abstract
:1. Introduction
2. Results
2.1. Genome Analyses of New Ape Adenoviruses
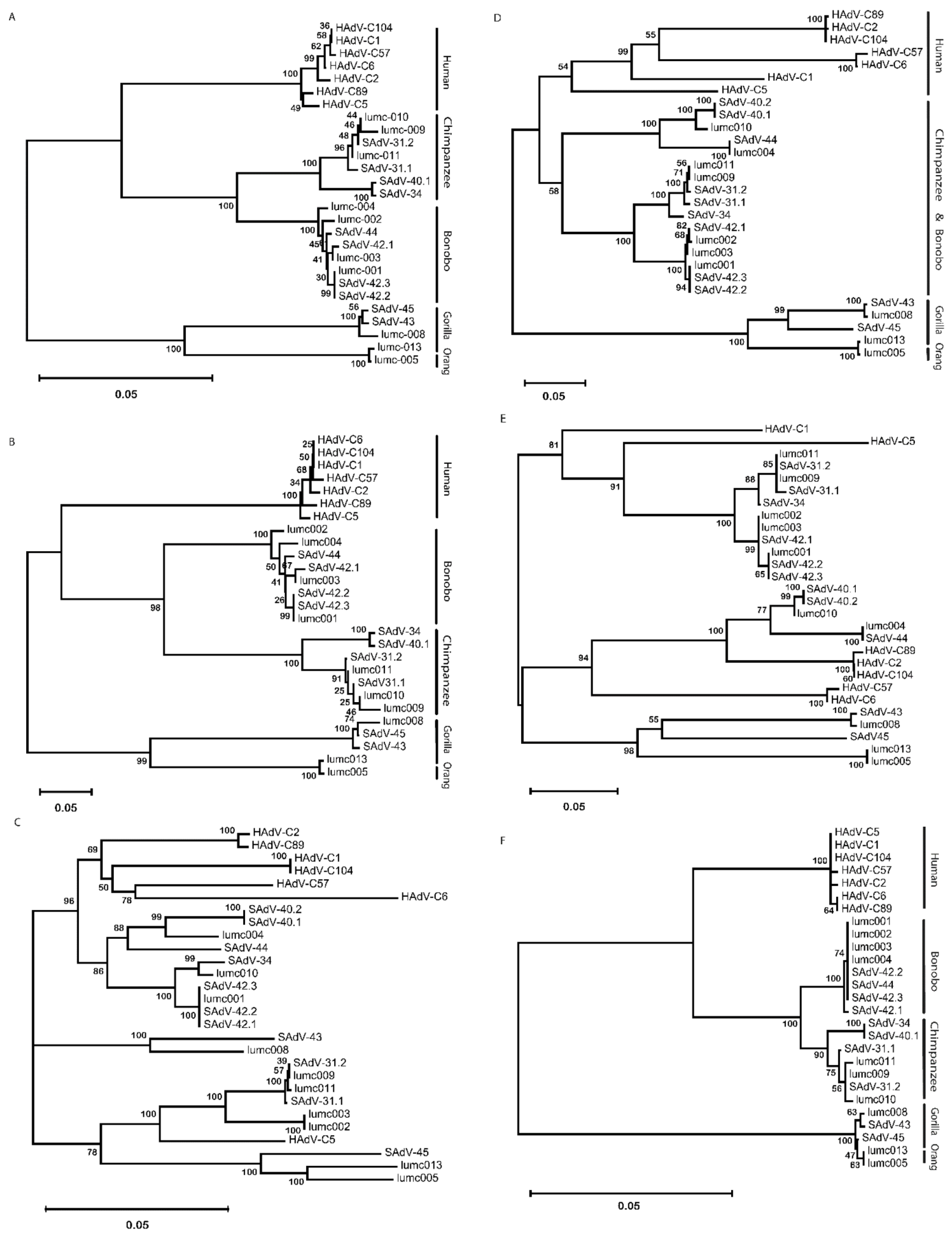
2.2. Molecular Phylogeny of AdV Proteins
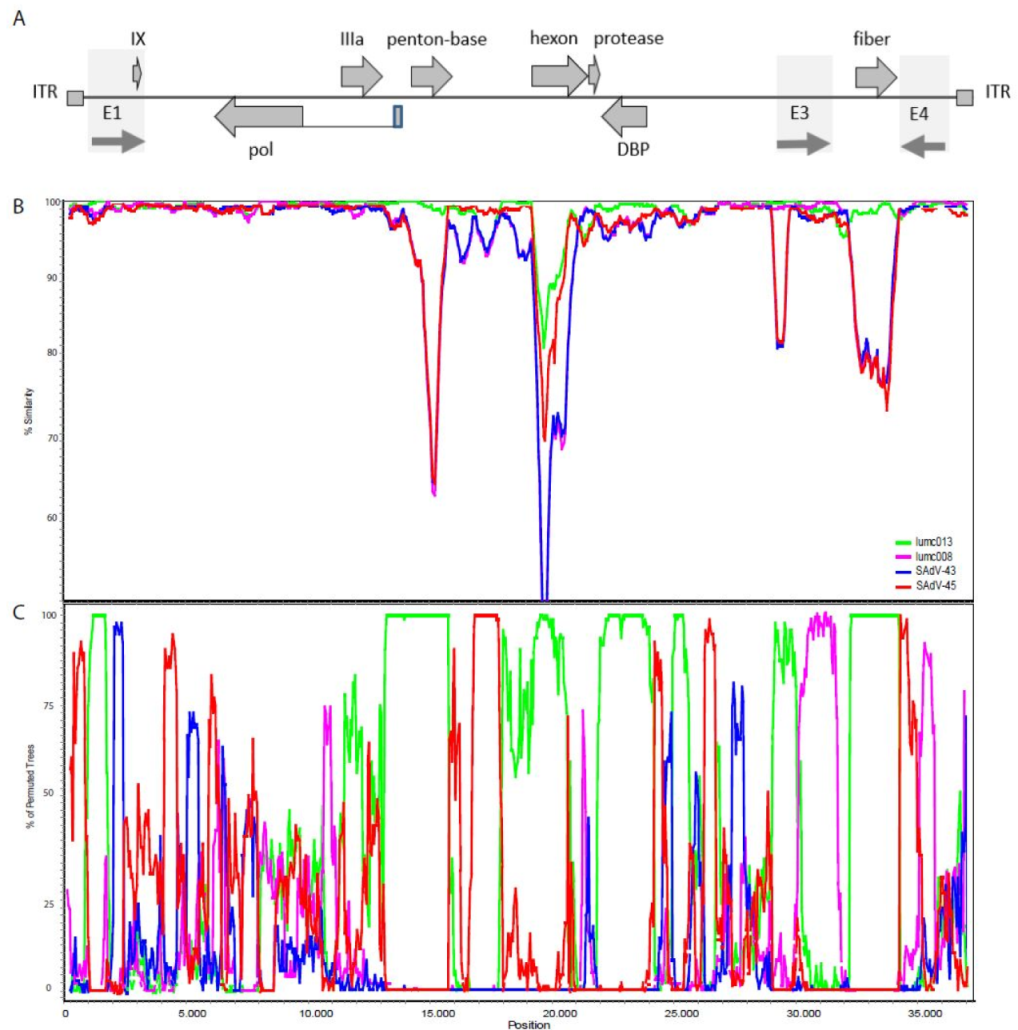
2.3. Recombination
2.4. The Origin of the AdV Isolate from Orangutan
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Virus Isolation
4.3. Hexon Sequencing
4.4. Next Generation Sequencing
4.5. Genome Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoeben, R.C.; Uil, T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a013003. [Google Scholar] [CrossRef] [PubMed]
- Khanal, S.; Ghimire, P.; Dhamoon, A.S. The Repertoire of Adenovirus in Human Disease: The Innocuous to the Deadly. Biomedicines 2018, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Vogels, R.; Zuijdgeest, D.; van Rijnsoever, R.; Hartkoorn, E.; Damen, I.; de Bethune, M.P.; Kostense, S.; Penders, G.; Helmus, N.; Koudstaal, W.; et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: Efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 2003, 77, 8263–8271. [Google Scholar] [CrossRef]
- Fong, T.T.; Phanikumar, M.S.; Xagoraraki, I.; Rose, J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010, 76, 715–723. [Google Scholar] [CrossRef]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef]
- Ip, W.H.; Dobner, T. Cell transformation by the adenovirus oncogenes E1 and E4. FEBS Lett. 2020, 594, 1848–1860. [Google Scholar] [CrossRef] [PubMed]
- Tessier, T.M.; Dodge, M.J.; MacNeil, K.M.; Evans, A.M.; Prusinkiewicz, M.A.; Mymryk, J.S. Almost famous: Human adenoviruses (and what they have taught us about cancer). Tumour Virus Res. 2021, 12, 200225. [Google Scholar] [CrossRef]
- Greber, U.F.; Gomez-Gonzalez, A. Adenovirus—A blueprint for gene delivery. Curr. Opin. Virol. 2021, 48, 49–56. [Google Scholar] [CrossRef]
- Harrach, B.; Tarjan, Z.L.; Benko, M. Adenoviruses across the animal kingdom: A walk in the zoo. FEBS Lett. 2019, 593, 3660–3673. [Google Scholar] [CrossRef]
- Aoki, K.; Benko, M.; Davison, A.J.; Echavarria, M.; Erdman, D.D.; Harrach, B.; Kajon, A.E.; Schnurr, D.; Wadell, G.; Members of the Adenovirus Research Community. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J. Virol. 2011, 85, 5703–5704. [Google Scholar] [CrossRef] [Green Version]
- Seto, D.; Chodosh, J.; Brister, J.R.; Jones, M.S.; Members of the Adenovirus Research Community. Using the whole-genome sequence to characterize and name human adenoviruses. J. Virol. 2011, 85, 5701–5702. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Bottcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A.N.; Ivanova, O.E.; Eremeeva, T.P.; Iggo, R.D. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 2008, 89, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Rivailler, P.; Mao, N.; Zhu, Z.; Xu, W. Recombination analysis of Human mastadenovirus C whole genomes. Sci. Rep. 2019, 9, 2182. [Google Scholar] [CrossRef]
- Dehghan, S.; Seto, J.; Liu, E.B.; Walsh, M.P.; Dyer, D.W.; Chodosh, J.; Seto, D. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 2013, 443, 197–207. [Google Scholar] [CrossRef]
- Bots, S.T.F.; Hoeben, R.C. Non-Human Primate-Derived Adenoviruses for Future Use as Oncolytic Agents? Int. J. Mol. Sci. 2020, 21, 4821. [Google Scholar] [CrossRef]
- Podgorski, I.I.; Panto, L.; Papp, T.; Harrach, B.; Benko, M. Genome analysis of four Old World monkey adenoviruses supports the proposed species classification of primate adenoviruses and reveals signs of possible homologous recombination. J. Gen. Virol. 2016, 97, 1604–1614. [Google Scholar] [CrossRef]
- Hoppe, E.; Pauly, M.; Gillespie, T.R.; Akoua-Koffi, C.; Hohmann, G.; Fruth, B.; Karhemere, S.; Madinda, N.F.; Mugisha, L.; Muyembe, J.J.; et al. Multiple Cross-Species Transmission Events of Human Adenoviruses (HAdV) during Hominine Evolution. Mol. Biol. Evol. 2015, 32, 2072–2084. [Google Scholar] [CrossRef]
- Moffatt, S.; Hays, J.; HogenEsch, H.; Mittal, S.K. Circumvention of vector-specific neutralizing antibody response by alternating use of human and non-human adenoviruses: Implications in gene therapy. Virology 2000, 272, 159–167. [Google Scholar] [CrossRef]
- Bots, S.T.; Kemp, V.; Cramer, S.J.; van den Wollenberg, D.J.; Hornsveld, M.; Lamfers, M.; van der Pluijm, G.; Hoeben, R.C. Non-human primate adenoviruses of the Human Adenovirus B species are potent and broadly-acting oncolytic vector candidates. Hum. Gene Ther. 2022, 33, 275–289. [Google Scholar] [CrossRef]
- Madisch, I.; Hofmayer, S.; Moritz, C.; Grintzalis, A.; Hainmueller, J.; Pring-Akerblom, P.; Heim, A. Phylogenetic analysis and structural predictions of human adenovirus penton proteins as a basis for tissue-specific adenovirus vector design. J. Virol. 2007, 81, 8270–8281. [Google Scholar] [CrossRef] [PubMed]
- Han, J.W.; La, T.M.; Kim, J.H.; Choi, I.S.; Song, C.S.; Park, S.Y.; Lee, J.B.; Lee, S.W. The possible origin of human adenovirus type 3: Evidence of natural genetic recombination between human and simian adenovirus. Infect. Genet. Evol. 2018, 65, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, S.; Seto, J.; Liu, E.B.; Ismail, A.M.; Madupu, R.; Heim, A.; Jones, M.S.; Dyer, D.W.; Chodosh, J.; Seto, D. A zoonotic adenoviral human pathogen emerged through genomic recombination among human and nonhuman simian hosts. J. Virol. 2019, 93, e00564-19. [Google Scholar] [CrossRef]
- Kilbourn, A.M.; Karesh, W.B.; Wolfe, N.D.; Bosi, E.J.; Cook, R.A.; Andau, M. Health evaluation of free-ranging and semi-captive orangutans (Pongo pygmaeus pygmaeus) in Sabah, Malaysia. J. Wildl. Dis. 2003, 39, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Vandenberghe, L.H.; Kryazhimskiy, S.; Grant, R.; Calcedo, R.; Yuan, X.; Keough, M.; Sandhu, A.; Wang, Q.; Medina-Jaszek, C.A.; et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009, 5, e1000503. [Google Scholar] [CrossRef]
- Mathias, P.; Wickham, T.; Moore, M.; Nemerow, G. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 1994, 68, 6811–6814. [Google Scholar] [CrossRef]
- Nemerow, G.R.; Stewart, P.L. Insights into Adenovirus Uncoating from Interactions with Integrins and Mediators of Host Immunity. Viruses 2016, 8, 337. [Google Scholar] [CrossRef]
- Wickham, T.J.; Mathias, P.; Cheresh, D.A.; Nemerow, G.R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 1993, 73, 309–319. [Google Scholar] [CrossRef]
- Lindert, S.; Silvestry, M.; Mullen, T.M.; Nemerow, G.R.; Stewart, P.L. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J. Virol. 2009, 83, 11491–11501. [Google Scholar] [CrossRef]
- Panto, L.; Podgorski, I.I.; Janoska, M.; Marko, O.; Harrach, B. Taxonomy proposal for Old World monkey adenoviruses: Characterisation of several non-human, non-ape primate adenovirus lineages. Arch. Virol. 2015, 160, 3165–3177. [Google Scholar] [CrossRef] [Green Version]
- Pina, M.; Green, M. Biochemical studies on adenovirus multiplication. IX. Chemical and base composition analysis of 28 human adenoviruses. Proc. Natl. Acad. Sci. USA 1965, 54, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Ismail, A.M.; Dehghan, S.; Rajaiya, J.; Allard, M.W.; Lim, H.C.; Dyer, D.W.; Chodosh, J.; Seto, D. Genomics-based re-examination of the taxonomy and phylogeny of human and simian Mastadenoviruses: An evolving whole genomes approach, revealing putative zoonosis, anthroponosis, and amphizoonosis. Cladistics 2020, 36, 358–373. [Google Scholar] [CrossRef] [PubMed]
- Fallaux, F.J.; Kranenburg, O.; Cramer, S.J.; Houweling, A.; van Ormondt, H.; Hoeben, R.C.; Van der Eb, A.J. Characterization of 911: A new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 1996, 7, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Tu, S.L.; Staheli, J.P.; McClay, C.; McLeod, K.; Rose, T.M.; Upton, C. Base-By-Base Version 3: New Comparative Tools for Large Virus Genomes. Viruses 2018, 10, 637. [Google Scholar] [CrossRef] [Green Version]


| AdV Species | Type | Accession Number |
|---|---|---|
| SAdV-A | SAdV-3 | NC_006144 |
| SAdV-B | SAdV-08 | NC_028113 |
| SAdV-C | BaAdV-3 | KC693023 |
| SAdV-D | SAdV-13 | NC_028103 |
| SAdV-E | SAdV-16 | NC_028105 |
| SAdV-F | SAdV-18 | FJ025931 |
| SAdV-G | SAdV-20 | NC_020485 |
| SAdV-H | SAdV-DM2014 | NC_025678 |
| SAdV-I | SAdV-WIV19 | KX505867 |
| HAdV-A | HAdV-A12 | KX868289 |
| HAdV-B | HAdV-B3 | NC011203 |
| HAdV-C | HAdV-C2 | AC_000007 |
| HAdV-D | HAdV-D8 | KP016723 |
| HAdV-E | HAdV-E4 | AY458656 |
| HAdV-F | HAdV-F40 | KU162869 |
| HAdV-G | HAdV-G52 | DQ923122 |
| Virus Isolate | Genome Length | Percentage G + C Basepairs | Hexon Length | Penton-Base Length | Penton-Base RGD-Loop | Penton-Base HVR1 Loop | Penton-Base Proline-Rich Region | Host Species | GenBank Accession Number |
|---|---|---|---|---|---|---|---|---|---|
| SAdV-42.2 | 37,820 | 60.4 | 957 | 586 | 120 | 11 | 27 | Bonobo | FJ025902 |
| Ad-lumc001 | 37,820 | 60.5 | 957 | 587 | 120 | 11 | 27 | Bonobo | MZ882379 |
| SAdV-42.3 | 37,820 | 60.6 | 957 | 586 | 120 | 11 | 27 | Bonobo | FJ025925 |
| Ad-lumc002 | 37,702 | 60.4 | 955 | 584 | 118 | 11 | 27 | Bonobo | MZ882389 |
| Ad-lumc003 | 37,724 | 60.4 | 955 | 582 | 116 | 11 | 27 | Bonobo | MZ882380 |
| SAdV-42.1 | 37,786 | 60.3 | 957 | 581 | 115 | 11 | 27 | Bonobo | FJ025903 |
| Ad-lumc004 | 37,647 | 60.6 | 963 | 581 | 115 | 11 | 27 | Bonobo | MZ882381 |
| SAdV-44 | 37,711 | 60.6 | 962 | 581 | 115 | 11 | 27 | Bonobo | FJ025899 |
| Ad-lumc009 | 37,885 | 60.6 | 954 | 592 | 128 | 11 | 25 | Chimpanzee | MZ882384 |
| SAdV-40.2 | 37,729 | 60.3 | 960 | 596 | 126 | 11 | 31 | Chimpanzee | FJ025926 |
| SAdV-31.1 | 37,828 | 60.6 | 954 | 589 | 125 | 11 | 25 | Chimpanzee | FJ025906 |
| SAdV-31.2 | 37,860 | 60.6 | 954 | 589 | 124 | 11 | 25 | Chimpanzee | FJ025904 |
| SAdV-34 | 37,799 | 60.2 | 958 | 593 | 123 | 11 | 31 | Chimpanzee | FJ025905 |
| SAdV-40.1 | 37,718 | 60.3 | 960 | 593 | 123 | 11 | 31 | Chimpanzee | FJ025907 |
| Ad-lumc010 | 37,738 | 60.7 | 958 | 581 | 117 | 11 | 25 | Chimpanzee | MZ882385 |
| Ad-lumc011 | 37,780 | 60.6 | 954 | 579 | 115 | 11 | 25 | Chimpanzee | MZ882386 |
| Ad-lumc008 | 37,200 | 58.0 | 959 | 656 | 199 | 8 | 23 | Gorilla | MZ882383 |
| SAdV-45 | 37,152 | 57.6 | 953 | 651 | 194 | 11 | 27 | Gorilla | FJ025901 |
| SAdV-43 | 37,188 | 58.0 | 955 | 651 | 194 | 6 | 23 | Gorilla | FJ025900 |
| Ad-lumc013 | 37,087 | 57.6 | 947 | 622 | 164 | 7 | 23 | Gorilla | OM807070 |
| HAdV-C57 | 35,818 | 55.2 | 959 | 574 | 102 | 11 | 32 | Human | HQ003817 |
| HAdV-C1 | 36,001 | 55.3 | 964 | 574 | 102 | 11 | 32 | Human | AC_000017 |
| HAdV-C104 | 35,933 | 55.3 | 964 | 574 | 102 | 11 | 32 | Human | MH558113 |
| HAdV-C6 | 35,758 | 55.4 | 978 | 574 | 102 | 11 | 32 | Human | FJ349096 |
| HAdV-C2 | 35,937 | 55.2 | 968 | 571 | 99 | 11 | 32 | Human | AC_000007 |
| HAdV-C5 | 35,938 | 55.2 | 952 | 571 | 99 | 11 | 32 | Human | AC_000008 |
| HAdV-C89 | 35,923 | 55.2 | 969 | 570 | 98 | 11 | 32 | Human | MH121114 |
| Ad-lumc005 | 37,034 | 56.8 | 947 | 621 | 163 | 7 | 23 | Orang utan | MZ882384 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bots, S.T.F.; Kemp, V.; Dautzenberg, I.J.C.; Hoeben, R.C. Genome Analyses of Ten New Ape Adenoviruses with Similarity to Human Mastadenovirus C. Int. J. Mol. Sci. 2022, 23, 9832. https://doi.org/10.3390/ijms23179832
Bots STF, Kemp V, Dautzenberg IJC, Hoeben RC. Genome Analyses of Ten New Ape Adenoviruses with Similarity to Human Mastadenovirus C. International Journal of Molecular Sciences. 2022; 23(17):9832. https://doi.org/10.3390/ijms23179832
Chicago/Turabian StyleBots, Selas T. F., Vera Kemp, Iris J. C. Dautzenberg, and Rob C. Hoeben. 2022. "Genome Analyses of Ten New Ape Adenoviruses with Similarity to Human Mastadenovirus C" International Journal of Molecular Sciences 23, no. 17: 9832. https://doi.org/10.3390/ijms23179832
APA StyleBots, S. T. F., Kemp, V., Dautzenberg, I. J. C., & Hoeben, R. C. (2022). Genome Analyses of Ten New Ape Adenoviruses with Similarity to Human Mastadenovirus C. International Journal of Molecular Sciences, 23(17), 9832. https://doi.org/10.3390/ijms23179832








