NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth
Abstract
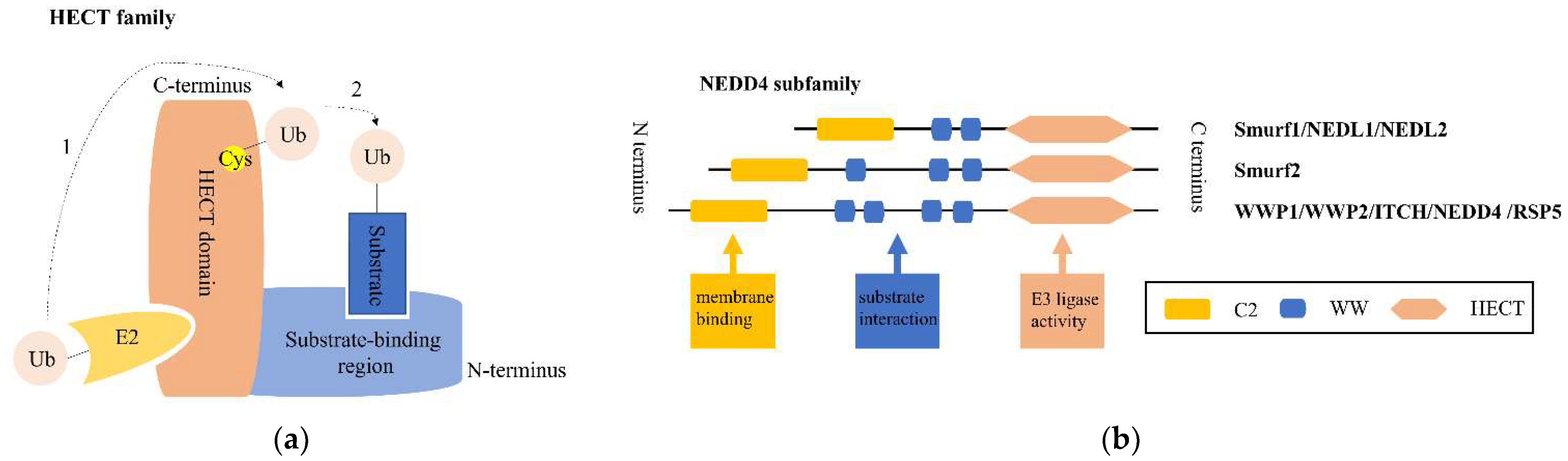
1. Introduction
2. NEDD4 E3 Ligases and Bone
2.1. Smurf1
2.1.1. Smurf1 Is a Negative Regulatory Factor in Bone Phenotype
2.1.2. Smurf1 as an E3 Ligase Ubiquitylates Important Molecules Involved in Osteogenesis
2.1.3. Regulation of Smurf1–Substrate Interactions
Expression Regulation of Smurf1
Structural Modification of Smurf1
Induced Degradation of Smurf1
Modification of Substrates—Smad1 and RUNX2
2.1.4. Smurf1 and Clinical Application of rhBMPs
2.2. Smurf2
2.3. WWPs and Other NEDD4s
3. NEDD4 E3 Ligases and Tooth
4. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Xu, Y.; Lin, S.; Guo, Y.; Deng, W.; Zhang, Y.; Guo, A.; Xue, Y. iUUCD 2.0: An update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res. 2018, 46, D447–D453. [Google Scholar] [CrossRef] [PubMed]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Maspero, E.; Valentini, E.; Mari, S.; Cecatiello, V.; Soffientini, P.; Pasqualato, S.; Polo, S. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 2013, 20, 696–701. [Google Scholar] [CrossRef]
- Kamadurai, H.B.; Qiu, Y.; Deng, A.; Harrison, J.S.; Macdonald, C.; Actis, M.; Rodrigues, P.; Miller, D.J.; Souphron, J.; Lewis, S.M.; et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife 2013, 2, e00828. [Google Scholar] [CrossRef]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Wang, Z.W.; Hu, X.; Ye, M.; Lin, M.; Chu, M.; Shen, X. NEDD4 E3 ligase: Functions and mechanism in human cancer. Semin. Cancer Biol. 2020, 67, 92–101. [Google Scholar] [CrossRef]
- Koganti, P.; Levy-Cohen, G.; Blank, M. Smurfs in Protein Homeostasis, Signaling, and Cancer. Front. Oncol. 2018, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, H.; Wu, B.; You, S.; Wu, S.; Lu, S.; Wang, P.; Cao, L.; Zhang, N.; Sun, Y. E3 Ubiquitin ligase NEDD4 familyregulatory network in cardiovascular disease. Int. J. Biol. Sci. 2020, 16, 2727–2740. [Google Scholar] [CrossRef]
- He, H.; Huang, C.; Chen, Z.; Huang, H.; Wang, X.; Chen, J. An outlined review for the role of Nedd4-1 and Nedd4-2 in lung disorders. Biomed. Pharmacother. 2020, 125, 109983. [Google Scholar] [CrossRef]
- Wang, Y.; Argiles-Castillo, D.; Kane, E.I.; Zhou, A.; Spratt, D.E. HECT E3 ubiquitin ligases—Emerging insights into their biological roles and disease relevance. J. Cell Sci. 2020, 133, jcs228072. [Google Scholar] [CrossRef]
- Kovacs, C.S.; Chaussain, C.; Osdoby, P.; Brandi, M.L.; Clarke, B.; Thakker, R.V. The role of biomineralization in disorders of skeletal development and tooth formation. Nat. Rev. Endocrinol. 2021, 17, 336–349. [Google Scholar] [CrossRef]
- Kular, J.; Tickner, J.; Chim, S.M.; Xu, J. An overview of the regulation of bone remodelling at the cellular level. Clin. Biochem. 2012, 45, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.I.; Bruzzaniti, A. Molecular signaling in bone cells: Regulation of cell differentiation and survival. Adv. Protein Chem. Struct. Biol. 2019, 116, 237–281. [Google Scholar]
- Shen, F.; Shi, Y. Recent Advances in Single-Cell View of Mesenchymal Stem Cell in Osteogenesis. Front. Cell Dev. Biol. 2021, 9, 809918. [Google Scholar] [CrossRef]
- He, J.; Yan, J.; Wang, J.; Zhao, L.; Xin, Q.; Zeng, Y.; Sun, Y.; Zhang, H.; Bai, Z.; Li, Z.; et al. Dissecting human embryonic skeletal stem cell ontogeny by single-cell transcriptomic and functional analyses. Cell Res. 2021, 31, 742–757. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.W.; Tan, Z.; To, M.K.T.; Chan, D. Regulation and Role of Transcription Factors in Osteogenesis. Int. J. Mol. Sci. 2021, 22, 5445. [Google Scholar] [CrossRef]
- Gomathi, K.; Akshaya, N.; Srinaath, N.; Rohini, M.; Selvamurugan, N. Histone acetyl transferases and their epigenetic impact on bone remodeling. Int. J. Biol. Macromol. 2021, 170, 326–335. [Google Scholar] [CrossRef]
- Zou, M.L.; Chen, Z.H.; Teng, Y.Y.; Liu, S.Y.; Jia, Y.; Zhang, K.W.; Sun, Z.L.; Wu, J.J.; Yuan, Z.D.; Feng, Y.; et al. The Smad Dependent TGF-beta and BMP Signaling Pathway in Bone Remodeling and Therapies. Front. Mol. Biosci. 2021, 8, 593310. [Google Scholar] [CrossRef]
- Kim, S.-M.; Yuen, T.; Iqbal, J.; Rubin, M.R.; Zaidi, M. The NO-cGMP-PKG pathway in skeletal remodeling. Ann. N. Y. Acad. Sci. 2021, 1487, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984. [Google Scholar] [CrossRef]
- Mari, S.; Ruetalo, N.; Maspero, E.; Stoffregen, M.C.; Pasqualato, S.; Polo, S.; Wiesner, S. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure 2014, 22, 1639–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Qiao, M.; Harris, S.E.; Oyajobi, B.O.; Mundy, G.R.; Chen, D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J. Biol. Chem. 2004, 279, 12854–12859. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Ying, S.X.; Zhang, G.M.; Li, C.; Cheng, S.Y.; Deng, C.X.; Zhang, Y.E. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 2005, 121, 101–113. [Google Scholar] [CrossRef]
- Al-Rawi, R.; Al-Beshri, A.; Mikhail, F.M.; McCormick, K. Fragile Bones Secondary to SMURF1 Gene Duplication. Calcif. Tissue Int. 2020, 106, 567–573. [Google Scholar] [CrossRef]
- Lu, K.; Li, P.; Zhang, M.; Xing, G.; Li, X.; Zhou, W.; Bartlam, M.; Zhang, L.; Rao, Z.; He, F. Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection. J. Biol. Chem. 2011, 286, 16861–16870. [Google Scholar] [CrossRef]
- Sangadala, S.; Metpally, R.P.R.; Reddy, B.V.B. Molecular interaction between Smurf1 WW2 domain and PPXY motifs of Smad1, Smad5, and Smad6--modeling and analysis. J. Biomol. Struct. Dyn. 2007, 25, 11–23. [Google Scholar] [CrossRef]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef]
- Murakami, G.; Watabe, T.; Takaoka, K.; Miyazono, K.; Imamura, T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 2003, 14, 2809–2817. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.-X.; Hussain, Z.J.; Zhang, Y.E. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J. Biol. Chem. 2003, 278, 39029–39036. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Qiao, M.; Oyajobi, B.O.; Mundy, G.R.; Chen, D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J. Biol. Chem. 2003, 278, 27939–27944. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, J.; Guo, R.; Wang, Y.; Chen, D.; Xing, L. Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. J. Bone Miner. Res. 2010, 25, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Liu, F.; Lee, M.; Wu, B.; Ting, K.; Zara, J.N.; Soo, C.; Al Hezaimi, K.; Zou, W.; Chen, X.; et al. NF-κB inhibits osteogenic differentiation of mesenchymal stem cells by promoting β-catenin degradation. Proc. Natl. Acad. Sci. USA 2013, 110, 9469–9474. [Google Scholar] [CrossRef]
- Boonanantanasarn, K.; Lee, H.L.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Baek, J.H.; Kim, G.S. EGF Inhibits Wnt/β-Catenin-Induced Osteoblast Differentiation by Promoting β-Catenin Degradation. J. Cell Biochem. 2015, 116, 2849–2857. [Google Scholar] [CrossRef]
- Nam, B.; Park, H.; Lee, Y.L.; Oh, Y.; Park, J.; Kim, S.Y.; Weon, S.; Choi, S.H.; Yang, J.H.; Jo, S.; et al. TGFβ1 Suppressed Matrix Mineralization of Osteoblasts Differentiation by Regulating SMURF1-C/EBPβ-DKK1 Axis. Int. J. Mol. Sci. 2020, 21, 9771. [Google Scholar] [CrossRef]
- Andrews, P.S.; Schneider, S.; Yang, E.; Michaels, M.; Chen, H.; Tang, J.; Emkey, R. Identification of substrates of SMURF1 ubiquitin ligase activity utilizing protein microarrays. Assay Drug Dev. Technol. 2010, 8, 471–487. [Google Scholar] [CrossRef]
- O’Connor, H.F.; Huibregtse, J.M. Enzyme-substrate relationships in the ubiquitin system: Approaches for identifying substrates of ubiquitin ligases. Cell. Mol. Life Sci. 2017, 74, 3363–3375. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Park, H.J.; Kwon, A.; Baek, K.; Woo, K.M.; Ryoo, H.M.; Kim, G.S.; Baek, J.H. Smurf1 plays a role in EGF inhibition of BMP2-induced osteogenic differentiation. Exp. Cell Res. 2014, 323, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Yamashita, M.; Ikegami, K.; Nakamura, T.; Yanagita, M.; Yamada, S.; Kitamura, M.; Murakami, S. TGF-Beta Negatively Regulates the BMP2-Dependent Early Commitment of Periodontal Ligament Cells into Hard Tissue Forming Cells. PLoS ONE 2015, 10, e0125590. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xie, Z.; Ma, Y.; Pan, X.; Wang, J.; Chen, Z.; Shi, P. TGF-β inhibits osteogenesis by upregulating the expression of ubiquitin ligase SMURF1 via MAPK-ERK signaling. J. Cell. Physiol. 2018, 233, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Kaneki, H.; Guo, R.; Chen, D.; Yao, Z.; Schwarz, E.M.; Zhang, Y.E.; Boyce, B.F.; Xing, L. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J. Biol. Chem. 2006, 281, 4326–4333. [Google Scholar] [CrossRef]
- Guo, R.; Yamashita, M.; Zhang, Q.; Zhou, Q.; Chen, D.; Reynolds, D.G.; Awad, H.A.; Yanoso, L.; Zhao, L.; Schwarz, E.M.; et al. Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. J. Biol. Chem. 2008, 283, 23084–23092. [Google Scholar] [CrossRef]
- Lee, H.L.; Yi, T.; Baek, K.; Kwon, A.; Hwang, H.R.; Qadir, A.S.; Park, H.J.; Woo, K.M.; Ryoo, H.M.; Kim, G.S.; et al. Tumor necrosis factor-α enhances the transcription of Smad ubiquitination regulatory factor 1 in an activating protein-1- and Runx2-dependent manner. J. Cell. Physiol. 2013, 228, 1076–1086. [Google Scholar] [CrossRef]
- Jang, W.G.; Jeong, B.C.; Kim, E.J.; Choi, H.; Oh, S.H.; Kim, D.K.; Koo, S.H.; Choi, H.S.; Koh, J.T. Cyclic AMP Response Element-binding Protein H (CREBH) Mediates the Inhibitory Actions of Tumor Necrosis Factor α in Osteoblast Differentiation by Stimulating Smad1 Degradation. J. Biol. Chem. 2015, 290, 13556–13566. [Google Scholar] [CrossRef]
- Kim, K.M.; Jeon, W.J.; Kim, E.J.; Jang, W.G. CRTC2 suppresses BMP2-induced osteoblastic differentiation via Smurf1 expression in MC3T3-E1 cells. Life Sci. 2018, 214, 70–76. [Google Scholar] [CrossRef]
- Lian, C.; Wu, Z.; Gao, B.; Peng, Y.; Liang, A.; Xu, C.; Liu, L.; Qiu, X.; Huang, J.; Zhou, H.; et al. Melatonin reversed tumor necrosis factor-alpha-inhibited osteogenesis of human mesenchymal stem cells by stabilizing SMAD1 protein. J. Pineal Res. 2016, 61, 317–327. [Google Scholar] [CrossRef]
- Guo, J.; Qiu, X.; Zhang, L.; Wei, R. Smurf1 regulates macrophage proliferation, apoptosis and migration via JNK and p38 MAPK signaling pathways. Mol. Immunol. 2018, 97, 20–26. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, W.; Hu, C.; Xue, Z.; Wang, G.; Ding, B.; Luo, H.; Tang, L.; Kong, X.; Chen, X.; et al. MiR-17 modulates osteogenic differentiation through a coherent feed-forward loop in mesenchymal stem cells isolated from periodontal ligaments of patients with periodontitis. Stem Cells 2011, 29, 1804–1816. [Google Scholar] [CrossRef]
- Liu, W.; Qi, M.; Konermann, A.; Zhang, L.; Jin, F.; Jin, Y. The p53/miR-17/Smurf1 pathway mediates skeletal deformities in an age-related model via inhibiting the function of mesenchymal stem cells. Aging 2015, 7, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Partridge, N.; Selvamurugan, N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J. Cell. Physiol. 2014, 229, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xu, J.; Xu, L.; Zhang, J.; Chan, K.; Pan, X.; Li, G. MiR-503 Promotes Bone Formation in Distraction Osteogenesis through Suppressing Smurf1 Expression. Sci. Rep. 2017, 7, 409. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Kushwaha, P.; Karvande, A.; Tripathi, A.K.; Kothari, P.; Adhikary, S.; Khedgikar, V.; Mishra, V.K.; Trivedi, R. MicroRNA-672-5p Identified during Weaning Reverses Osteopenia and Sarcopenia in Ovariectomized Mice. Mol. Ther. Nucleic Acids 2019, 14, 536–549. [Google Scholar] [CrossRef]
- Ye, L.C.; Qian, L.F.; Liang, L.; Jiang, L.J.; Che, Z.Y.; Guo, Y.H. Overexpression of miR-195-5p reduces osteoporosis through activating BMP-2/SMAD/Akt/RUNX2 pathway via targeting SMURF1. J. Biol. Regul. Homeost. Agents 2021, 35. [Google Scholar] [CrossRef]
- Liu, C.; Gao, X.; Li, Y.; Sun, W.; Xu, Y.; Tan, Y.; Du, R.; Zhong, G.; Zhao, D.; Liu, Z.; et al. The mechanosensitive lncRNA Neat1 promotes osteoblast function through paraspeckle-dependent Smurf1 mRNA retention. Bone Res. 2022, 10, 18. [Google Scholar] [CrossRef]
- Huang, S.; Li, Y.; Wu, P.; Xiao, Y.; Duan, N.; Quan, J.; Du, W. microRNA-148a-3p in extracellular vesicles derived from bone marrow mesenchymal stem cells suppresses SMURF1 to prevent osteonecrosis of femoral head. J. Cell. Mol. Med. 2020, 24, 11512–11523. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, J.; Li, Z.; Jia, G. Bone Marrow Mesenchymal Stem Cell-Derived Exosomal miR-25 Regulates the Ubiquitination and Degradation of Runx2 by SMURF1 to Promote Fracture Healing in Mice. Front. Med. 2020, 7, 577578. [Google Scholar] [CrossRef]
- Xiong, A.; He, Y.; Gao, L.; Li, G.; Weng, J.; Kang, B.; Wang, D.; Zeng, H. Smurf1-targeting miR-19b-3p-modified BMSCs combined PLLA composite scaffold to enhance osteogenic activity and treat critical-sized bone defects. Biomater. Sci. 2020, 8, 6069–6081. [Google Scholar] [CrossRef] [PubMed]
- Bellido, T.; Ali, A.A.; Plotkin, L.I.; Fu, Q.; Gubrij, I.; Roberson, P.K.; Weinstein, R.S.; O’Brien, C.A.; Manolagas, S.C.; Jilka, R.L. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts: A putative explanation for why intermittent administration is needed for bone anabolism. J. Biol. Chem. 2003, 278, 50259–50272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, X.; Li, D.; Cai, L.; Xu, Q. METTL3 Regulates Osteoblast Differentiation and Inflammatory Response via Smad Signaling and MAPK Signaling. Int. J. Mol. Sci. 2019, 21, 199. [Google Scholar] [CrossRef]
- Sangadala, S.; Boden, S.D.; Viggeswarapu, M.; Liu, Y.; Titus, L. LIM mineralization protein-1 potentiates bone morphogenetic protein responsiveness via a novel interaction with Smurf1 resulting in decreased ubiquitination of Smads. J. Biol. Chem. 2006, 281, 17212–17219. [Google Scholar] [CrossRef]
- Sangadala, S.; Boden, S.D.; Metpally, R.P.; Reddy, B.V. Modeling and analysis of molecularinteraction between Smurf1-WW2 domain and various isoforms of LIM mineralization protein. Proteins 2007, 68, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Sangadala, S.; Yoshioka, K.; Enyo, Y.; Liu, Y.; Titus, L.; Boden, S.D. Characterization of a unique motif in LIM mineralization protein-1 that interacts with jun activation-domain-binding protein 1. Mol. Cell. Biochem. 2014, 385, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xue, F.; Zhang, C.; Li, G. LMCD1 promotes osteogenic differentiation of human bone marrow stem cells by regulating BMP signaling. Cell Death Dis. 2019, 10, 647. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, R.; Zhang, K.; Yuan, J.; Wang, J.; Wang, X.; Ma, J.; Wu, C. A PINCH-1-Smurf1 signaling axis mediates mechano-regulation of BMPR2 and stem cell differentiation. J. Cell Biol. 2019, 218, 3773–3794. [Google Scholar] [CrossRef]
- Lu, K.; Yin, X.; Weng, T.; Xi, S.; Li, L.; Xing, G.; Cheng, X.; Yang, X.; Zhang, L.; He, F. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat. Cell Biol. 2008, 10, 994–1002. [Google Scholar] [CrossRef]
- Liu, J.; Liang, C.; Guo, B.; Wu, X.; Li, D.; Zhang, Z.; Zheng, K.; Dang, L.; He, X.; Lu, C.; et al. Increased PLEKHO1 within osteoblasts suppresses Smad-dependent BMP signaling to inhibit bone formation during aging. Aging Cell 2017, 16, 360–376. [Google Scholar] [CrossRef]
- Wan, L.; Zou, W.; Gao, D.; Inuzuka, H.; Fukushima, H.; Berg, A.H.; Drapp, R.; Shaik, S.; Hu, D.; Lester, C.; et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol. Cell 2011, 44, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Shimazu, J.; Wei, J.; Karsenty, G. Smurf1 Inhibits Osteoblast Differentiation, Bone Formation, and Glucose Homeostasis through Serine 148. Cell Rep. 2016, 15, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.; Nguyen, P.H.; Davis, B.N.; Ohoka, N.; Hayashi, H.; Du, K.; Lagna, G.; Hata, A. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol. Cell. Biol. 2007, 27, 5776–5789. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; He, S.; Xing, C.; Lu, K.; Wang, J.; Xing, G.; Meng, A.; Jia, S.; He, F.; Zhang, L. SCFFBXL¹⁵ regulates BMP signalling by directing the degradation of HECT-type ubiquitin ligase Smurf1. EMBO J. 2011, 30, 2675–2689. [Google Scholar] [CrossRef]
- Li, H.; Cui, Y.; Wei, J.; Liu, C.; Chen, Y.; Cui, C.P.; Li, L.; Zhang, X.; Zhang, L. VCP/p97 increases BMP signaling by accelerating ubiquitin ligase Smurf1 degradation. FASEB J. 2019, 33, 2928–2943. [Google Scholar] [CrossRef]
- Jeon, E.J.; Lee, K.Y.; Choi, N.S.; Lee, M.H.; Kim, H.N.; Jin, Y.H.; Ryoo, H.M.; Choi, J.Y.; Yoshida, M.; Nishino, N.; et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006, 281, 16502–16511. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, X.; Zhu, T.; Zhang, M.; Shen, R.; Xing, L.; O’Keefe, R.J.; Chen, D. Bone morphogenetic protein 2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J. Biol. Chem. 2007, 282, 10742–10748. [Google Scholar] [CrossRef]
- Yan, X.; Wang, H.; Li, Y.; Jiang, Y.; Shao, Q.; Xu, W. MicroRNA-92a overexpression promotes the osteogenic differentiation of bone mesenchymal stem cells by impeding Smad6-mediated runt-related transcription factor 2 degradation. Mol. Med. Rep. 2018, 17, 7821–7826. [Google Scholar]
- Sapkota, G.; Alarcón, C.; Spagnoli, F.M.; Brivanlou, A.H.; Massagué, J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell 2007, 25, 441–454. [Google Scholar] [CrossRef]
- Yoon, W.-J.; Islam, R.; Cho, Y.-D.; Woo, K.-M.; Baek, J.-H.; Uchida, T.; Komori, T.; van Wijnen, A.; Stein, J.L.; Lian, J.B.; et al. Pin1-mediated Runx2 modification is critical for skeletal development. J. Cell. Physiol. 2013, 228, 2377–2385. [Google Scholar] [CrossRef]
- Yoon, W.J.; Islam, R.; Cho, Y.D.; Ryu, K.M.; Shin, H.R.; Woo, K.M.; Baek, J.H.; Ryoo, H.M. Pin1 plays a critical role as a molecular switch in canonical BMP signaling. J. Cell. Physiol. 2015, 230, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.C.; Wang, M.Y.; Zhang, S.W.; Wu, Y.S.; Zhou, C.C.; Zheng, R.X.; Shao, B.; Wang, Y.; Xie, L.; Liu, W.Q.; et al. Ubiquitin-specific protease USP34 controls osteogenic differentiation and bone formation by regulating BMP2 signaling. EMBO J. 2018, 37, e99398, published correction appears in EMBO J. 2020, 39, e105578. [Google Scholar]
- Cao, Y.; Wang, C.; Zhang, X.; Xing, G.; Lu, K.; Gu, Y.; He, F.; Zhang, L. Selective small molecule compounds increase BMP-2 responsiveness by inhibiting Smurf1-mediated Smad1/5 degradation. Sci. Rep. 2014, 4, 4965. [Google Scholar] [CrossRef]
- Okada, M.; Sangadala, S.; Liu, Y.; Yoshida, M.; Reddy, B.V.; Titus, L.; Boden, S.D. Development and optimization of a cell-based assay for the selection of synthetic compounds that potentiate bone morphogenetic protein-2 activity. Cell Biochem. Funct. 2009, 27, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Peng, S.; Li, J.; Lu, J.; Guan, D.; Jiang, F.; Lu, C.; Li, F.; He, X.; Zhu, H.; et al. Inhibition of osteoblastic Smurf1 promotes bone formation in mouse models of distinctive age-related osteoporosis. Nat. Commun. 2018, 9, 3428. [Google Scholar] [CrossRef]
- Hsu, C.W.; Liu, S.; Hsu, E.; Hollinger, J.O. Inhibition of rhBMP-2-induced ALP activity by intracellular delivery of SMURF1 in murine calvarial preosteoblast cells. J. Biomed. Mater. Res. A 2014, 102, 4037–4043. [Google Scholar] [CrossRef]
- Rodríguez-Évora, M.; García-Pizarro, E.; del Rosario, C.; Pérez-López, J.; Reyes, R.; Delgado, A.; Rodríguez-Rey, J.C.; Évora, C. Smurf1 knocked-down, mesenchymal stem cells and BMP-2 in an electrospun system for bone regeneration. Biomacromolecules 2014, 15, 1311–1322. [Google Scholar] [CrossRef]
- García-García, P.; Ruiz, M.; Reyes, R.; Delgado, A.; Évora, C.; Riancho, J.A.; Rodríguez-Rey, J.C.; Pérez-Campo, F.M. Smurf1 Silencing Using a LNA-ASOs/Lipid Nanoparticle System to Promote Bone Regeneration. Stem Cells Transl. Med. 2019, 8, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Zeng, T.; Liu, M.; Han, S.; Lin, H.; Lin, Q.; Li, L.; Jiang, T.; Li, G.; Lin, H.; et al. A cell-based high-throughput screening method based on a ubiquitin-reference technique for identifying modulators of E3 ligases. J. Biol. Chem. 2019, 294, 2880–2891. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, S.; Ogunjimi, A.A.; Wang, H.R.; Rotin, D.; Sicheri, F.; Wrana, J.L.; Forman-Kay, J.D. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell 2007, 130, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Ruetalo, N.; Anders, S.; Stollmaier, C.; Jäckl, M.; Schütz-Stoffregen, M.C.; Stefan, N.; Wolf, C.; Wiesner, S. The WW1 Domain Enhances Autoinhibition in Smurf Ubiquitin Ligases. J. Mol. Biol. 2019, 431, 4834–4847. [Google Scholar] [CrossRef]
- Córdova, L.A.; Loi, F.; Lin, T.H.; Gibon, E.; Pajarinen, J.; Nabeshima, A.; Lu, L.; Yao, Z.; Goodman, S.B. CCL2, CCL5, and IGF-1 participate in the immunomodulation of osteogenesis during M1/M2 transition in vitro. J. Biomed. Mater. Res. A 2017, 105, 3069–3076. [Google Scholar] [CrossRef]
- Khedgikar, V.; Kushwaha, P.; Gautam, J.; Verma, A.; Changkija, B.; Kumar, A.; Sharma, S.; Nagar, G.K.; Singh, D.; Trivedi, P.K.; et al. Withaferin A: A proteasomal inhibitor promotes healing after injury and exerts anabolic effect on osteoporotic bone. Cell Death Dis. 2013, 4, e778. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; He, H.; Wang, M.; Liang, J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 2019, 52, e12688. [Google Scholar] [CrossRef]
- Hu, F.; Jiang, C.; Bu, G.; Fu, Y.; Yu, Y. Silencing long noncoding RNA colon cancer-associated transcript-1 upregulates microRNA-34a-5p to promote proliferation and differentiation of osteoblasts in osteoporosis. Cancer Gene Ther. 2021, 28, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Vishal, M.; Vimalraj, S.; Ajeetha, R.; Gokulnath, M.; Keerthana, R.; He, Z.; Partridge, N.C.; Selvamurugan, N. MicroRNA-590-5p Stabilizes Runx2 by Targeting Smad7 During Osteoblast Differentiation. J. Cell. Physiol. 2017, 232, 371–380. [Google Scholar] [CrossRef]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, Y.J.; Jeong, H.M.; Jin, Y.H.; Yeo, C.Y.; Lee, K.Y. Akt enhances Runx2 protein stability by regulating Smurf2 function during osteoblast differentiation. FEBS J. 2014, 281, 3656–3666. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, P.; Xie, Z.; Wang, S.; Cen, S.; Li, M.; Liu, W.; Tang, S.; Ye, G.; Zheng, G.; et al. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019, 26, 2652–2666. [Google Scholar] [CrossRef]
- Xu, Z.; Greenblatt, M.B.; Yan, G.; Feng, H.; Sun, J.; Lotinun, S.; Brady, N.; Baron, R.; Glimcher, L.H.; Zou, W. SMURF2 regulates bone homeostasis by disrupting SMAD3 interaction with vitamin D receptor in osteoblasts. Nat. Commun. 2017, 8, 14570. [Google Scholar] [CrossRef]
- Kushioka, J.; Kaito, T.; Okada, R.; Ishiguro, H.; Bal, Z.; Kodama, J.; Chijimatsu, R.; Pye, M.; Narimatsu, M.; Wrana, J.L.; et al. A novel negative regulatory mechanism of Smurf2 in BMP/Smad signaling in bone. Bone Res. 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Obri, A.; Makinistoglu, M.P.; Zhang, H.; Karsenty, G. HDAC4 integrates PTH and sympathetic signaling in osteoblasts. J. Cell Biol. 2014, 205, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, T.L.; Wang, L.; Meng, X.H.; Zhu, W.; Zeng, Y.; Zhu, J.Q.; Zhou, Y.; Xiao, H.M.; Deng, H.W. Network-based Transcriptome-wide Expression Study for Postmenopausal Osteoporosis. J. Clin. Endocrinol. Metab. 2020, 105, 2678–2691. [Google Scholar] [CrossRef]
- Bonni, S.; Wang, H.R.; Causing, C.G.; Kavsak, P.; Stroschein, S.L.; Luo, K.; Wrana, J.L. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 2001, 3, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liang, M.; Feng, X.H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 2000, 275, 36818–36822. [Google Scholar] [CrossRef]
- Nakano, A.; Koinuma, D.; Miyazawa, K.; Uchida, T.; Saitoh, M.; Kawabata, M.; Hanai, J.; Akiyama, H.; Abe, M.; Miyazono, K.; et al. Pin1 down-regulates transforming growth factor-beta (TGF-beta) signaling by inducing degradation of Smad proteins. J. Biol. Chem. 2009, 284, 6109–6115. [Google Scholar] [CrossRef]
- Zhang, Y.; Chang, C.; Gehling, D.J.; Hemmati-Brivanlou, A.; Derynck, R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2001, 98, 974–979. [Google Scholar] [CrossRef]
- Jones, D.C.; Wein, M.N.; Glimcher, L.H. Schnurri-3, a Key Regulator of Postnatal Skeletal Remodeling. In Osteoimmunology; Choi, Y., Ed.; Advances in Experimental Medicine and Biology Series 602; Springer: Boston, MA, USA, 2007; pp. 1–13. [Google Scholar]
- Glimcher, L.H.; Jones, D.C.; Wein, M.N. Control of postnatal bone mass by the zinc finger adapter protein Schnurri-3. Ann. N. Y. Acad. Sci. 2007, 1116, 174–181. [Google Scholar] [CrossRef]
- Jones, D.C.; Wein, M.N.; Oukka, M.; Hofstaetter, J.G.; Glimcher, M.J.; Glimcher, L.H. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 2006, 312, 1223–1227. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, J.; Zhang, H.; Wang, Y.; Matesic, L.E.; Takahata, M.; Awad, H.; Chen, D.; Xing, L. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells 2011, 29, 1601–1610. [Google Scholar] [CrossRef]
- Shu, L.; Zhang, H.; Boyce, B.F.; Xing, L. Ubiquitin E3 ligase Wwp1 negatively regulates osteoblast function by inhibiting osteoblast differentiation and migration. J. Bone Miner. Res. 2013, 28, 1925–1935. [Google Scholar] [CrossRef]
- Wang, Y.; Malcolm, D.W.; Benoit, D.S.W. Controlled and sustained delivery of siRNA/NPs from hydrogels expedites bone fracture healing. Biomaterials 2017, 139, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Tang, J.; He, H.; Cheng, P.; Chen, C. MiR-142-5p promotes bone repair by maintaining osteoblast activity. J. Bone Miner. Metab. 2017, 35, 255–264. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Ma, Y.; Du, W.; Feng, H.; Feng, K.; Li, G.; Wang, S. MicroRNA-15b shuttled by bone marrow mesenchymal stem cell-derived extracellular vesicles binds to WWP1 and promotes osteogenic differentiation. Arthritis Res. Ther. 2020, 22, 269. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Y.; Feng, S.; He, P.; Sheng, B.; Ni, J. miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/β-catenin signaling pathway. Exp. Mol. Med. 2021, 53, 973–985. [Google Scholar] [CrossRef]
- Tucker, W.O.; Kinghorn, A.B.; Fraser, L.A.; Cheung, Y.W.; Tanner, J.A. Selection and Characterization of a DNA Aptamer Specifically Targeting Human HECT Ubiquitin Ligase WWP1. Int. J. Mol. Sci. 2018, 19, 763. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, X.; Shim, J.H.; Huang, Z.; Brady, N.; Hu, D.; Drapp, R.; Sigrist, K.; Glimcher, L.H.; Jones, D. The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 2011, 13, 59–65. [Google Scholar] [CrossRef]
- Shao, R.; Liu, J.; Yan, G.; Zhang, J.; Han, Y.; Guo, J.; Xu, Z.; Yuan, Z.; Liu, J.; Malumbres, M.; et al. Cdh1 regulates craniofacial development via APC-dependent ubiquitination and activation of Goosecoid. Cell Res. 2016, 26, 699–712. [Google Scholar] [CrossRef][Green Version]
- Mokuda, S.; Nakamichi, R.; Matsuzaki, T.; Ito, Y.; Sato, T.; Miyata, K.; Inui, M.; Olmer, M.; Sugiyama, E.; Lotz, M.; et al. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat. Commun. 2019, 10, 2429. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; He, X.; Hua, Y.; Li, Q.; Wang, J.; Gan, X. The E3 ubiquitin ligase WWP2 facilitates RUNX2 protein transactivation in a mono-ubiquitination manner during osteogenic differentiation. J. Biol. Chem. 2017, 292, 11178–11188. [Google Scholar] [CrossRef]
- Zhang, H.; Xing, L. Ubiquitin e3 ligase itch negatively regulates osteoblast differentiation from mesenchymal progenitor cells. Stem Cells 2013, 31, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, C.; Matesic, L.E.; Li, X.; Wang, Z.; Boyce, B.F.; Xing, L. Ubiquitin E3 ligase Itch negatively regulates osteoclast formation by promoting deubiquitination of tumor necrosis factor (TNF) receptor-associated factor 6. J. Biol. Chem. 2013, 288, 22359–22368. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, W.; Li, J.; Wang, M.; Zhang, H.; Pei, L.; Boyce, B.F.; Wang, Z.; Xing, L. Clomipramine causes osteoporosis by promoting osteoclastogenesis via E3 ligase Itch, which is prevented by Zoledronic acid. Sci. Rep. 2017, 7, 41358. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Zhang, H.; Gu, R.; Wang, Z.; Gao, Z.; Xing, L. Ubiquitin E3 ligase Itch negatively regulates osteoblast function by promoting proteasome degradation of osteogenic proteins. Bone Jt. Res. 2017, 6, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-P.; Deng, F.-Y.; Yang, T.-L.; Zhang, F.; Chen, X.-D.; Shen, H.; Zhu, X.-Z.; Tian, Q.; Deng, H.-W. Genome-wide association study identified CNP12587 region underlying height variation in Chinese females. PLoS ONE 2012, 7, e44292. [Google Scholar]
- Liang, C.; Liang, G.; Zheng, X.; Huang, Y.; Huang, S.; Yin, D. RSP5 Positively Regulates the Osteogenic Differentiation of Mesenchymal Stem Cells by Activating the K63-Linked Ubiquitination of Akt. Stem Cells Int. 2020, 2020, 7073805. [Google Scholar] [PubMed]
- Butler, W.T.; Brunn, J.C.; Qin, C. Dentin extracellular matrix (ECM) proteins: Comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect. Tissue Res. 2003, 44 (Suppl. S1), 171–178. [Google Scholar] [CrossRef]
- Fisher, L.W.; Fedarko, N.S. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect. Tissue Res. 2003, 44 (Suppl. S1), 33–40. [Google Scholar] [CrossRef]
- Vijaykumar, A.; Dyrkacz, P.; Vidovic-Zdrilic, I.; Maye, P.; Mina, M. Expression of BSP-GFPtpz Transgene during Osteogenesis and Reparative Dentinogenesis. J. Dent. Res. 2020, 99, 89–97. [Google Scholar] [CrossRef]
- Opsahl Vital, S.; Gaucher, C.; Bardet, C.; Rowe, P.S.; George, A.; Linglart, A.; Chaussain, C. Tooth dentin defects reflect genetic disorders affecting bone mineralization. Bone 2012, 50, 989–997. [Google Scholar] [CrossRef]
- Jung, J.K.; Gwon, G.J.; Neupane, S.; Sohn, W.J.; Kim, K.R.; Kim, J.Y.; An, S.Y.; Kwon, T.Y.; An, C.H.; Lee, Y.; et al. Bortezomib Facilitates Reparative Dentin Formation after Pulp Access Cavity Preparation in Mouse Molar. J. Endod. 2017, 43, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Yoon, W.J.; Cho, E.S.; Kim, H.J.; Gronostajski, R.M.; Cho, M.I.; Park, J.C. Crosstalk between nuclear factor I-C and transforming growth factor-beta1 signaling regulates odontoblast differentiation and homeostasis. PLoS ONE 2011, 6, e29160. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xu, N.; Li, D.; Guan, L.; He, Y.; Zhang, Y.; Lu, Q.; Zhang, X. A feedback loop between RUNX2 and the E3 ligase SMURF1 in regulation of differentiation of human dental pulp stem cells. J. Endod. 2014, 40, 1579–1586. [Google Scholar] [CrossRef]
- Zhao, L.-D.; Xu, W.-C.; Cui, J.; Liang, Y.-C.; Cheng, W.-Q.; Xin, B.-C.; Song, J. Long non-coding RNA maternally expressed gene 3 inhibits osteogenic differentiation of human dental pulp stem cells via microRNA-543/smad ubiquitin regulatory factor 1/runt-related transcription factor 2 axis. Arch. Oral Biol. 2020, 118, 104838. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Song, M.; Du, J.; Yu, J.; Zheng, W.; Zhang, C.; Wang, Y. MiR-497-5p Regulates Osteo/Odontogenic Differentiation of Stem Cells from Apical Papilla via the Smad Signaling Pathway by Targeting Smurf2. Front. Genet. 2020, 11, 582366. [Google Scholar] [CrossRef]
- Fu, J.; Zheng, H.; Xue, Y.; Jin, R.; Yang, G.; Chen, Z.; Yuan, G. WWP2 Promotes Odontoblastic Differentiation by Monoubiquitinating KLF5. J. Dent. Res. 2021, 100, 432–439. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, X.; Zheng, H.; Yang, G.; Chen, Z.; Yuan, G. A WWP2-PTEN-KLF5 signaling axis regulates odontoblast differentiation and dentinogenesis in mice. J. Biol. Chem. 2022, 298, 102220. [Google Scholar] [CrossRef]
- Zheng, H.; Fu, J.; Chen, Z.; Yang, G.; Yuan, G. Mdm2 Promotes Odontoblast-like Differentiation by Ubiquitinating Dlx3 and p53. J. Dent. Res. 2020, 99, 320–328. [Google Scholar] [CrossRef]
- Zheng, H.; Fu, J.; Chen, Z.; Yang, G.; Yuan, G. Dlx3 Ubiquitination by Nuclear Mdm2 Is Essential for Dentinogenesis in Mice. J. Dent. Res. 2022, 101, 1064–1074. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Jiang, S.; Sheng, R.; Qi, X.; Wang, J.; Guo, Y.; Yuan, Q. USP34 regulates tooth root morphogenesis by stabilizing NFIC. Int. J. Oral Sci. 2021, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, K.; Kim, E.J.; Tyagi, A.; Karapurkar, J.K.; Haq, S.; Jung, H.S.; Kim, K.S.; Ramakrishna, S. Genome-wide screening for deubiquitinase subfamily identifies ubiquitin-specific protease 49 as a novel regulator of odontogenesis. Cell Death Differ. 2022, 29, 1689–1704. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Michon, F.; Yamada, A.; Inuzuka, H.; Yamaguchi, S.; Fukumoto, E.; Yoshizaki, K.; Nakamura, T.; Arakaki, M.; Chiba, Y.; et al. Sox21 Regulates Anapc10 Expression and Determines the Fate of Ectodermal Organ. IScience 2020, 23, 101329. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Chu, Y.; Liu, Q.; Fan, W.; He, H.; Huang, F. NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth. Int. J. Mol. Sci. 2022, 23, 9937. https://doi.org/10.3390/ijms23179937
Xu K, Chu Y, Liu Q, Fan W, He H, Huang F. NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth. International Journal of Molecular Sciences. 2022; 23(17):9937. https://doi.org/10.3390/ijms23179937
Chicago/Turabian StyleXu, Ke, Yanhao Chu, Qin Liu, Wenguo Fan, Hongwen He, and Fang Huang. 2022. "NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth" International Journal of Molecular Sciences 23, no. 17: 9937. https://doi.org/10.3390/ijms23179937
APA StyleXu, K., Chu, Y., Liu, Q., Fan, W., He, H., & Huang, F. (2022). NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth. International Journal of Molecular Sciences, 23(17), 9937. https://doi.org/10.3390/ijms23179937




