The Deubiquitinating Enzyme USP48 Interacts with the Retinal Degeneration-Associated Proteins UNC119a and ARL3
Abstract
1. Introduction
2. Results
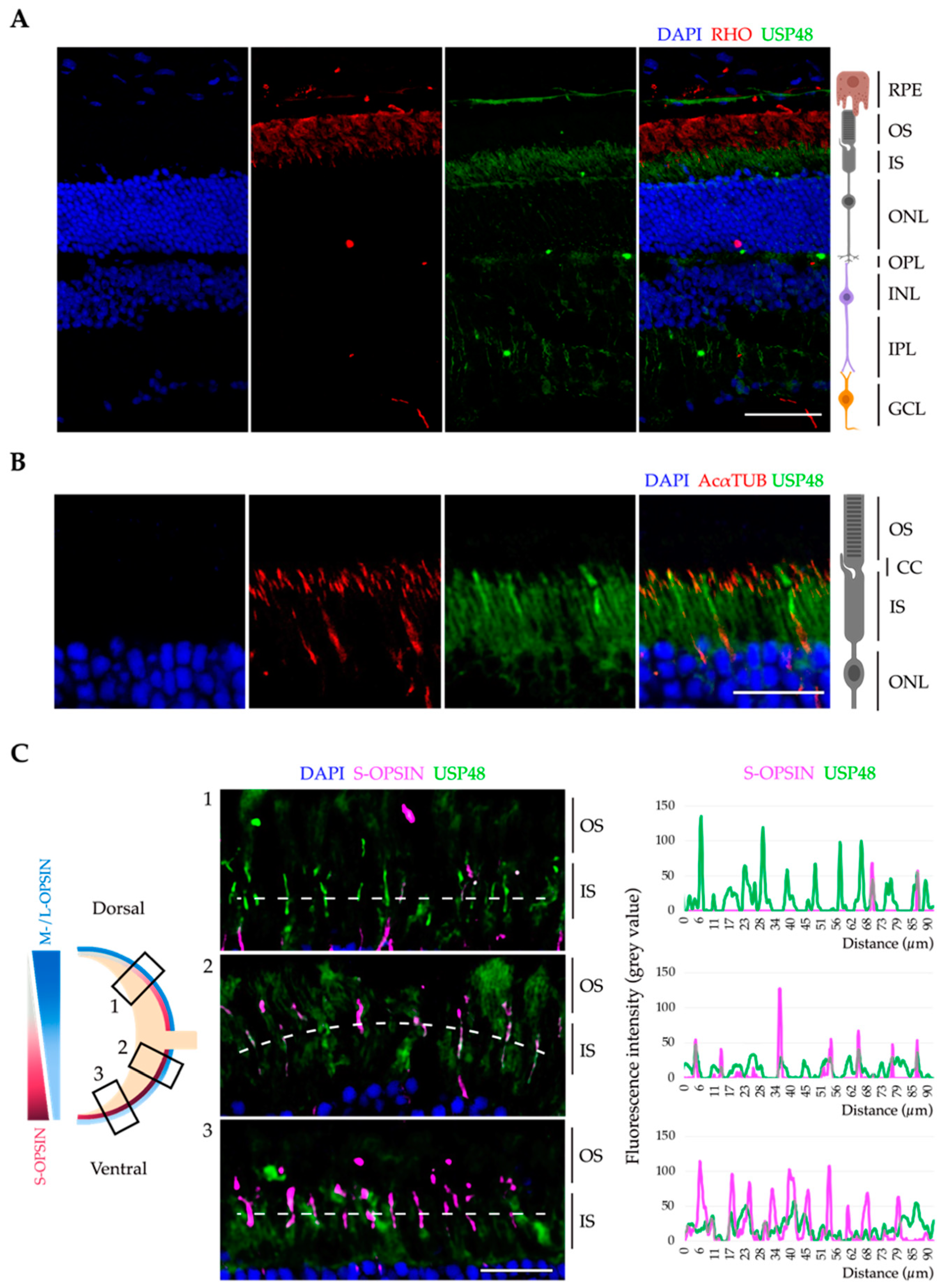
2.1. USP48 Is Highly Expressed in Cone Photoreceptors
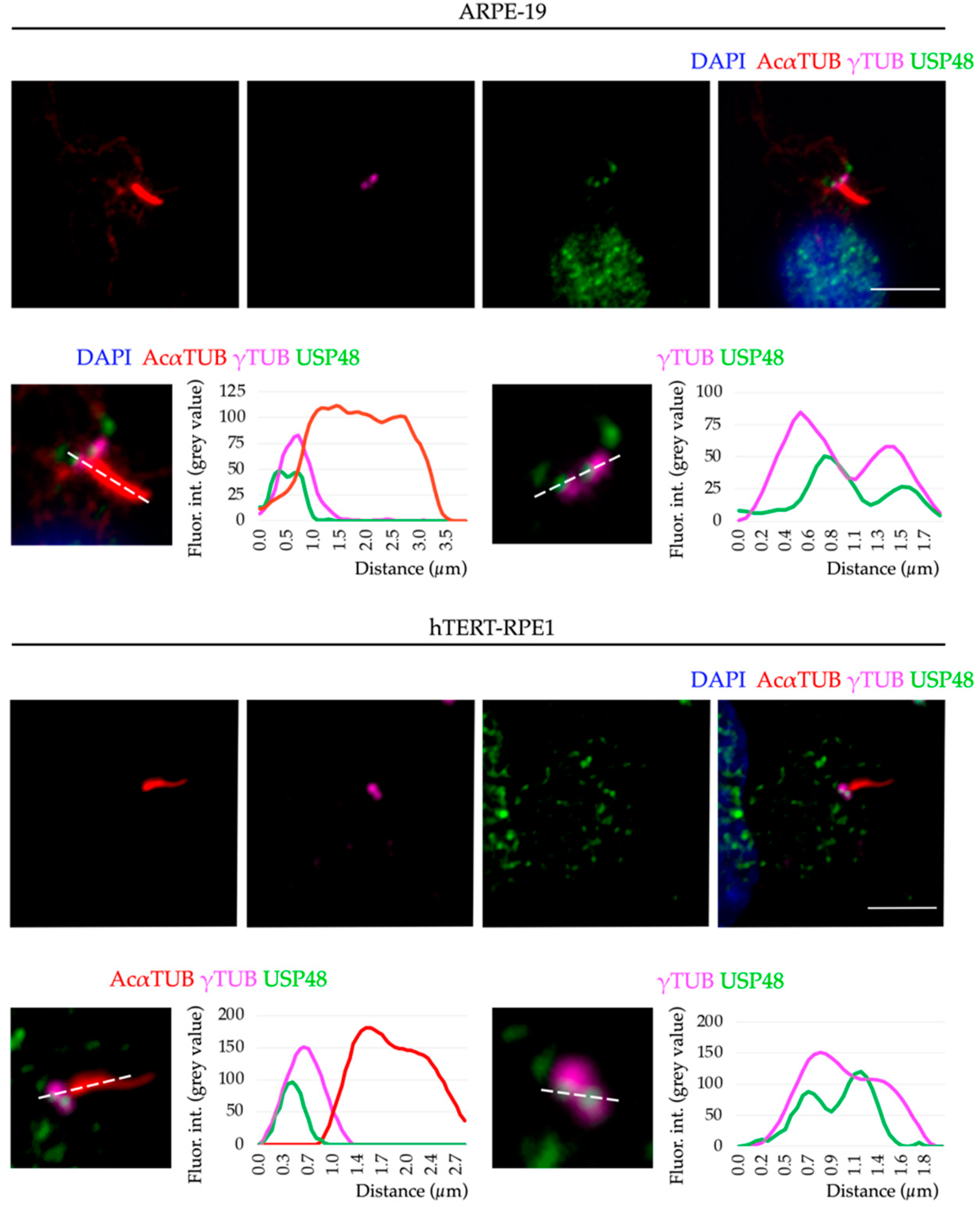
2.2. USP48 Is a Basal Body-Associated Protein
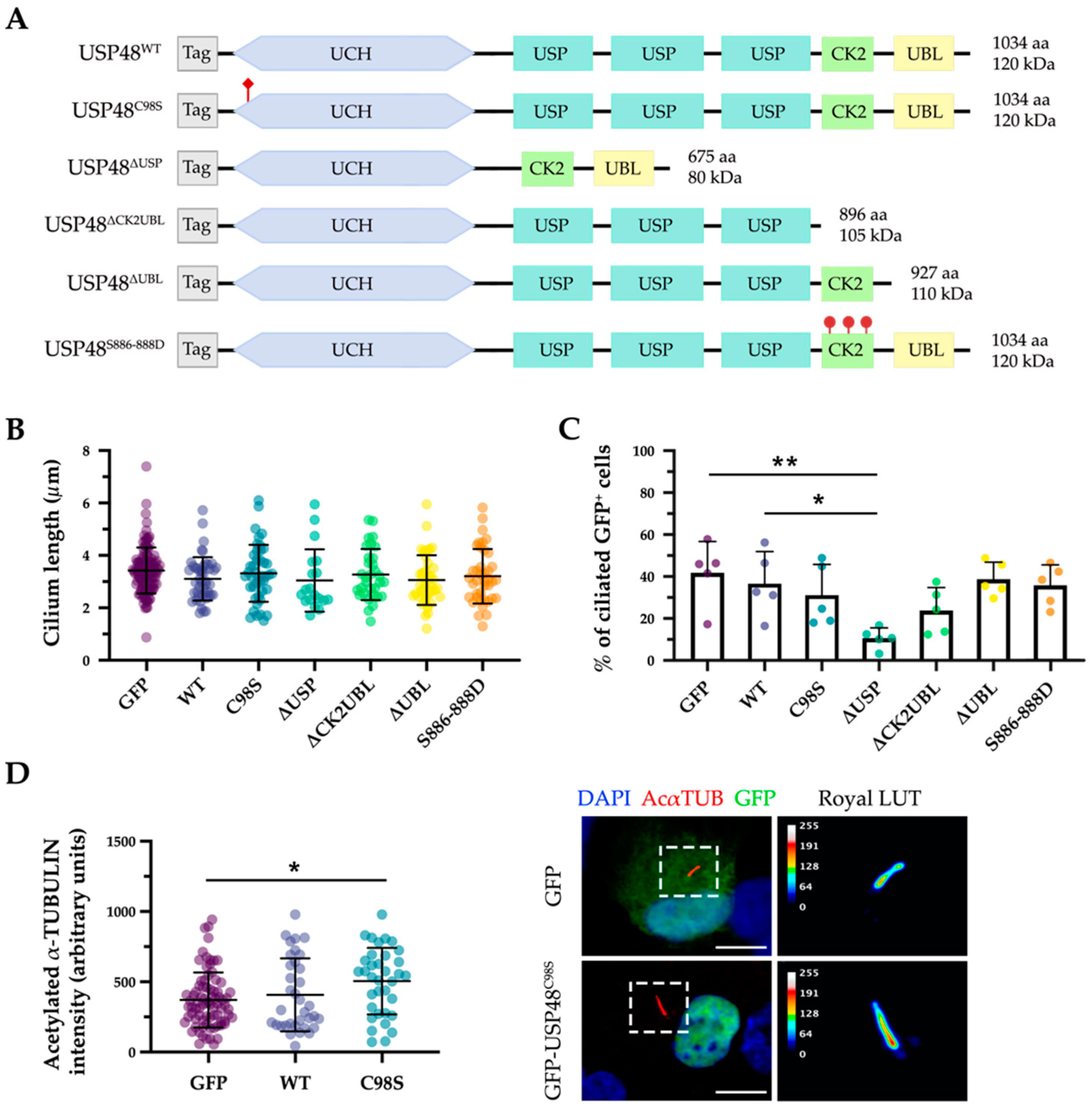
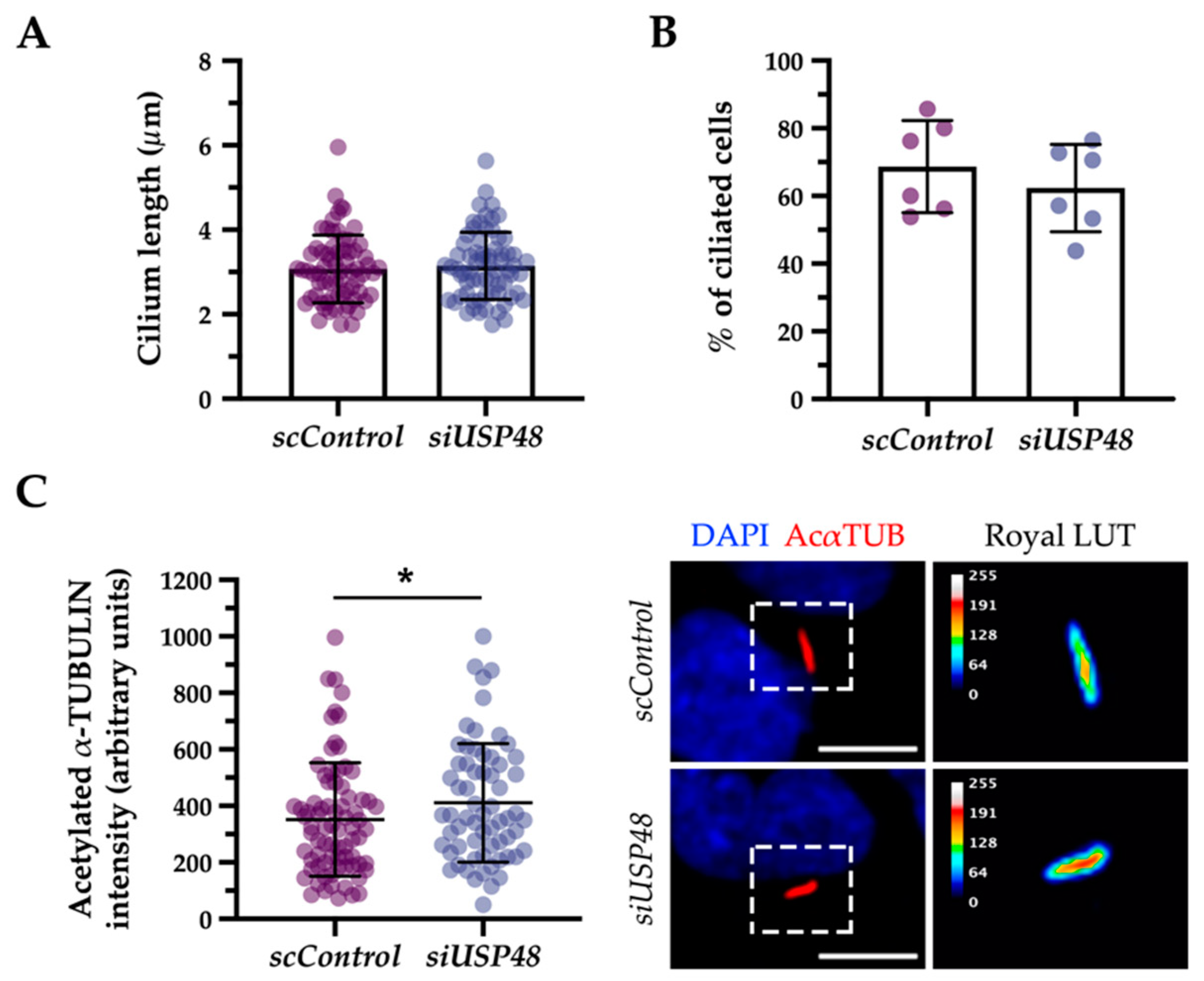
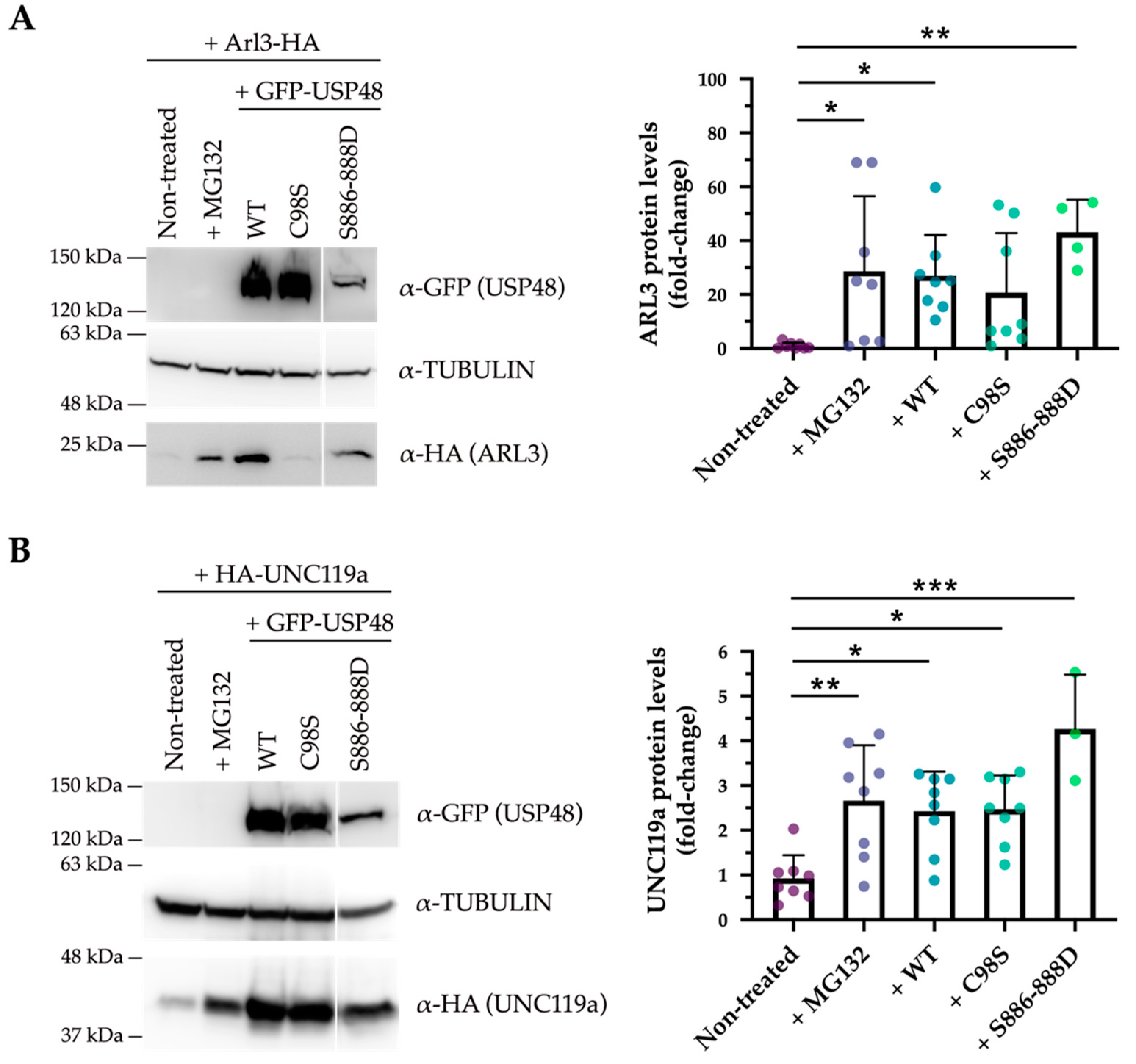
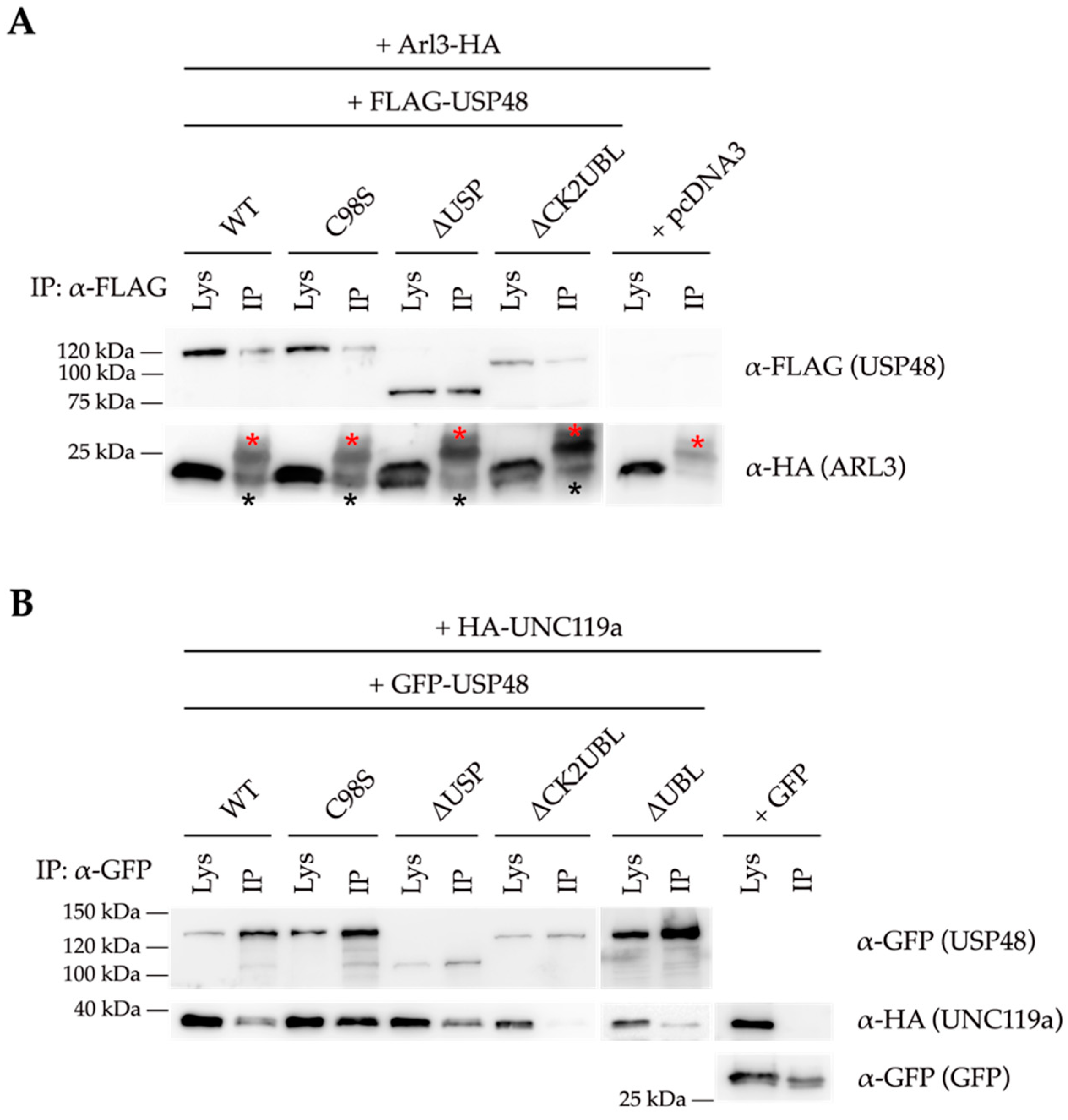
2.3. USP48 Interacts with ARL3 and UNC119a
3. Discussion
4. Materials and Methods
4.1. Animal Handling
4.2. Cell Culture and Transfections
4.3. Plasmid Vectors and Plasmid Constructions
4.4. Immunohistochemistry on Mouse Retinal Cryosections
4.5. Immunocytochemistry
4.6. Microscope Image Acquisition and Analysis
4.7. Co-Immunoprecipitation Assays
4.8. Western Blot
4.9. Mass Spectrometry and Proteomic Analysis
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoon, M.; Okawa, H.; Della Santina, L.; Wong, R.O.L. Functional Architecture of the Retina: Development and Disease. Prog. Retin. Eye Res. 2014, 42, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Khanna, H. Photoreceptor Sensory Cilium: Traversing the Ciliary Gate. Cells 2015, 4, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Bujakowska, K.M.; Liu, Q.; Pierce, E.A. Photoreceptor Cilia and Retinal Ciliopathies. Cold Spring Harb. Perspect. Biol. 2017, 9, a028274. [Google Scholar] [CrossRef]
- Heath Jeffery, R.C.; Aqif Mukhtar, S.; McAllister, I.L.; Morgan, W.H.; Mackey, D.A.; Chen, F.K. Inherited Retinal Diseases Are the Most Common Cause of Blindness in the Working-Age Population in Australia. Ophthalmic Genet. 2021, 42, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Liew, G.; Michaelides, M.; Bunce, C. A Comparison of the Causes of Blindness Certifications in England and Wales in Working Age Adults (16–64 Years), 1999–2000 with 2009–2010. BMJ Open 2014, 4, e004015. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Welby, E.; Li, T.; Swaroop, A. Retinal Disease in Ciliopathies: Recent Advances with a Focus on Stem Cell-Based Therapies. Transl. Sci. Rare Dis. 2019, 4, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Wheway, G.; Lord, J.; Baralle, D. Splicing in the Pathogenesis, Diagnosis and Treatment of Ciliopathies. Biochim. Biophys. Acta-Gene Regul. Mech. 2019, 1862, 194433. [Google Scholar] [CrossRef]
- Tatour, Y.; Ben-yosef, T. Syndromic Inherited Retinal Diseases: Genetic, Clinical and Diagnostic Aspects. Diagnostics 2020, 10, 779. [Google Scholar] [CrossRef]
- Wu, D.; Huang, J.; Zhu, H.; Chen, Z.; Chai, Y.; Ke, J.; Lei, K.; Peng, Z.; Zhang, R.; Li, X.; et al. Ciliogenesis Requires Sphingolipid-Dependent Membrane and Axoneme Interaction. Proc. Natl. Acad. Sci. USA 2022, 119, e2201096119. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Bellver, L.; Toulis, V.; Marfany, G. On the Wrong Track: Alterations of Ciliary Transport in Inherited Retinal Dystrophies. Front. Cell Dev. Biol. 2021, 9, 623734. [Google Scholar] [CrossRef]
- Murga-Zamalloa, C.A.; Swaroop, A.; Khanna, H. RPGR-Containing Protein Complexes in Syndromic and Non-Syndromic Retinal Degeneration Due to Ciliary Dysfunction. J. Genet. 2009, 88, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Kelley, R.A.; Li, T.; Swaroop, A. Primary Cilia Biogenesis and Associated Retinal Ciliopathies. Semin. Cell Dev. Biol. 2021, 110, 70–88. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, O.; Khanna, H. Ciliary Signaling Cascades in Photoreceptors. Vision Res. 2012, 75, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Shiromizu, T.; Yuge, M.; Kasahara, K.; Yamakawa, D.; Matsui, T.; Bessho, Y.; Inagaki, M.; Nishimura, Y. Targeting E3 Ubiquitin Ligases and Deubiquitinases in Ciliopathy and Cancer. Int. J. Mol. Sci. 2020, 21, 5962. [Google Scholar] [CrossRef] [PubMed]
- Hossain, D.; Tsang, W.Y. The Role of Ubiquitination in the Regulation of Primary Cilia Assembly and Disassembly. Semin. Cell Dev. Biol. 2019, 93, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.P.; Beck, J.S.; Yen, H.J.; Tayeh, M.K.; Scheetz, T.E.; Swiderski, R.E.; Nishimura, D.Y.; Braun, T.A.; Kim, K.Y.A.; Huang, J.; et al. Homozygosity Mapping with SNP Arrays Identifies TRIM32, an E3 Ubiquitin Ligase, as a Bardet-Biedl Syndrome Gene (BBS11). Proc. Natl. Acad. Sci. USA 2006, 103, 6287–6292. [Google Scholar] [CrossRef]
- Chakarova, C.F.; Khanna, H.; Shah, A.Z.; Patil, S.B.; Sedmak, T.; Murga-Zamalloa, C.A.; Papaioannou, M.G.; Nagel-Wolfrum, K.; Lopez, I.; Munro, P.; et al. TOPORS, Implicated in Retinal Degeneration, Is a Cilia-Centrosomal Protein. Hum. Mol. Genet. 2011, 20, 975–987. [Google Scholar] [CrossRef]
- Yi, Z.; Ouyang, J.; Sun, W.; Xiao, X.; Li, S.; Jia, X.; Wang, P.; Zhang, Q. Biallelic Mutations in USP45, Encoding a Deubiquitinating Enzyme, Are Associated with Leber Congenital Amaurosis. J. Med. Genet. 2019, 56, 325–331. [Google Scholar] [CrossRef]
- Toulis, V.; García-Monclús, S.; de la Peña-Ramírez, C.; Arenas-Galnares, R.; Abril, J.F.; Todi, S.V.; Khan, N.; Garanto, A.; do Carmo Costa, M.; Marfany, G. The Deubiquitinating Enzyme Ataxin-3 Regulates Ciliogenesis and Phagocytosis in the Retina. Cell Rep. 2020, 33, 108360. [Google Scholar] [CrossRef]
- Toulis, V.; Casaroli-Marano, R.; Camós-Carreras, A.; Figueras-Roca, M.; Sánchez-Dalmau, B.; Muñoz, E.; Ashraf, N.S.; Ferreira, A.F.; Khan, N.; Marfany, G.; et al. Altered Retinal Structure and Function in Spinocerebellar Ataxia Type 3. Neurobiol. Dis. 2022, 170, 105774. [Google Scholar] [CrossRef]
- Schweitzer, K.; Naumann, M. CSN-Associated USP48 Confers Stability to Nuclear NF-κB/RelA by Trimming K48-Linked Ub-Chains. Biochim. Biophys. Acta 2015, 1853, 453–469. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, P.J.; Hulihan, M.; Lincoln, S.; Hussey, J.; Skipper, L.; Bisceglio, G.; Wilkes, K.; Farrer, M.J. Identification of the Human Ubiquitin Specific Protease 31 (USP31) Gene: Structure, Sequence and Expression Analysis. DNA Seq. 2004, 15, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Lin, K.; Zhang, S.; Ma, L.; Xue, J.; Morris, S.; Aldape, K.D.; Huang, S. Gli1-Induced Deubiquitinase USP48 Aids Glioblastoma Tumorigenesis by Stabilizing Gli1. EMBO Rep. 2017, 18, 1318–1330. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, D.; Zhao, J.; Weathington, N.M.; Shang, D.; Zhao, Y. The Deubiquitinating Enzyme USP48 Stabilizes TRAF2 and Reduces E-Cadherin-Mediated Adherens Junctions. FASEB J. 2019, 32, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Faesen, A.C.; Luna-Vargas, M.P.A.; Sixma, T.K. The Role of UBL Domains in Ubiquitin-Specific Proteases. Biochem. Soc. Trans. 2012, 40, 539–545. [Google Scholar] [CrossRef]
- Velimezi, G.; Robinson-Garcia, L.; Muñoz-Martínez, F.; Wiegant, W.W.; Ferreira, J.; Owusu, M.; Moder, M.; Wiedner, M.; Rosenthal, S.B.; Fisch, K.M.; et al. Map of Synthetic Rescue Interactions for the Fanconi Anemia DNA Repair Pathway Identifies USP48. Nat. Commun. 2018, 9, 2280. [Google Scholar] [CrossRef]
- Uckelmann, M.; Densham, R.M.; Baas, R.; Winterwerp, H.H.K.; Fish, A.; Sixma, T.K.; Morris, J.R. USP48 Restrains Resection by Site-Specific Cleavage of the BRCA1 Ubiquitin Mark from H2A. Nat. Commun. 2018, 9, 229. [Google Scholar] [CrossRef]
- Antao, A.M.; Kaushal, K.; Das, S.; Singh, V.; Suresh, B.; Kim, K.S.; Ramakrishna, S. USP48 Governs Cell Cycle Progression by Regulating the Protein Level of Aurora B. Int. J. Mol. Sci. 2021, 22, 8508. [Google Scholar] [CrossRef]
- Mirra, S.; Sánchez-Bellver, L.; Casale, C.; Pescatore, A.; Marfany, G. Ubiquitin Specific Protease USP48 Destabilizes NF-κB/P65 in Retinal Pigment Epithelium Cells. Int. J. Mol. Sci. 2022, 23, 9682. [Google Scholar] [CrossRef]
- Tzimas, C.; Michailidou, G.; Arsenakis, M.; Kieff, E.; Mosialos, G.; Hatzivassiliou, E.G. Human Ubiquitin Specific Protease 31 Is a Deubiquitinating Enzyme Implicated in Activation of Nuclear Factor-κB. Cell. Signal. 2006, 18, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, A.; Schweitzer, K.; Naumann, M. Catalytic Domain of Deubiquitinylase USP48 Directs Interaction with Rel Homology Domain of Nuclear Factor KappaB Transcription Factor RelA. Mol. Biol. Rep. 2019, 46, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, Y.; Kang, M.; Feng, M.; Ren, Y.; Dai, H.; Wang, Y.; Wang, Y.; Tang, B. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021, 81, 3822–3834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, L.; Wang, B.; Ran, X.; Yang, S.; Luo, Y.; Li, Y.; Wang, Z.; Liu, Y.; Zhu, B. MiR-489-3p Promotes Malignant Progression of Non-Small Cell Lung Cancer through the Inactivation of Wnt/β-Catenin Signaling Pathway via Regulating USP48. Respir. Res. 2022, 23, 93. [Google Scholar] [CrossRef] [PubMed]
- Bassani, S.; van Beelen, E.; Rossel, M.; Voisin, N.; Morgan, A.; Arribat, Y.; Chatron, N.; Chrast, J.; Cocca, M.; Delprat, B.; et al. Variants in USP48 Encoding Ubiquitin Hydrolase Are Associated with Autosomal Dominant Non-Syndromic Hereditary Hearing Loss. Hum. Mol. Genet. 2021, 30, 1785–1796. [Google Scholar] [CrossRef]
- Tian, Q.B.; Okano, A.; Nakayama, K.; Miyazawa, S.; Endo, S.; Suzuki, T. A Novel Ubiquitin-Specific Protease, SynUSP, Is Localized at the Post-Synaptic Density and Post-Synaptic Lipid Raft. J. Neurochem. 2003, 87, 665–675. [Google Scholar] [CrossRef]
- Armando, I.; Van Villar, A.M.; Jones, J.E.; Lee, H.; Wang, X.; Asico, L.D.; Yu, P.; Yang, J.; Escano, C.S.; Pascua-Crusan, A.M.; et al. Dopamine D3 Receptor Inhibits the Ubiquitin-Specific Peptidase 48 to Promote NHE3 Degradation. FASEB J. 2014, 28, 1422–1434. [Google Scholar] [CrossRef]
- Esquerdo-Barragán, M.; Brooks, M.J.; Toulis, V.; Swaroop, A.; Marfany, G. Expression of Deubiquitinating Enzyme Genes in the Developing Mammal Retina. Mol. Vis. 2019, 25, 800–813. [Google Scholar]
- Tse, W.K.F.; Eisenhaber, B.; Ho, S.H.K.; Ng, Q.; Eisenhaber, F.; Jiang, Y.J. Genome-Wide Loss-of-Function Analysis of Deubiquitylating Enzymes for Zebrafish Development. BMC Genom. 2009, 10, 637. [Google Scholar] [CrossRef]
- Roberts, M.R.; Srinivas, M.; Forrest, D.; De Escobar, G.M.; Reh, T.A. Making the Gradient: Thyroid Hormone Regulates Cone Opsin Expression in the Developing Mouse Retina. Proc. Natl. Acad. Sci. USA 2006, 103, 6218. [Google Scholar] [CrossRef]
- Liu, Y.L.; Yan, J.; Zheng, J.; Tian, Q.B. 938RHRK941 Is Responsible for Ubiquitin Specific Protease 48 Nuclear Translocation Which Can Stabilize NF-κB (P65) in the Nucleus. Gene 2018, 669, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.J.; Baye, L.M.; Olivier-Mason, A.; Mukhopadhyay, S.; Sang, L.; Kwong, M.; Wang, W.; Pretorius, P.R.; Sheffield, V.C.; Sengupta, P.; et al. An ARL3-UNC119-RP2 GTPase Cycle Targets Myristoylated NPHP3 to the Primary Cilium. Genes Dev. 2011, 25, 2347–2360. [Google Scholar] [CrossRef] [PubMed]
- Hanke-Gogokhia, C.; Wu, Z.; Gerstner, C.D.; Frederick, J.M.; Zhang, H.; Baehr, W. Arf-like Protein 3 (ARL3) Regulates Protein Trafficking and Ciliogenesis in Mouse Photoreceptors. J. Biol. Chem. 2016, 291, 7142–7155. [Google Scholar] [CrossRef] [PubMed]
- Toulis, V.; Marfany, G. By the Tips of Your Cilia: Ciliogenesis in the Retina and the Ubiquitin-Proteasome System. In Proteostasis and Disease. Advances in Experimental Medicine and Biology; Barrio, R., Sutherland, J., Rodriguez, M., Eds.; Springer: Cham, Switzerland, 2020; Volume 1233, pp. 303–310. [Google Scholar]
- Kasahara, K.; Kawakami, Y.; Kiyono, T.; Yonemura, S.; Kawamura, Y.; Era, S.; Matsuzaki, F.; Goshima, N.; Inagaki, M. Ubiquitin-Proteasome System Controls Ciliogenesis at the Initial Step of Axoneme Extension. Nat. Commun. 2014, 5, 5081. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.R.; Nager, A.R.; Nachury, M.V. Ubiquitin Chains Earmark GPCRs for BBSome-Mediated Removal from Cilia. J. Cell Biol. 2020, 219, e202003020. [Google Scholar] [CrossRef]
- Esquerdo, M.; Grau-Bové, X.; Garanto, A.; Toulis, V.; Garcia-Monclús, S.; Millo, E.; López-Iniesta, M.J.; Abad-Morales, V.; Ruiz-Trillo, I.; Marfany, G. Expression Atlas of the Deubiquitinating Enzymes in the Adult Mouse Retina, Their Evolutionary Diversification and Phenotypic Roles. PLoS ONE 2016, 11, e0150364. [Google Scholar] [CrossRef]
- Bauskis, A.; Strange, C.; Molster, C.; Fisher, C. The Diagnostic Odyssey: Insights from Parents of Children Living with an Undiagnosed Condition. Orphanet J. Rare Dis. 2022, 17, 233. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Zhang, X.; Song, G.; Guo, Q.; Zhang, Z.; Diao, Y.; Yin, H.; Liu, H.; Jiang, G. Deubiquitinase USP48 Promotes ATRA-Induced Granulocytic Differentiation of Acute Promyelocytic Leukemia Cells. Int. J. Oncol. 2018, 53, 895–903. [Google Scholar] [CrossRef]
- Aísa-Marín, I.; García-Arroyo, R.; Mirra, S.; Marfany, G. The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease. Int. J. Mol. Sci. 2021, 22, 1855. [Google Scholar] [CrossRef]
- Murphy, D.; Singh, R.; Kolandaivelu, S.; Ramamurthy, V.; Stoilov, P. Alternative Splicing Shapes the Phenotype of a Mutation in BBS8 To Cause Nonsyndromic Retinitis Pigmentosa. Mol. Cell. Biol. 2015, 35, 1860–1870. [Google Scholar] [CrossRef]
- Vervoort, R.; Lennon, A.; Bird, A.C.; Tulloch, B.; Axton, R.; Miano, M.G.; Meindl, A.; Meitinger, T.; Ciccodicola, A.; Wright, A.F. Mutational Hot Spot within a New RPGR Exon in X-Linked Retinitis Pigmentosa. Nat. Genet. 2000, 25, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Ciampi, L.; Mantica, F.; López-Blanch, L.; Permanyer, J.; Rodriguez-Marín, C.; Zang, J.; Cianferoni, D.; Jiménez-Delgado, S.; Bonnal, S.; Miravet-Verde, S.; et al. Specialization of the Photoreceptor Transcriptome by Srrm3-Dependent Microexons Is Required for Outer Segment Maintenance and Vision. Proc. Natl. Acad. Sci. USA 2022, 119, e2117090119. [Google Scholar] [CrossRef] [PubMed]
- Janke, C.; Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 2017, 27, R1287–R1292. [Google Scholar] [CrossRef] [PubMed]
- Nekooki-Machida, Y.; Hagiwara, H. Role of Tubulin Acetylation in Cellular Functions and Diseases. Med. Mol. Morphol. 2020, 53, 191–197. [Google Scholar] [CrossRef]
- Villarroel-Campos, D.; Gonzalez-Billault, C. The MAP1B Case: An Old MAP That Is New Again. Dev. Neurobiol. 2014, 74, 953–971. [Google Scholar] [CrossRef]
- Goldberg, A.F.X.; Moritz, O.L.; Williams, D.S. Molecular Basis for Photoreceptor Outer Segment Architecture. Prog. Retin. Eye Res. 2016, 55, 52–81. [Google Scholar] [CrossRef]
- Malakhova, O.A.; Kim, K., II; Luo, J.K.; Zou, W.; Kumar, K.G.S.; Fuchs, S.Y.; Shuai, K.; Zhang, D.E. UBP43 Is a Novel Regulator of Interferon Signaling Independent of Its ISG15 Isopeptidase Activity. EMBO J. 2006, 25, 2358–2367. [Google Scholar] [CrossRef]
- Cetkovská, K.; Hana, Š.; Uldrijan, S. Ubiquitin-Specific Peptidase 48 Regulates Mdm2 Protein Levels Independent of Its Deubiquitinase Activity. Sci. Rep. 2017, 7, 43180. [Google Scholar] [CrossRef]
- Chaya, T.; Furukawa, T. Post-Translational Modification Enzymes as Key Regulators of Ciliary Protein Trafficking. J. Biochem. 2021, 169, 633–642. [Google Scholar] [CrossRef]
- Chaya, T.; Tsutsumi, R.; Varner, L.R.; Maeda, Y.; Yoshida, S.; Furukawa, T. Cul3-Klhl18 Ubiquitin Ligase Modulates Rod Transducin Translocation during Light-dark Adaptation. EMBO J. 2019, 38, e101409. [Google Scholar] [CrossRef]
- He, L.; Kooistra, R.; Das, R.; Oudejans, E.; van Leen, E.; Ziegler, J.; Portegies, S.; de Haan, B.; van Regteren Altena, A.S.; Stucchi, R.; et al. Cortical Anchoring of the Microtubule Cytoskeleton Is Essential for Neuron Polarity. Elife 2020, 9, e55111. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Yamada, M.; Arai, Y.; Nagai, T.; Hirotsune, S. Arl3 and LC8 Regulate Dissociation of Dynactin from Dynein. Nat. Commun. 2014, 5, 5295. [Google Scholar] [CrossRef] [PubMed]
- Suiwal, S.; Dembla, M.; Schwarz, K.; Katiyar, R.; Jung, M.; Carius, Y.; Maxeiner, S.; Lauterbach, M.A.; Lancaster, C.R.D.; Schmitz, F. Ciliary Proteins Repurposed by the Synaptic Ribbon: Trafficking Myristoylated Proteins at Rod Photoreceptor Synapses. Int. J. Mol. Sci. 2022, 23, 7135. [Google Scholar] [CrossRef]
- Kubota, S.; Kobayashi, A.; Mori, N.; Higashide, T.; McLaren, M.J.; Inana, G. Changes in Retinal Synaptic Proteins in the Transgenic Model Expressing a Mutant HRG4 (UNC119). Investig. Ophthalmol. Vis. Sci. 2002, 43, 308–313. [Google Scholar]
- Lange, S.M.; Armstrong, L.A.; Kulathu, Y. Deubiquitinases: From Mechanisms to Their Inhibition by Small Molecules. Mol. Carcinog. 2022, 82, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Holtan, J.P.; Teigen, K.; Aukrust, I.; Bragadóttir, R.; Houge, G. Dominant ARL3-Related Retinitis Pigmentosa. Ophthalmic Genet. 2019, 40, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Ratnapriya, R.; Jacobson, S.G.; Cideciyan, A.V.; English, M.A.; Roman, A.J.; Sumaroka, A.; Sheplock, R.; Swaroop, A. A Novel ARL3 Gene Mutation Associated with Autosomal Dominant Retinal Degeneration. Front. Cell Dev. Biol. 2021, 9, 720782. [Google Scholar] [CrossRef]
- Alkanderi, S.; Molinari, E.; Shaheen, R.; Elmaghloob, Y.; Stephen, L.A.; Sammut, V.; Ramsbottom, S.A.; Srivastava, S.; Cairns, G.; Edwards, N.; et al. ARL3 Mutations Cause Joubert Syndrome by Disrupting Ciliary Protein Composition. Am. J. Hum. Genet. 2018, 103, 612–620. [Google Scholar] [CrossRef]
- Toulis, V.; Garanto, A.; Marfany, G. Combining Zebrafish and Mouse Odels to Test the Function of Deubiquitinating Enzyme (DUBs) Genes in Development: Role of USP45 in the Retina. In Proteostasis. Methods in Molecular Biology; Matthiesen, R., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1449, pp. 85–101. ISBN 9781493937561. [Google Scholar]
- Bairoch, A.; Apweiler, R. The SWISS-PROT Protein Sequence Data Bank and Its Supplement TrEMBL in 1999. Nucleic Acids Res. 1999, 27, 49–54. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making Genome-Scale Phylogenetics Accessible to All. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for Large-Scale Genome and Gene Function Analysis with the PANTHER Classification System (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]

| Protein | Uniprot ID | Gene Name | Associated Disorders |
|---|---|---|---|
| Retinal-specific phospholipid-transporting ATPase ABCA4 | O35600 | Abca4 | CRD, MD, RP, STGD |
| ADP-ribosylation factor-like GTPase 3 | Q9WUL7 | Arl3 | RP, CRD, JBTS |
| Chaperonin-containing T-complex protein 1 subunit 2 | P80314 | Cct2 | LCA |
| Crystallin Mu | O54983 | Crym | NSD |
| Dynactin 2 | Q99KJ8 | Dctn2 | CMT |
| Cytoplasmic dynein 1 heavy chain 1 | Q9JHU4 | Dync1h1 | CMT |
| α-glucosidase II | Q8BHN3 | Ganab | PKD |
| G protein subunit α-transducin 2 | P50149 | Gnat2 | CRD |
| G protein subunit β-3 | Q61011 | Gnb3 | CRD |
| Guanylate cyclase 2E | P52785 | Gucy2e | CRD, LCA, RP, SLS, USH |
| Inosine Monophosphate dehydrogenase 1 | P50096 | Impdh1 | CRD, LCA, RP |
| Microtubule-associated protein 1B | P14873 | Map1b | NSD |
| N-myc downstream regulated 1 | Q62433 | Ndrg1 | CMT with deafness |
| S-opsin | P51491 | Opn1sw | Colour blindness |
| Phosphodiesterase 6C | Q91ZQ1 | Pde6c | Achromatopsia, CRD, LCA |
| Peripherin 2 | P15499 | Prph2 | CRD, LCA, MD, RP |
| Regulator of G protein signalling 9 | O54828 | Rgs9 | Oligocone trichromacy, bradyopsia |
| Unc-119 lipid binding chaperone | Q9Z2R6 | Unc119a | CRD, LCA, RP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Bellver, L.; Férriz-Gordillo, A.; Carrillo-Pz, M.; Rabanal, L.; Garcia-Gonzalo, F.R.; Marfany, G. The Deubiquitinating Enzyme USP48 Interacts with the Retinal Degeneration-Associated Proteins UNC119a and ARL3. Int. J. Mol. Sci. 2022, 23, 12527. https://doi.org/10.3390/ijms232012527
Sánchez-Bellver L, Férriz-Gordillo A, Carrillo-Pz M, Rabanal L, Garcia-Gonzalo FR, Marfany G. The Deubiquitinating Enzyme USP48 Interacts with the Retinal Degeneration-Associated Proteins UNC119a and ARL3. International Journal of Molecular Sciences. 2022; 23(20):12527. https://doi.org/10.3390/ijms232012527
Chicago/Turabian StyleSánchez-Bellver, Laura, Andrea Férriz-Gordillo, Marc Carrillo-Pz, Laura Rabanal, Francesc R. Garcia-Gonzalo, and Gemma Marfany. 2022. "The Deubiquitinating Enzyme USP48 Interacts with the Retinal Degeneration-Associated Proteins UNC119a and ARL3" International Journal of Molecular Sciences 23, no. 20: 12527. https://doi.org/10.3390/ijms232012527
APA StyleSánchez-Bellver, L., Férriz-Gordillo, A., Carrillo-Pz, M., Rabanal, L., Garcia-Gonzalo, F. R., & Marfany, G. (2022). The Deubiquitinating Enzyme USP48 Interacts with the Retinal Degeneration-Associated Proteins UNC119a and ARL3. International Journal of Molecular Sciences, 23(20), 12527. https://doi.org/10.3390/ijms232012527






