Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats
Abstract
:1. Introduction
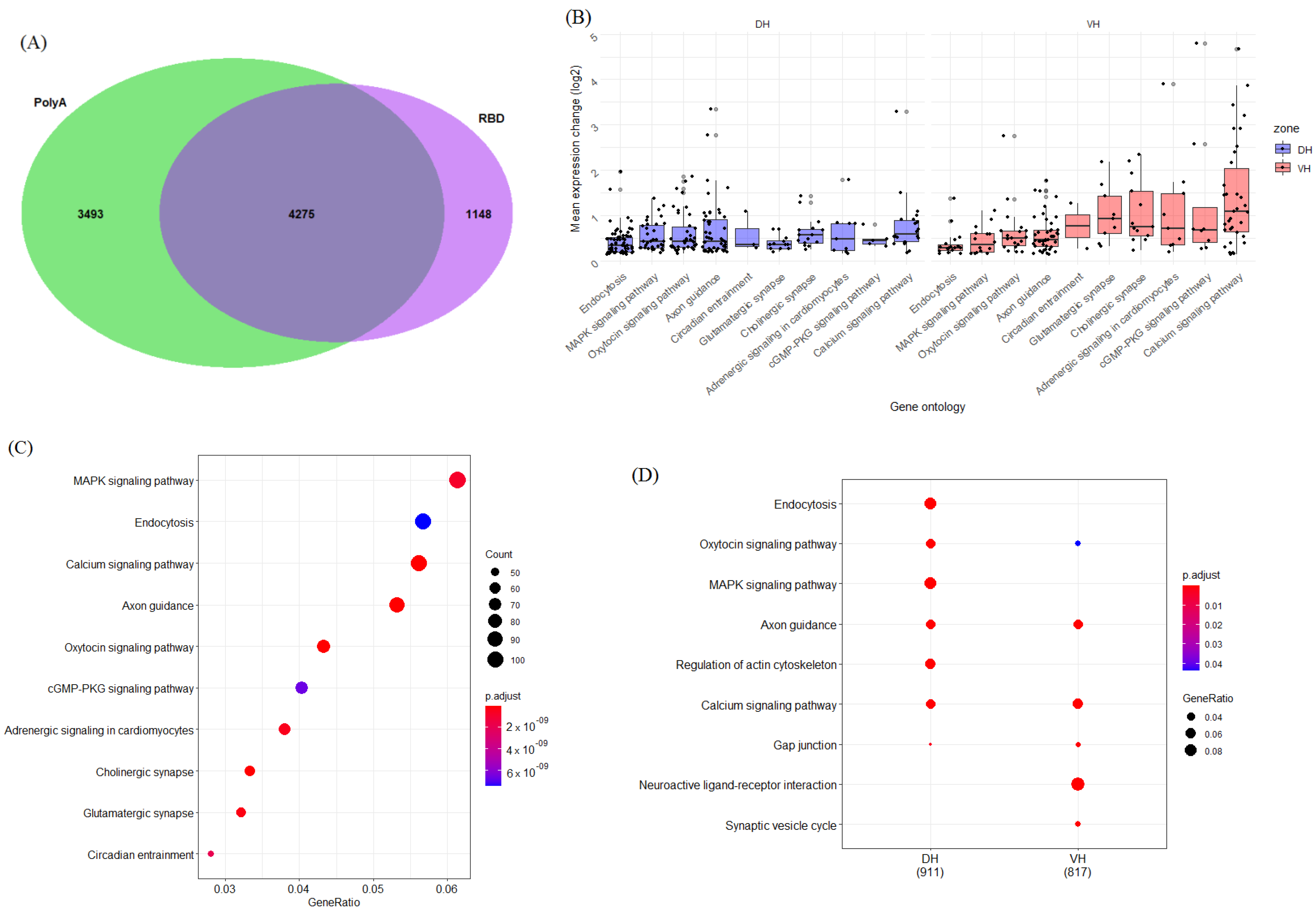
2. Results
2.1. Function-Related Differences in Gene Expression between the Dorsal and Ventral Hippocampus
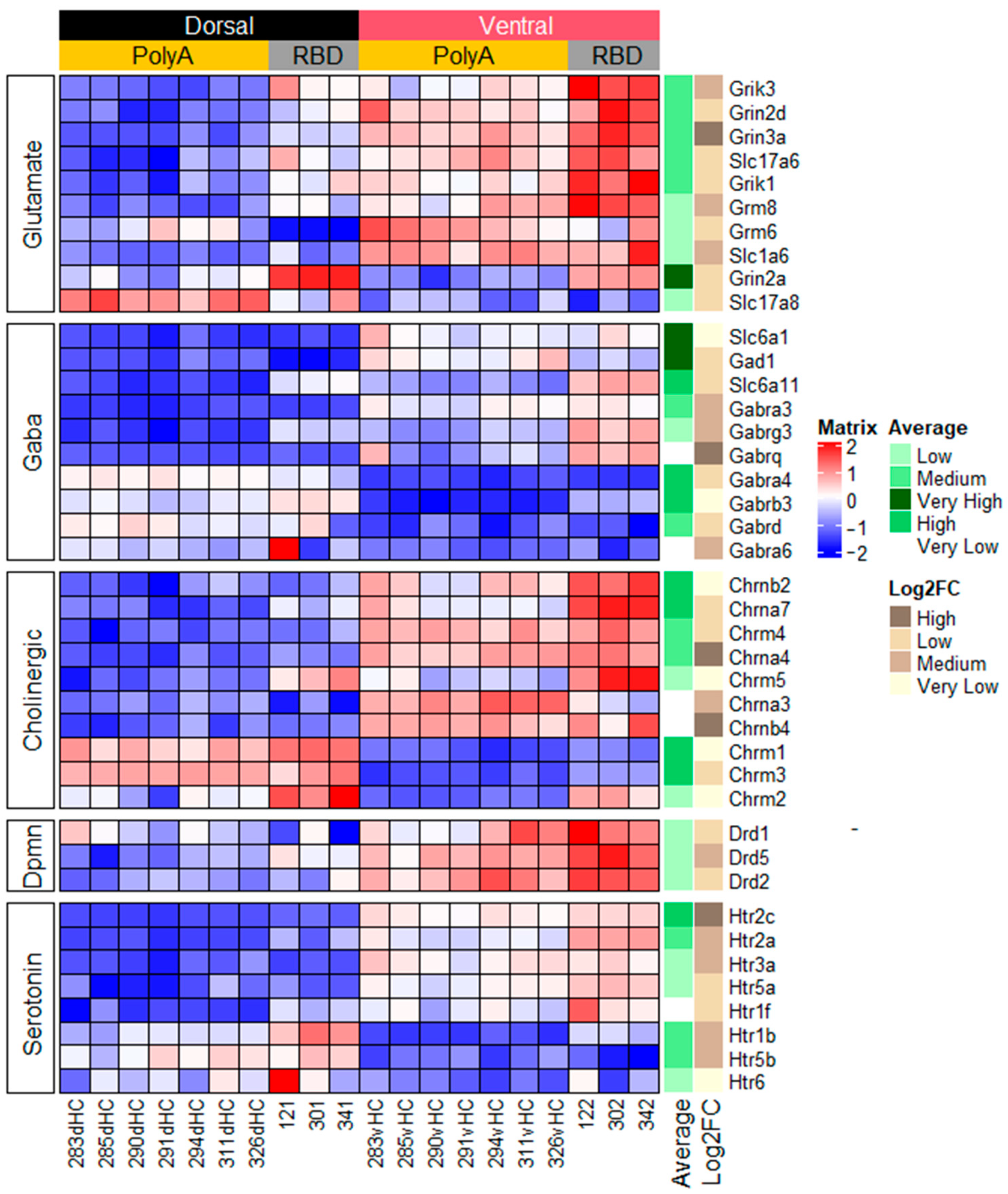
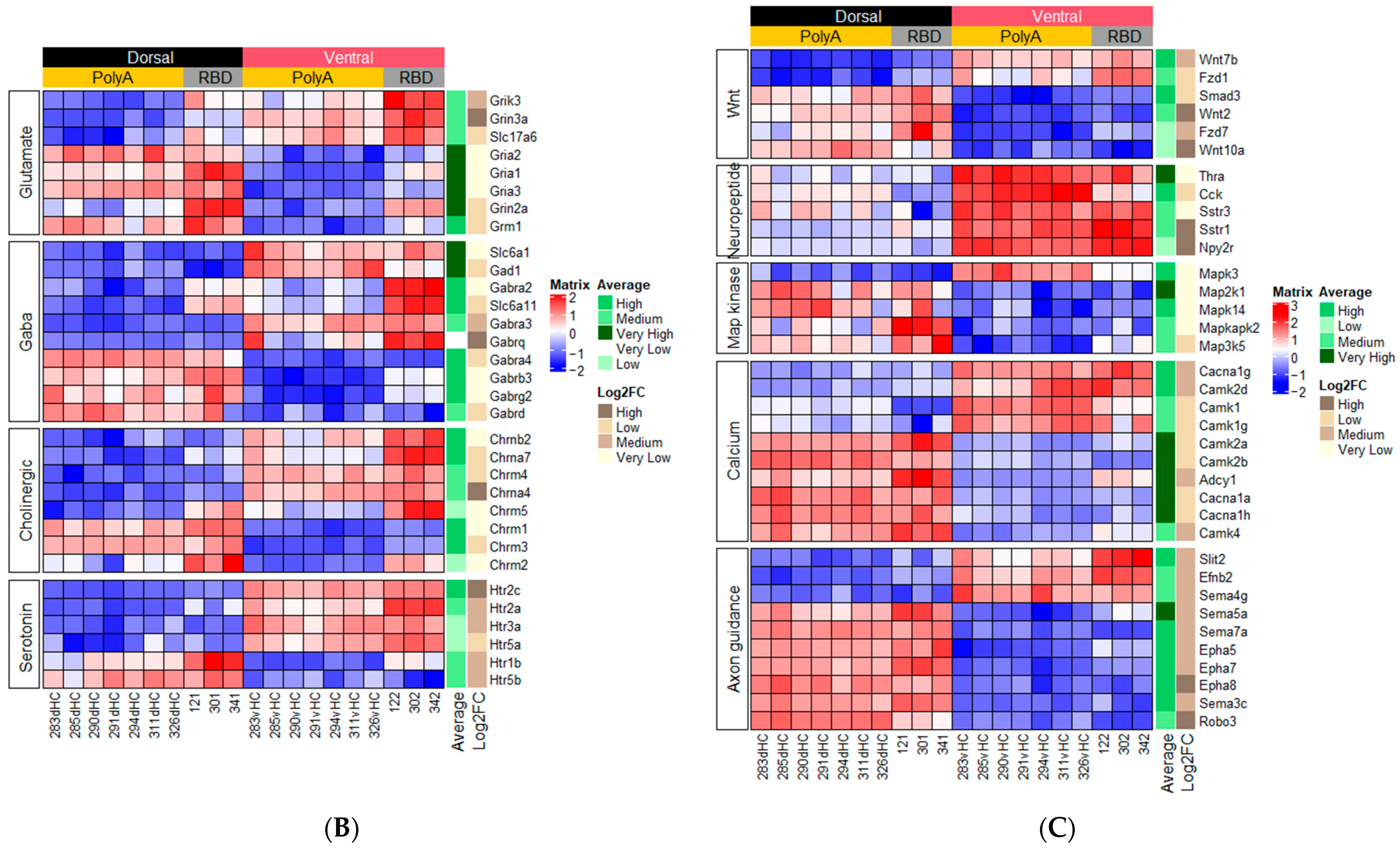
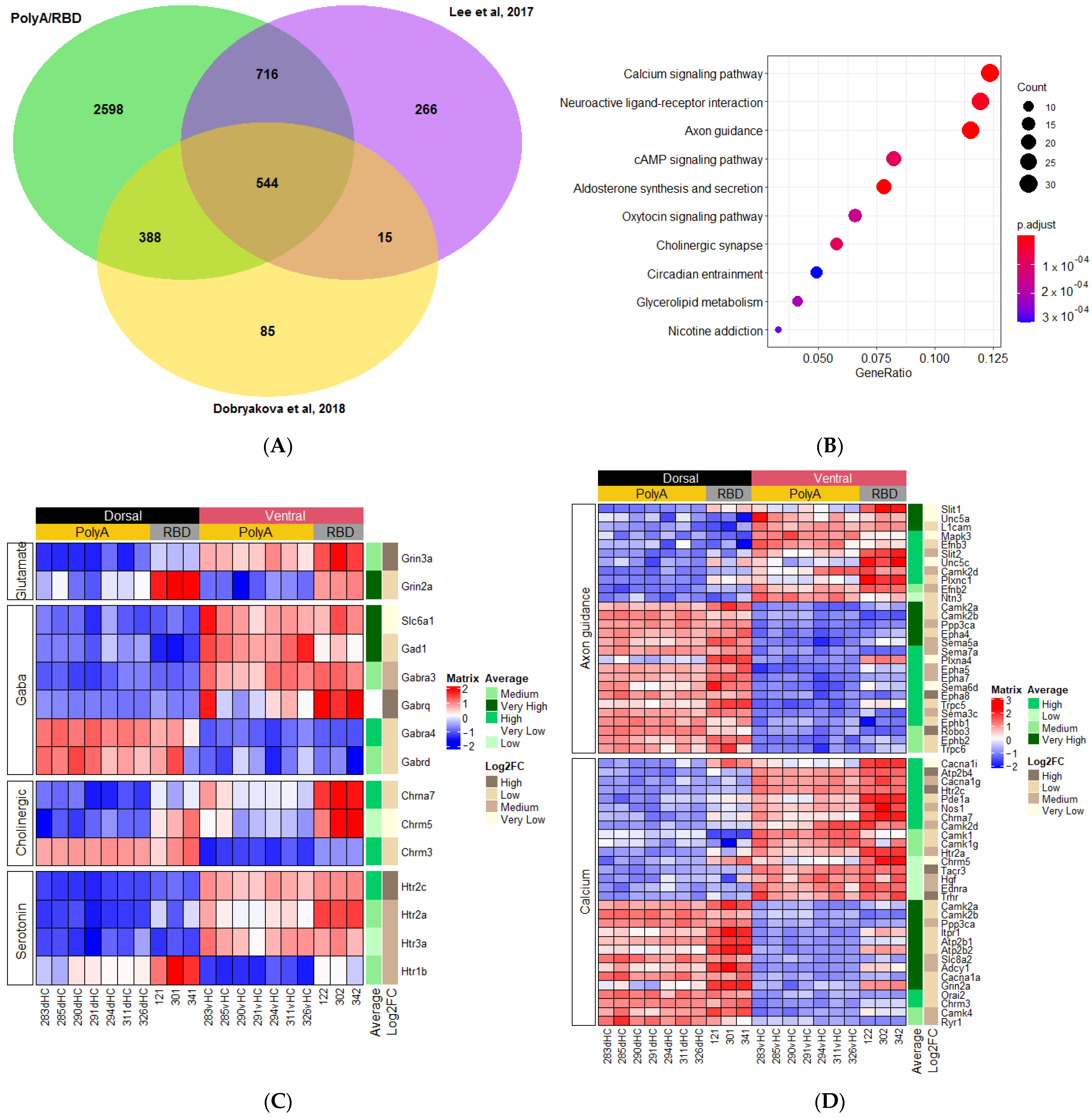
2.1.1. Genes Related to Classical Neurotransmitters
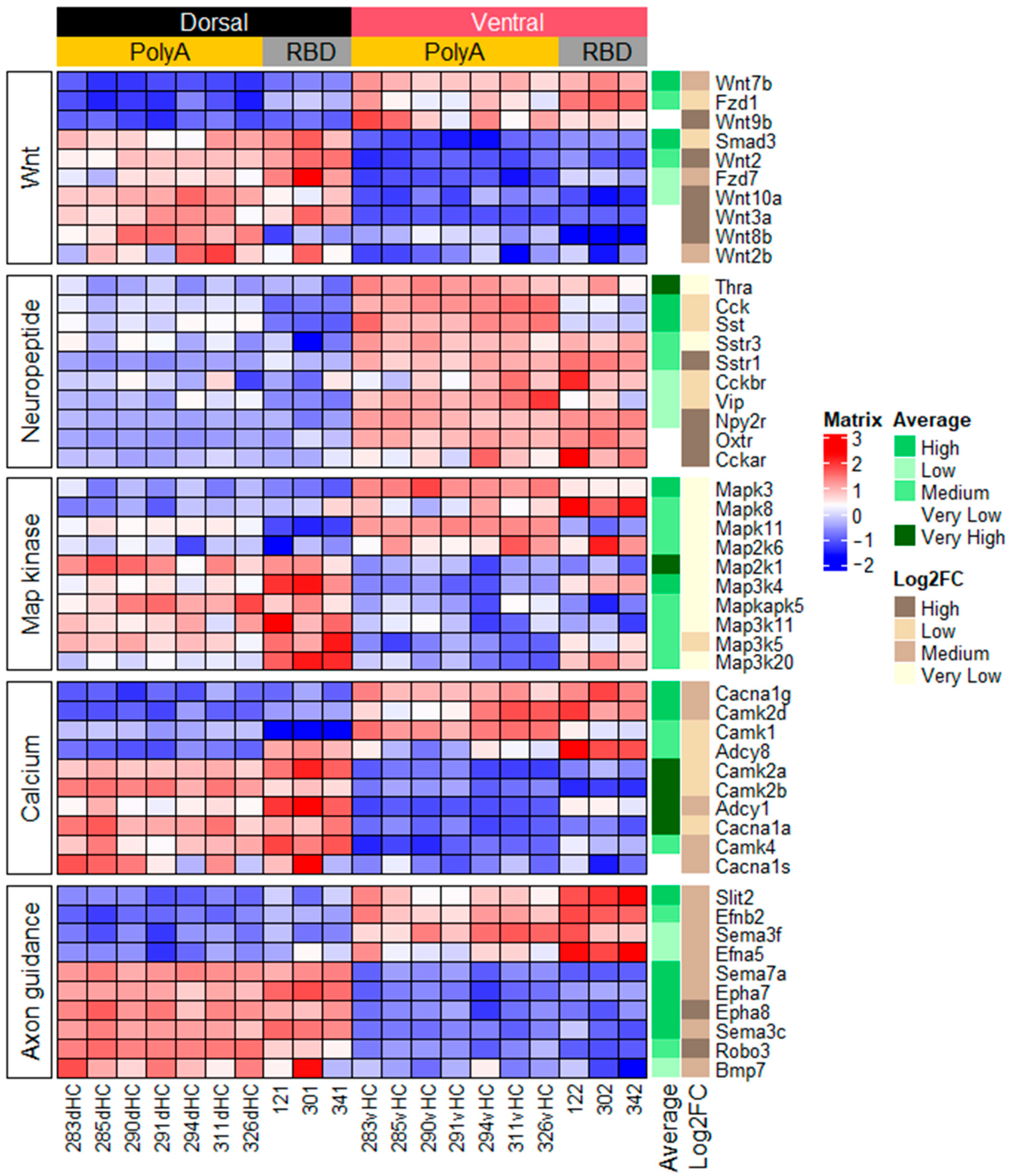
2.1.2. Genes Related to Peptide or Protein Ligands and Their Receptors
2.1.3. Genes Related to Other Signaling Pathways
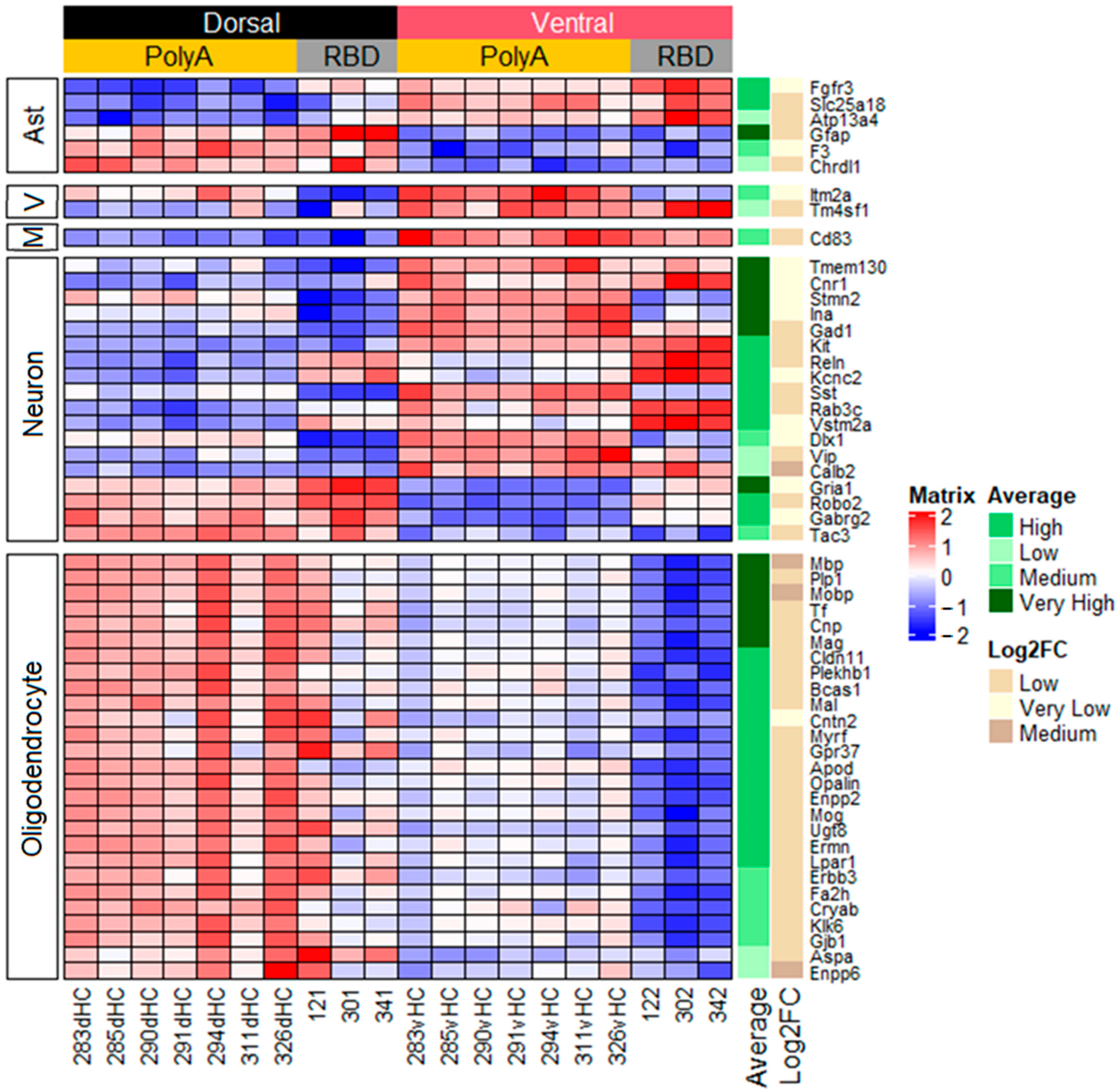
2.2. Cell-Specific Differences in Gene Expression between the Dorsal and Ventral Hippocampus
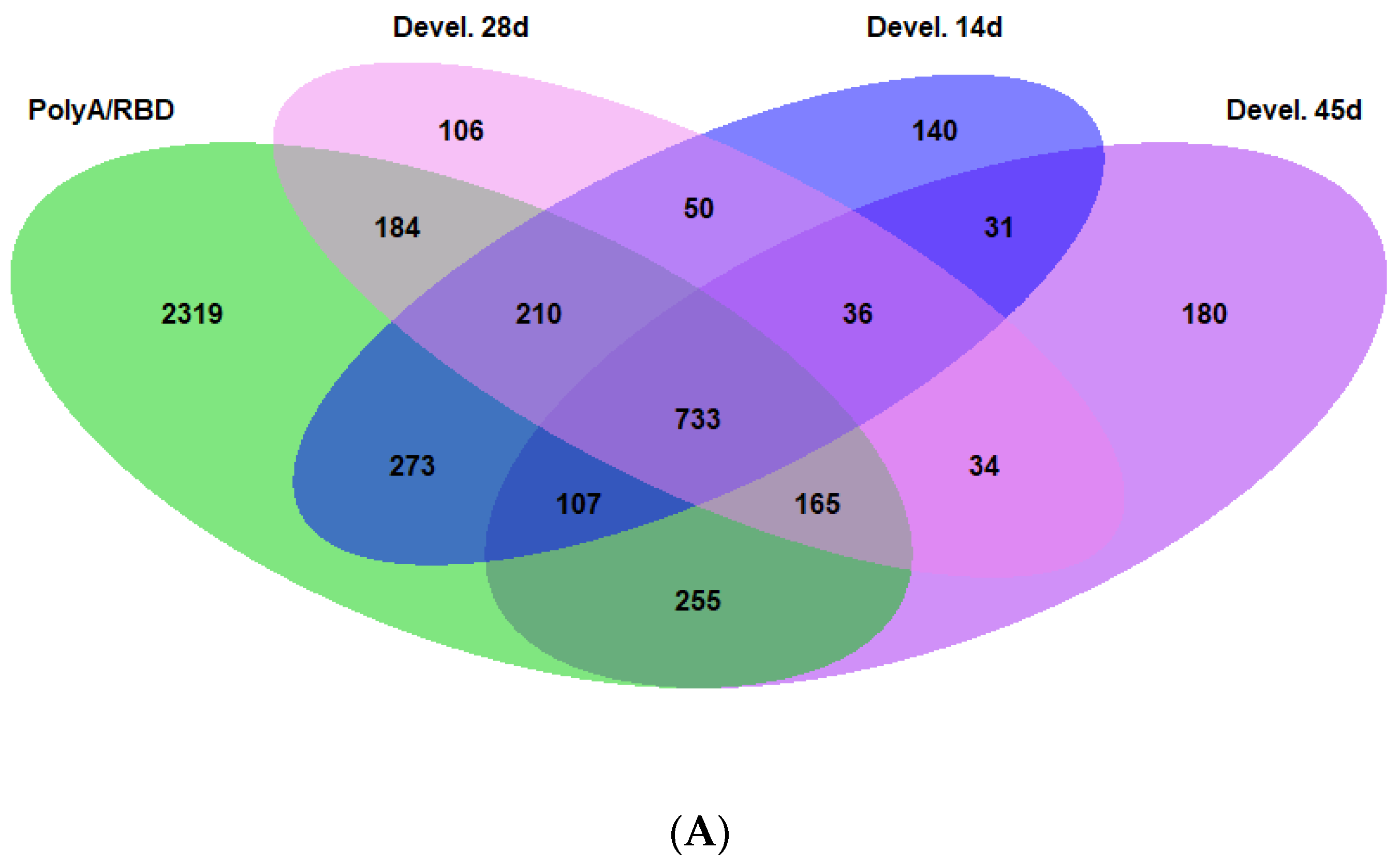
2.3. Differences in the Dorsoventral Gradient between Datasets Are Related to Genes Expressed in All Cell Types
2.4. Genes with Stable Dorsoventral Gradients of Expression
2.5. Differential Alternative Splicing between the Dorsal and Ventral Hippocampus
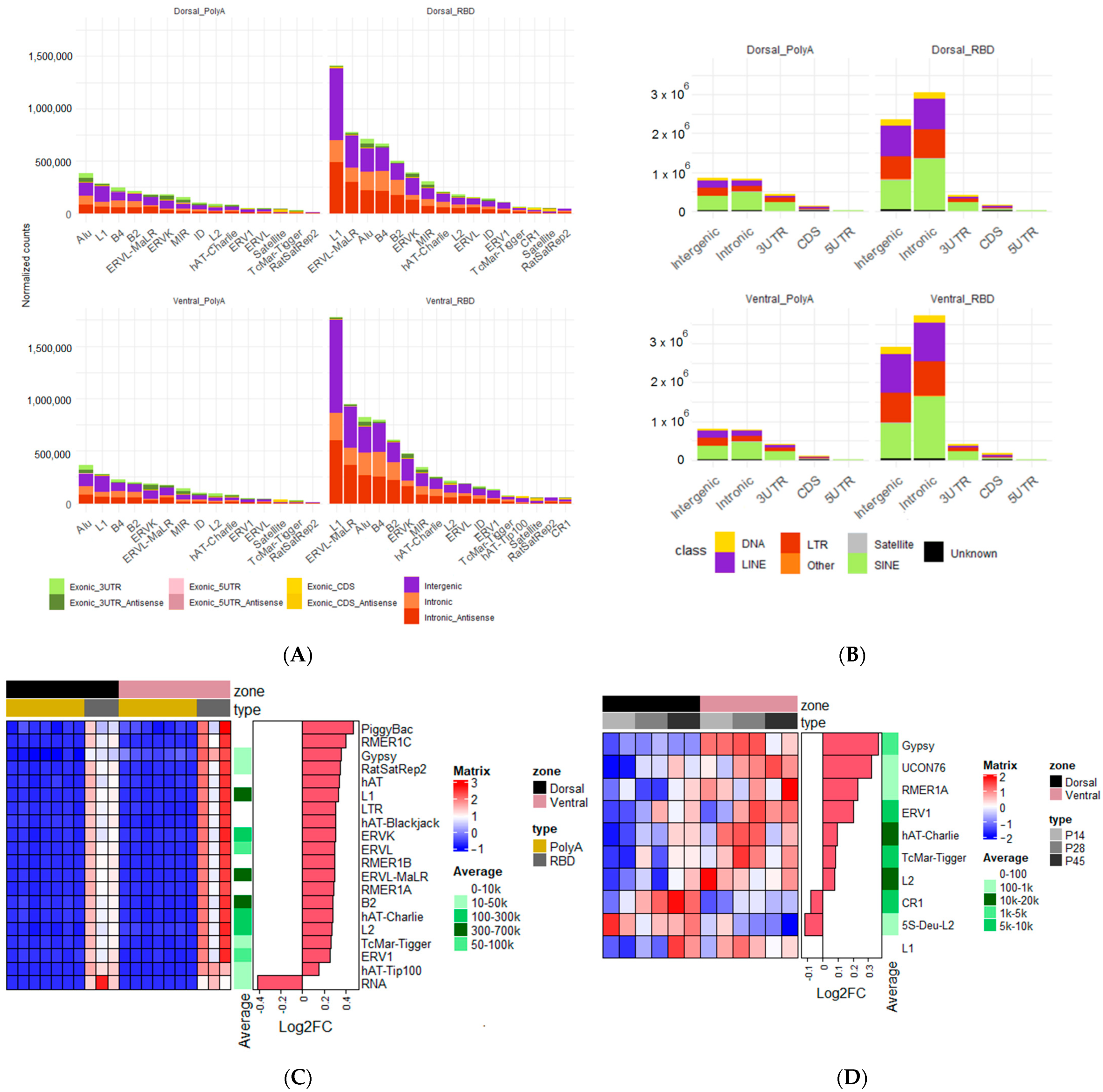
2.6. Analysis of Differential Expression of Transposable Elements
3. Discussion
3.1. Technical Issues
3.2. Functional Importance of Dorsoventral Gradients
3.3. Expression of Transposable Elements
4. Materials and Methods
4.1. Animals
4.2. Sample Acquisition for RNA-seq
4.3. RNA Extraction, cDNA Library Construction, RNA-seq and Data Analysis
4.4. Bioinformatic Analysis
4.4.1. Read Mapping and Counting
4.4.2. Gene Ontology and Visualisation
4.4.3. RNA-seq Data Access
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adey, W.R.; Dunlop, C.W.; Hendrix, C.E. Hippocampal Slow Waves. Distribution and Phase Relationships in the Course of Approach Learning. Arch. Neurol. 1960, 3, 74–90. [Google Scholar] [CrossRef] [PubMed]
- Elul, R. Regional Differences in the Hippocampus of the Cat. I. Specific Discharge Patterns of the Dorsal and Ventral Hippocampus and Their Role in Generalized Seizures. Electroencephalogr. Clin. Neurophysiol. 1964, 16, 470–488. [Google Scholar] [CrossRef]
- Elul, R. Regional Differences in the Hippocampus of the Cat. II. Projections of the Dorsal and Ventral Hippocampus. Electroencephalogr. Clin. Neurophysiol. 1964, 16, 489–502. [Google Scholar] [CrossRef]
- Nadel, L. Dorsal and Ventral Hippocampal Lesions and Behavior. Physiol. Behav. 1968, 3, 891–900. [Google Scholar] [CrossRef]
- Stevens, R.; Cowey, A. Effects of Dorsal and Ventral Hippocampal Lesions on Spontaneous Alternation, Learned Alternation and Probability Learning in Rats. Brain Res. 1973, 52, 203–224. [Google Scholar] [CrossRef]
- Strange, B.A.; Witter, M.P.; Lein, E.S.; Moser, E.I. Functional Organization of the Hippocampal Longitudinal Axis. Nat. Rev. Neurosci. 2014, 15, 655–669. [Google Scholar] [CrossRef]
- Caradonna, S.G.; Einhorn, N.R.; Saudagar, V.; Khalil, H.; Petty, G.H.; Lihagen, A.; LeFloch, C.; Lee, F.S.; Akil, H.; Guidotti, A.; et al. Corticosterone Induces Discrete Epigenetic Signatures in the Dorsal and Ventral Hippocampus That Depend upon Sex and Genotype: Focus on Methylated Nr3c1 Gene. Transl. Psychiatry 2022, 12, 109. [Google Scholar] [CrossRef]
- Moser, M.B.; Moser, E.I. Functional Differentiation in the Hippocampus. Hippocampus 1998, 8, 608–619. [Google Scholar] [CrossRef]
- Fanselow, M.; Dong, H.-W. Are the Dorsal and Ventral Hippocampus Functionally Distinct Structures. Neuron 2010, 65, 7–19. [Google Scholar] [CrossRef]
- Gulyaeva, N.V. Functional Neurochemistry of the Ventral and Dorsal Hippocampus: Stress, Depression, Dementia and Remote Hippocampal Damage. Neurochem. Res. 2019, 44, 1306–1322. [Google Scholar] [CrossRef]
- Bannerman, D.M.; Rawlins, J.N.P.; McHugh, S.B.; Deacon, R.M.J.; Yee, B.K.; Bast, T.; Zhang, W.N.; Pothuizen, H.H.J.; Feldon, J. Regional Dissociations within the Hippocampus—Memory and Anxiety. Neurosci. Biobehav. Rev. 2004, 28, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Maggio, N.; Segal, M. Corticosteroid Regulation of Synaptic Plasticity in the Hippocampus. Sci. World J. 2010, 10, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Dobryakova, Y.V.; Kasianov, A.; Zaichenko, M.I.; Stepanichev, M.Y.; Chesnokova, E.A.; Kolosov, P.M.; Markevich, V.A.; Bolshakov, A.P. Intracerebroventricular Administration of 192IgG-Saporin Alters Expression of Microglia-Associated Genes in the Dorsal But Not Ventral Hippocampus. Front. Mol. Neurosci. 2018, 10, 429. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.R.; Kim, J.H.; Cho, E.; Kim, M.; Park, M. Dorsal and Ventral Hippocampus Differentiate in Functional Pathways and Differentially Associate with Neurological Disease-Related Genes during Postnatal Development. Front. Mol. Neurosci. 2017, 10, 331. [Google Scholar] [CrossRef] [PubMed]
- Cembrowski, M.S.; Wang, L.; Sugino, K.; Shields, B.C.; Spruston, N. Hipposeq: A Comprehensive RNA-Seq Database of Gene Expression in Hippocampal Principal Neurons. eLife 2016, 5, e14997. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A Survey of Best Practices for RNA-Seq Data Analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Bodea, G.O.; McKelvey, E.G.Z.; Faulkner, G.J. Retrotransposon-Induced Mosaicism in the Neural Genome. Open Biol. 2018, 8, 180074. [Google Scholar] [CrossRef]
- Chesnokova, E.; Beletskiy, A.; Kolosov, P. The Role of Transposable Elements of the Human Genome in Neuronal Function and Pathology. Int. J. Mol. Sci. 2022, 23, 5847. [Google Scholar] [CrossRef]
- McKenzie, A.T.; Wang, M.; Hauberg, M.E.; Fullard, J.F.; Kozlenkov, A.; Keenan, A.; Hurd, Y.L.; Dracheva, S.; Casaccia, P.; Roussos, P.; et al. Brain Cell Type Specific Gene Expression and Co-Expression Network Architectures. Sci. Rep. 2018, 8, 8868. [Google Scholar] [CrossRef]
- Zeisel, A.; Muñoz-Manchado, A.B.; Codeluppi, S.; Lönnerberg, P.; La Manno, G.; Juréus, A.; Marques, S.; Munguba, H.; He, L.; Betsholtz, C.; et al. Cell Types in the Mouse Cortex and Hippocampus Revealed by Single-Cell RNA-Seq. Science 2015, 347, 1138–1142. [Google Scholar] [CrossRef]
- Love, M.I.; Soneson, C.; Patro, R. Swimming Downstream: Statistical Analysis of Differential Transcript Usage Following Salmon Quantification. F1000Research 2018, 7, 952. [Google Scholar] [CrossRef] [PubMed]
- Carulli, D.; de Winter, F.; Verhaagen, J. Semaphorins in Adult Nervous System Plasticity and Disease. Front. Synaptic Neurosci. 2021, 13, 672891. [Google Scholar] [CrossRef] [PubMed]
- Hruska, M.; Dalva, M.B. Ephrin Regulation of Synapse Formation, Function and Plasticity. Mol. Cell. Neurosci. 2012, 50, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Gonda, Y.; Namba, T.; Hanashima, C. Beyond Axon Guidance: Roles of Slit-Robo Signaling in Neocortical Formation. Front. Cell Dev. Biol. 2020, 8, 607415. [Google Scholar] [CrossRef] [PubMed]
- Czéh, B.; Varga, Z.K.K.; Henningsen, K.; Kovács, G.L.; Miseta, A.; Wiborg, O. Chronic Stress Reduces the Number of GABAergic Interneurons in the Adult Rat Hippocampus, Dorsal-Ventral and Region-Specific Differences. Hippocampus 2015, 25, 393–405. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Jin, Y.; Tam, O.H.; Paniagua, E.; Hammell, M. TEtranscripts: A Package for Including Transposable Elements in Differential Expression Analysis of RNA-Seq Datasets. Bioinformatics 2015, 31, 3593–3599. [Google Scholar] [CrossRef]
- Bendall, M.L.; De Mulder, M.; Iñiguez, L.P.; Lecanda-Sánchez, A.; Pérez-Losada, M.; Ostrowski, M.A.; Jones, R.B.; Mulder, L.C.F.; Reyes-Terán, G.; Crandall, K.A.; et al. Telescope: Characterization of the Retrotranscriptome by Accurate Estimation of Transposable Element Expression. PLoS Comput. Biol. 2019, 15, e1006453. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Reyes, A.; Huber, W. Detecting Differential Usage of Exons from RNA-Seq Data. Genome Res. 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G.; Brouwer, C. Pathview Web: User Friendly Pathway Visualization and Data Integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A.; Berger, B. IsoformSwitchAnalyzeR: Analysis of Changes in Genome-Wide Patterns of Alternative Splicing and Its Functional Consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beletskiy, A.; Positselskaya, E.; Vinarskaya, A.K.; Spivak, Y.S.; Dobryakova, Y.V.; Tyulenev, I.; Markevich, V.A.; Bolshakov, A.P. Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats. Int. J. Mol. Sci. 2022, 23, 9948. https://doi.org/10.3390/ijms23179948
Beletskiy A, Positselskaya E, Vinarskaya AK, Spivak YS, Dobryakova YV, Tyulenev I, Markevich VA, Bolshakov AP. Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats. International Journal of Molecular Sciences. 2022; 23(17):9948. https://doi.org/10.3390/ijms23179948
Chicago/Turabian StyleBeletskiy, Alexander, Ekaterina Positselskaya, Aliya Kh. Vinarskaya, Yulia S. Spivak, Yulia V. Dobryakova, Iliya Tyulenev, Vladimir A. Markevich, and Alexey P. Bolshakov. 2022. "Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats" International Journal of Molecular Sciences 23, no. 17: 9948. https://doi.org/10.3390/ijms23179948
APA StyleBeletskiy, A., Positselskaya, E., Vinarskaya, A. K., Spivak, Y. S., Dobryakova, Y. V., Tyulenev, I., Markevich, V. A., & Bolshakov, A. P. (2022). Detailed Analysis of Dorsal-Ventral Gradients of Gene Expression in the Hippocampus of Adult Rats. International Journal of Molecular Sciences, 23(17), 9948. https://doi.org/10.3390/ijms23179948








