Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin
Abstract
:1. Introduction
2. Result
2.1. Identification of the WRKY Protein Family in TKS
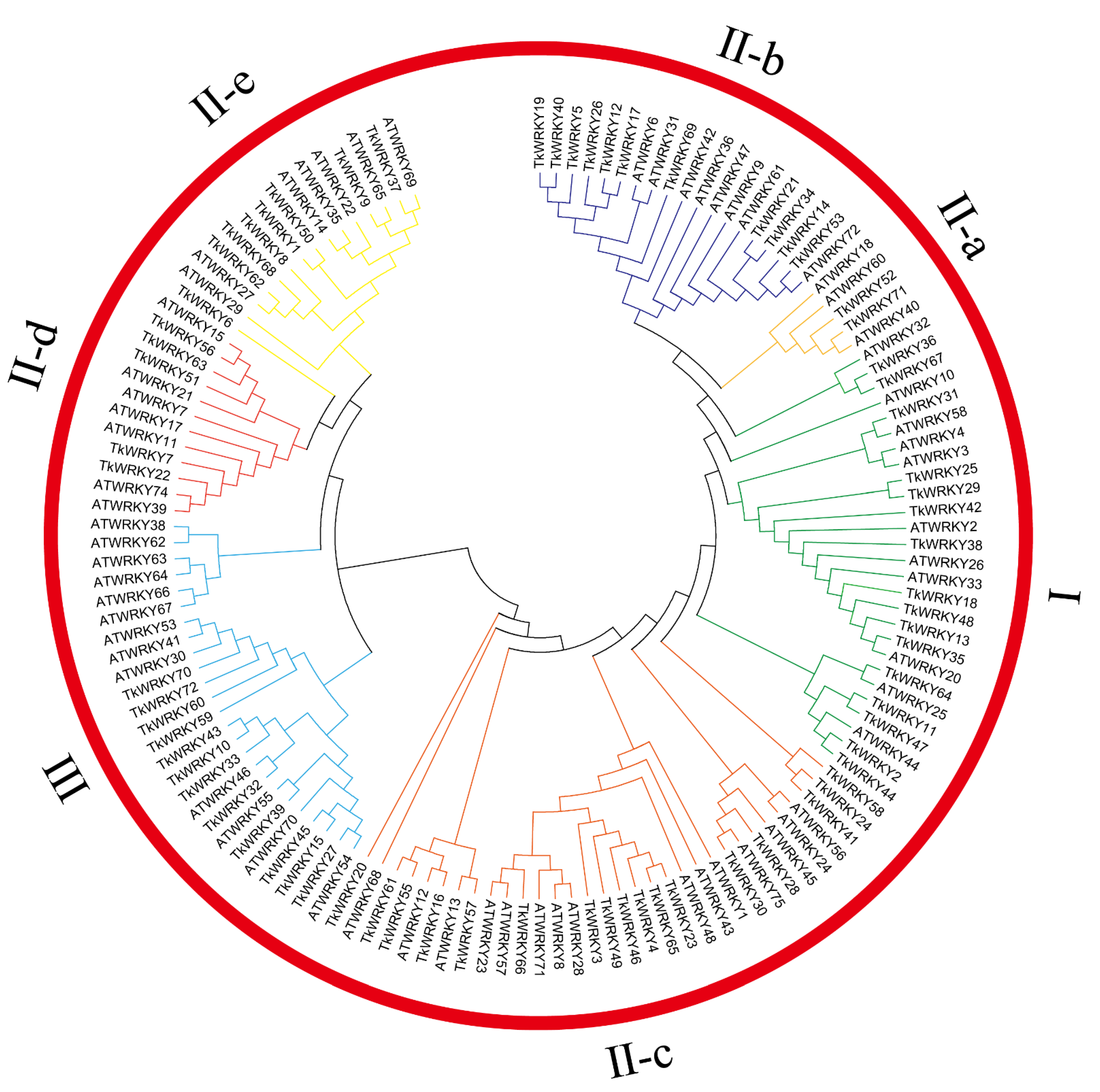
2.2. Multiple Sequence Alignment, Structural Analysis of the WRKY Variants, and Phylogenetic Analysis of the TkWRKY Genes
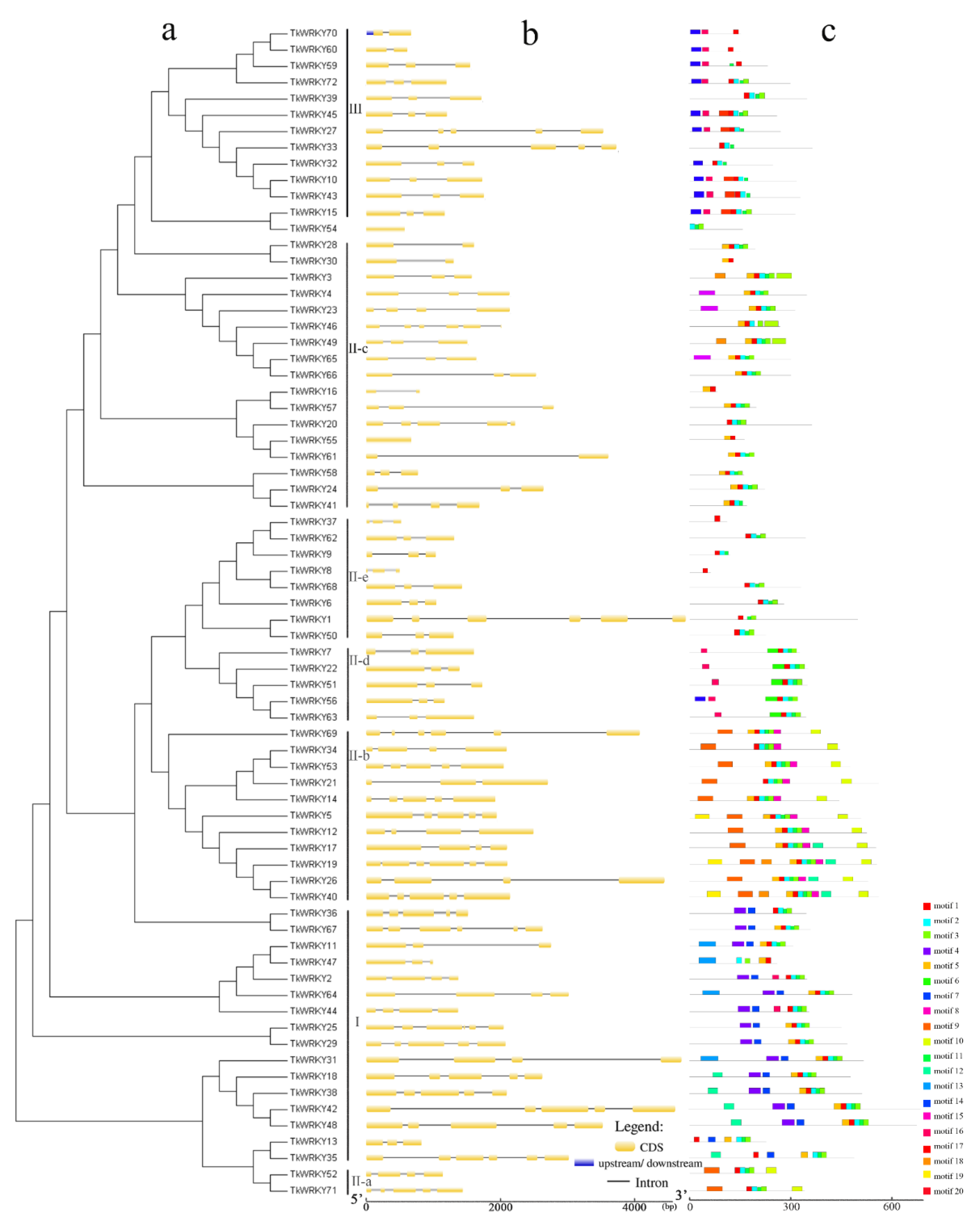
2.3. Gene Structure and MOTIF Composition of TKS WRKY Gene Family
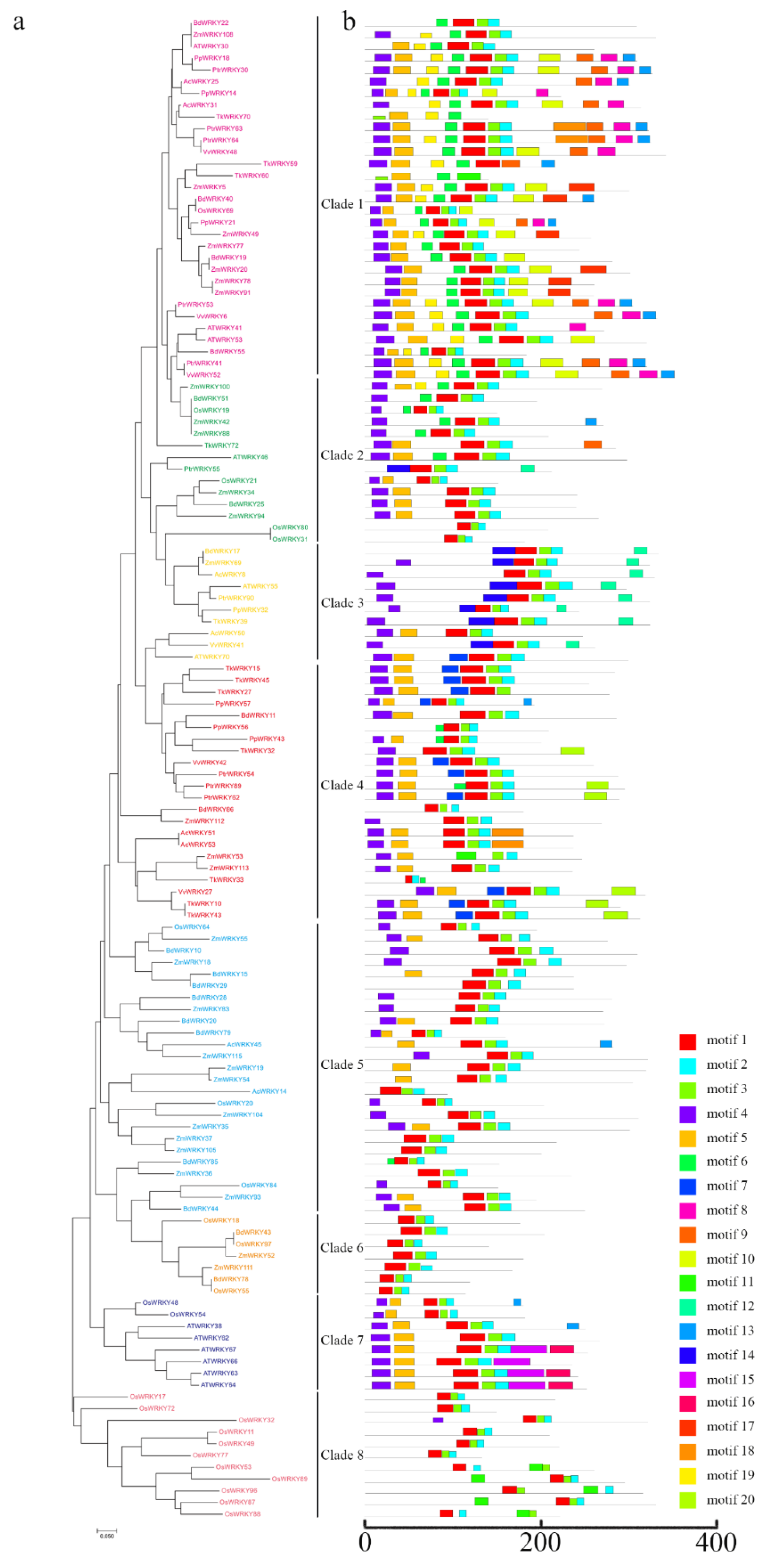
2.4. Evolutionary Analysis of Group III WRKY Genes in TKS and Several Different Species
2.5. Stress-Related Cis-Elements in TkWRKY Promoter Regions
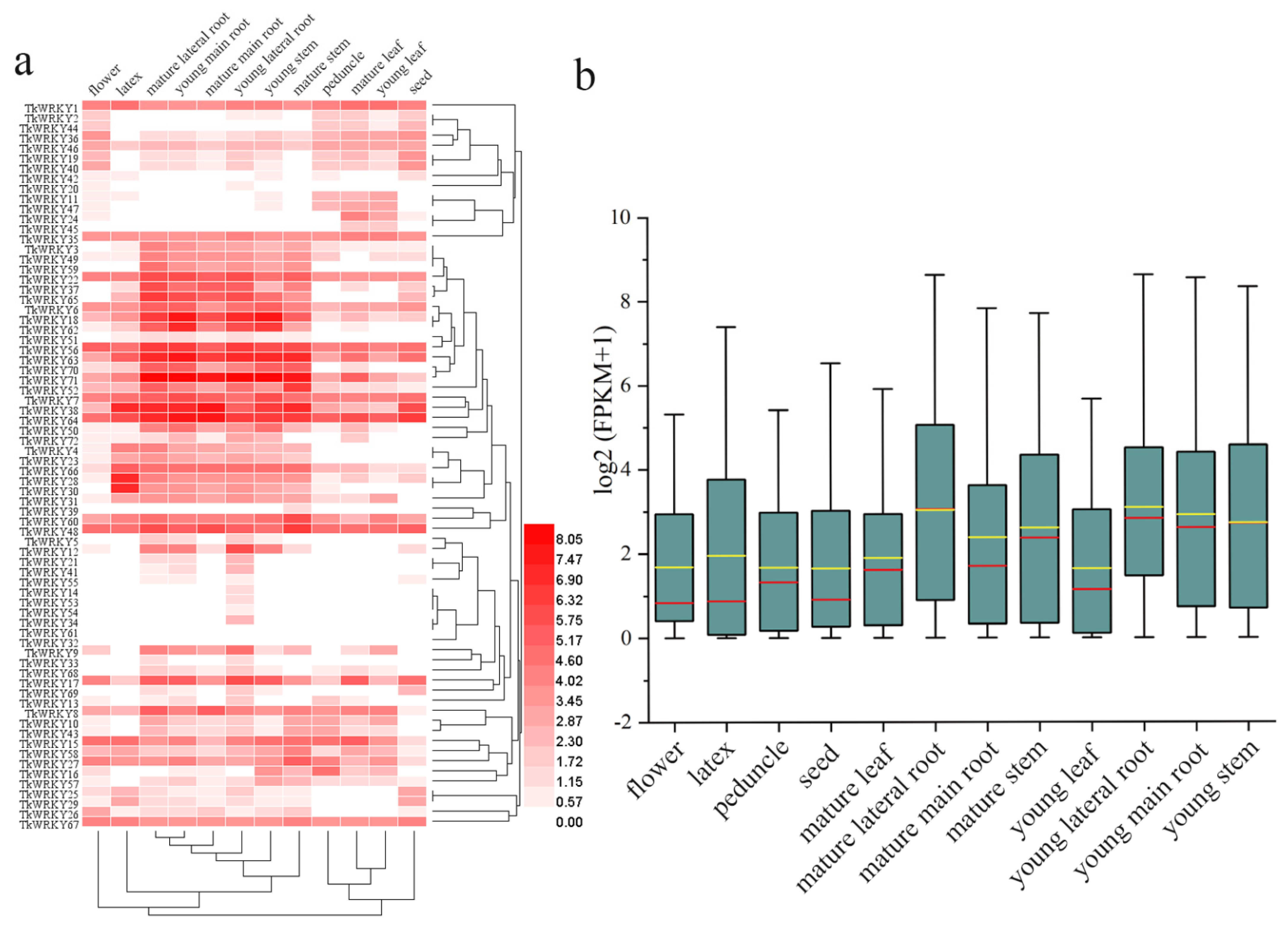
2.6. Expression Profiling of TKS WRKY Genes with RNA-Seq
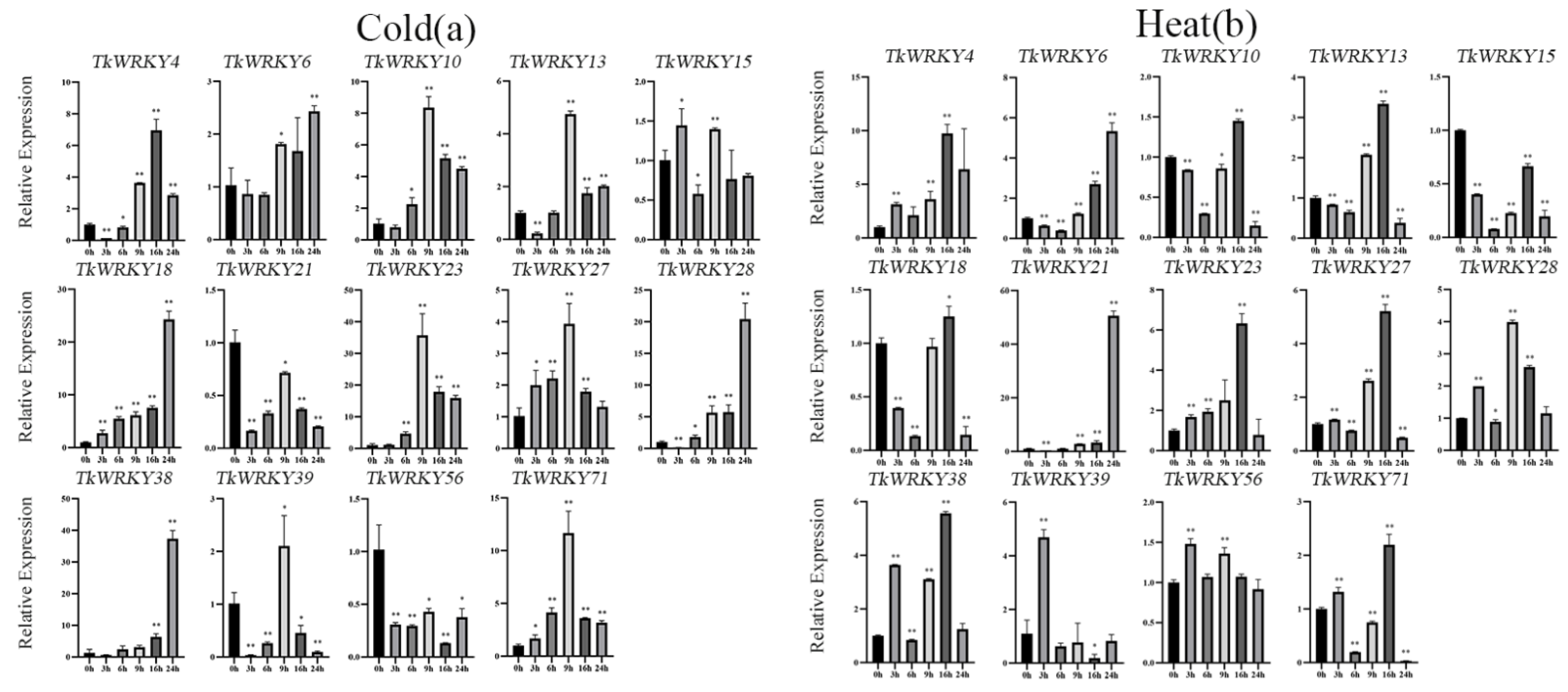
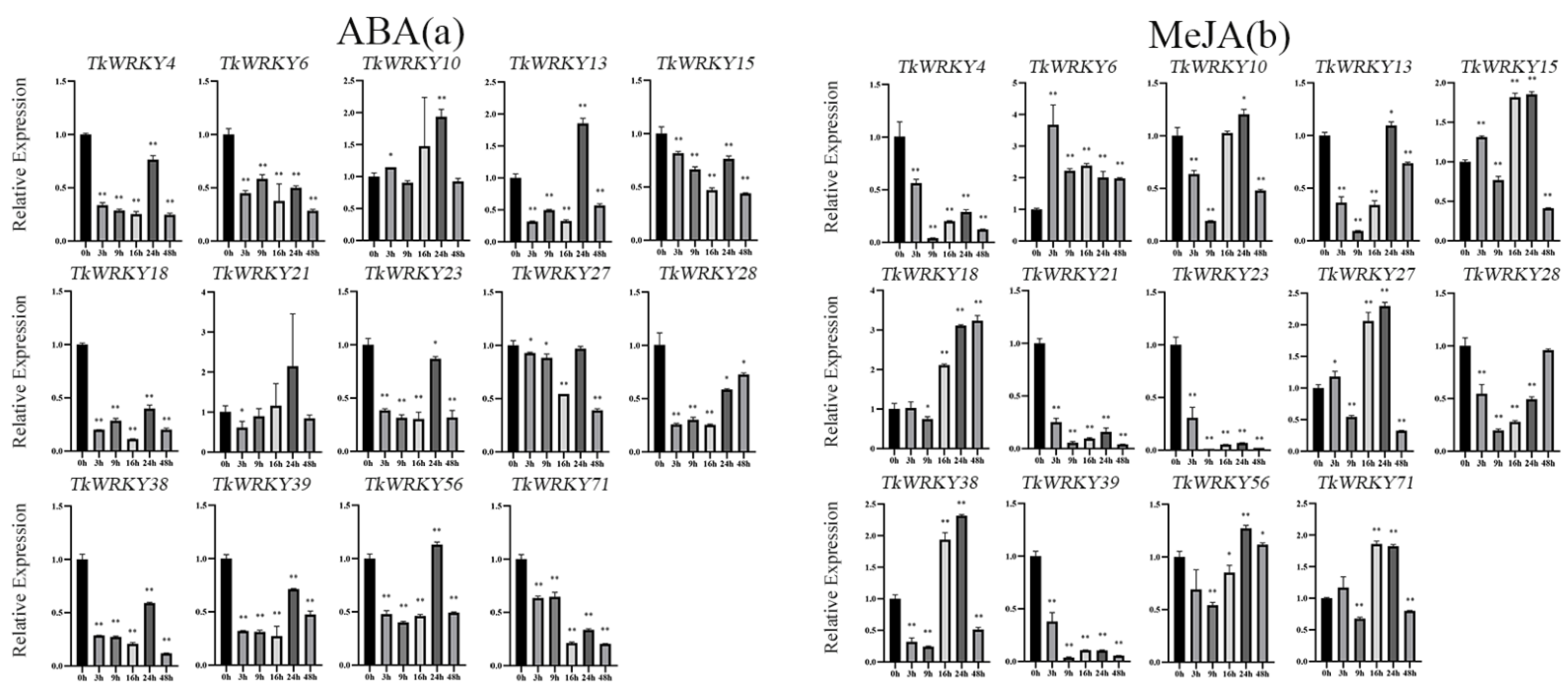
2.7. Expression Patterns of TKS WRKY Genes in Response to Different Treatments
3. Discussion
4. Materials and Methods
4.1. Gene Identification
4.2. Sequence Analysis and Alignment
4.3. Phylogenetic Analysis
4.4. Analysis of Cis-Acting Elements in TkWRKY Promoter Regions
4.5. Plant Materials and Treatments
4.6. RNA Extraction and Real-Time RT-PCR Analysis
4.7. Expression Profiling of TkWRKYs in Different Tissues Based on the Public RNA-seq Data Sets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.H.; Fischer, N.M.; Klaus, H.; Oliver, K.; Dierk, W. Elucidating the evolutionary conserved DNA–binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2015, 5, 1. [Google Scholar]
- Huang, Y.; Feng, C.; Ye, Q.; Wu, W.; Chen, Y. Arabidopsis WRKY6 Transcription Factor Acts as a Positive Regulator of Abscisic Acid Signaling during Seed Germination and Early Seedling Development. PLoS Genet. 2016, 15, e1005833. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Liu, X.; Miao, R.; Mou, C.; Nguyen, T.; Wang, J.; Zhang, X.; et al. WRKY Transcription Factor OsWRKY29 Represses Seed Dormancy in Rice by Weakening Abscisic Acid Response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- Grunewald, W.; De Smet, I.; De Rybel, B.; Robert, H.S.; van de Cotte, B.; Willemsen, V.; Gheysen, G.; Weijers, D.; Friml, J.; Beeckman, T. Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep. 2013, 14, 1136–1142. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, N.; Zhao, S. Functional characterization of a WRKY family gene involved in somatic embryogenesis in Panax ginseng. Protoplasma 2020, 257, 449–458. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002, 14, 1359–1375. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Yao, D.; Zhu, W.; Chen, L.; He, H.; Pan, J.; Cai, R. Transcriptome profiling of trichome–less reveals genes associated with multicellular trichome development in Cucumis sativus. Mol. Genet. Genom. 2015, 290, 2007–2018. [Google Scholar] [CrossRef]
- Guan, Y.; Meng, X.; Khanna, R.; Lamontagne, E.; Liu, Y.; Zhang, S. Phosphorylation of a WRKY Transcription Factor by MAPKs is Required for Pollen Development and Function in Arabidopsis. PLoS Genet. 2014, 10, e1004384. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, X.; Qu, A.; Zhang, M.; Tao, Y.; Yang, L.; Liu, Y.; Xu, J.; Zhang, S. Regulation of pollen lipid body biogenesis by MAP kinases and downstream WRKY transcription factors in Arabidopsis. PLoS Genet. 2018, 14, e1007880. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, J.; Zhang, J.; Liu, D.; Wang, Y.; Niu, Q.; Huang, D. Transcriptome revealed the molecular mechanism of Glycyrrhiza inflata root to maintain growth and development, absorb and distribute ions under salt stress. BMC Plant Biol. 2021, 21, 599. [Google Scholar] [CrossRef]
- Li, W.; Wang, H.; Yu, D. Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short–Day Conditions. Mol. Plant. 2016, 9, 1492–1503. [Google Scholar] [CrossRef]
- Kang, G.; Yan, D.; Chen, X.; Yang, L.; Zeng, R. HbWRKY82, a novel IIc WRKY transcription factor from Hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in Arabidopsis. Physiol. Plant. 2021, 171, 151–160. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, R.; Zheng, M.; Liu, X.; Meng, F.; Wu, H.; Yao, Y.; Xin, M.; Peng, H.; Ni, Z.; et al. TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). Plant J. 2018, 96, 372–388. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co–option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, F.; Zhu, C.; Li, X.; Dai, X.; Yang, B.; Zou, X.; Ma, Y. Identification, expression, alternative splicing and functional analysis of pepper WRKY gene family in response to biotic and abiotic stresses. PLoS ONE. 2019, 14, e0219775. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Li, X.; Jiang, M. CRISPR/Cas9–mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me–JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 2021, 48, 5821–5832. [Google Scholar] [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [Google Scholar] [CrossRef]
- Wang, C.; Ru, J.; Liu, Y.; Li, M.; Zhao, D.; Yang, J.; Fu, J.; Xu, Z. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef]
- Kuki, Y.; Ohno, R.; Yoshida, K.; Takumi, S. Heterologous expression of wheat WRKY transcription factor genes transcriptionally activated in hybrid necrosis strains alters abiotic and biotic stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 150, 71–79. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, J.; Yang, F.; Zhang, G.; Wang, D.; Zhang, L.; Ou, Y.; Yao, Y. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant. 2020, 168, 98–117. [Google Scholar] [CrossRef]
- Stolze, A.; Wanke, A.; van Deenen, N.; Geyer, R.; Prufer, D.; Gronover, C.S. Development of rubber–enriched dandelion varieties by metabolic engineering of the inulin pathway. Plant Biotechnol. J. 2017, 15, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Wieghaus, A.; Prufer, D.; Gronover, C.S. Loss of function mutation of the Rapid Alkalinization Factor (RALF1)–like peptide in the dandelion Taraxacum koksaghyz entails a high–biomass taproot phenotype. PLoS ONE. 2019, 14, e0217454. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.; Wu, S.; Shao, X.; Wang, X.; Gan, L.; Qin, B.; Yang, Y.; et al. Genome analysis of Taraxacum kok–saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2018, 5, 78–87. [Google Scholar] [CrossRef]
- Molinu, M.G.; Piluzza, G.; Campesi, G.; Sulas, L.; Re, G.A. Antioxidant Sources from Leaves of Russian Dandelion. Chem. Biodivers. 2019, 16, e1900250. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Poirier, Y. Guayule and Russian dandelion as alternative sources of natural rubber. Crit. Rev. Biotechnol. 2007, 27, 217–231. [Google Scholar] [CrossRef]
- Salehi, M.; Cornish, K.; Bahmankar, M.; Naghavi, M.R. Natural rubber–producing sources, systems, and perspectives for breeding and biotechnology studies of Taraxacum kok–saghyz. Ind. Crops Prod. 2021, 170, 113667. [Google Scholar] [CrossRef]
- Salehi, M.; Bahmankar, M.; Naghavi, M.R.; Cornish, K. Rubber and latex extraction processes for Taraxacum kok–saghyz. Ind. Crops Prod. 2022, 178, 114562. [Google Scholar] [CrossRef]
- Song, Y.; Cui, H.; Shi, Y.; Xue, J.; Ji, C.; Zhang, C.; Yuan, L.; Li, R. Genome–wide identification and functional characterization of Camelina sativa WRKY gene family in response to abiotic stress. BMC Genom. 2020, 21, 786. [Google Scholar] [CrossRef]
- Song, H.; Sun, W.; Yang, G.; Sun, J. WRKY transcription factors in legumes. BMC Plant Biol. 2018, 18, 243. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome–wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Wu, K.; Guo, Z.; Wang, H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, L.; Zhu, Y.; Li, Y.; Yan, H.; Xiang, Y. Comparative genomic analysis of the WRKY III gene family in populus, grape, Arabidopsis and rice. Biol. Direct. 2015, 10, 48. [Google Scholar] [CrossRef]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M.H.S.A. Genome–Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis–Elements in the Promoter, and Interaction Network. Int. J. Mol. Sci. 2018, 19, 1048. [Google Scholar] [CrossRef]
- Li, M.; Hou, L.; Liu, S.; Zhang, C.; Yang, W.; Pang, X.; Li, Y. Genome–wide identification and expression analysis of NAC transcription factors in Ziziphus jujuba Mill. reveal their putative regulatory effects on tissue senescence and abiotic stress responses. Ind. Crops Prod. 2021, 173, 114093. [Google Scholar] [CrossRef]
- Hu, W.; Ren, Q.; Chen, Y.; Xu, G.; Qian, Y. Genome–wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 2021, 21, 427. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Islam, F.; Huang, Q.; Wang, J.; Zhou, W.; Xu, L.; Yang, C. Genome–wide characterization of WRKY gene family in Helianthus annuus L. and their expression profiles under biotic and abiotic stresses. PLoS ONE. 2020, 15, e0241965. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Ouyang, Q.; Li, Y.; Shi, T.; Li, L.; Yang, X.; Ji, K.; Wang, L.; Yue, Y. Genome–wide investigation of WRKY transcription factors in sweet osmanthus and their potential regulation role in aroma synthesis. Tree Physiol. 2020, 40, 557–572. [Google Scholar] [CrossRef]
- Huang, X.; Li, K.; Xu, X.; Yao, Z.; Jin, C.; Zhang, S. Genome–wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genom. 2015, 16, 1104. [Google Scholar] [CrossRef]
- Gu, L.; Dou, L.; Guo, Y.; Wang, H.; Li, L.; Wang, C.; Ma, L.; Wei, H.; Yu, S. The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2019, 19, 116. [Google Scholar] [CrossRef]
- Rinerson, C.I.; Scully, E.D.; Palmer, N.A.; Donze–Reiner, T.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Sattler, S.E.; Rohila, J.S.; Sarath, G.; et al. The WRKY transcription factor family and senescence in switchgrass. BMC Genom. 2015, 16, 912. [Google Scholar] [CrossRef]
- Guo, C.; Guo, R.; Xu, X.; Gao, M.; Li, X.; Song, J.; Zheng, Y.; Wang, X. Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 2014, 65, 1513–1528. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress–responsive WRKY genes in eggplant. PeerJ. 2020, 8, e8777. [Google Scholar] [CrossRef]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome–wide analysis of the WRKY gene family in the cucumber genome and transcriptome–wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Chen, Y.; Wu, L.; Xie, D. Molecular Phylogenetic and Expression Analysis of the Complete WRKY Transcription Factor Family in Maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2011, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yan, L.; Liu, Z.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.; Wang, X.; Du, S.; Jiang, T.; et al. The Mg–Chelatase H Subunit of Arabidopsis Antagonizes a Group of WRKY Transcription Repressors to Relieve ABA–Responsive Genes of Inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Dehesh, K. Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Dhatterwal, P.; Basu, S.; Mehrotra, S.; Mehrotra, R. Genome wide analysis of W–box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci. Rep. 2019, 9, 1681. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. 5′–upstream region of WRKY18 transcription factor from banana is a stress inducible promoter with strong expression in guard cells. Physiol. Plant. 2021, 4, 1335–1350. [Google Scholar] [CrossRef]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.; Chen, Z. Protein–protein Interactions in the Regulation of WRKY Transcription Factors. Mol. Plant. 2013, 6, 287–300. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Li, S.; Nayar, S.; Jia, H.; Kapoor, S.; Wu, J.; Yukawa, Y. The Arabidopsis Hypoxia Inducible AtR8 Long Non–Coding RNA also Contributes to Plant Defense and Root Elongation Coordinating with WRKY Genes under Low Levels of Salicylic Acid. Non–Coding RNA 2020, 6, 8. [Google Scholar] [CrossRef]
- Li, W.; Tian, Z.; Yu, D. WRKY13 acts in stem development in Arabidopsis thaliana. Plant Sci. 2015, 236, 205–213. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, G.; Bhattacharya, S.; Singh, A. Comparative transcriptome meta–analysis of Arabidopsis thaliana under drought and cold stress. PLoS ONE 2018, 13, e0203266. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors are Involved in Brassinosteroid–Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome–wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Shen, Q.J.; Yu, D.; Jeon, J.S.; Piffanelli, P.; Abbruscato, P.; Guo, Z.; Zhang, Y.; Itoh, T.; Lee, S.S.; Buell, C.R.; et al. Nomenclature report on rice WRKY’s—Conflict regarding gene names and its solution. Rice 2012, 5, 3. [Google Scholar]
- Wen, F.; Zhu, H.; Li, P.; Jiang, M.; Mao, W.; Ong, C.; Chu, Z. Genome–Wide Evolutionary Characterization and Expression Analyses of WRKY Family Genes in Brachypodium distachyon. DNA Res. 2014, 21, 327–339. [Google Scholar] [CrossRef]
- Gu, Y.; Ji, Z.; Chi, F.; Qiao, Z.; Xu, C.; Zhang, J.; Zhou, Z.; Dong, Q. Genome–wide identification and expression analysis of the WRKY gene family in peach. Acta Agric. Boreali–Sin. 2017, 38, 254–270. [Google Scholar]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis–acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Salehi, M.; Karimzadeh, G.; Naghavi, M.R.; Badi, H.N.; Monfared, S.R. Expression of key genes affecting artemisinin content in five Artemisia species. Sci. Rep. 2018, 8, 12659. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Wu, C.; Liu, H.; Hu, Z.; Sun, G. Cloning and Molecular Characterization of a SERK Gene Transcriptionally Induced During Somatic Embryogenesis in Ananas comosus cv. Shenwan. Plant Mol. Biol. Rep. 2012, 30, 195–203. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA–Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE. 2014, 9, e111988. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.; Luo, J.; Li, H.; Wei, F.; Zhang, Y.; Jiang, H.; Peng, X. Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin. Int. J. Mol. Sci. 2022, 23, 10270. https://doi.org/10.3390/ijms231810270
Cheng Y, Luo J, Li H, Wei F, Zhang Y, Jiang H, Peng X. Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin. International Journal of Molecular Sciences. 2022; 23(18):10270. https://doi.org/10.3390/ijms231810270
Chicago/Turabian StyleCheng, Yifeng, Jinxue Luo, Hao Li, Feng Wei, Yuqi Zhang, Haiyang Jiang, and Xiaojian Peng. 2022. "Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin" International Journal of Molecular Sciences 23, no. 18: 10270. https://doi.org/10.3390/ijms231810270




