Uba1: A Potential Ubiquitin-like Activator Protein of Urm1 in Toxoplasma gondii
Abstract
:1. Introduction
2. Results
2.1. Characterization of Uba1 in T. gondii
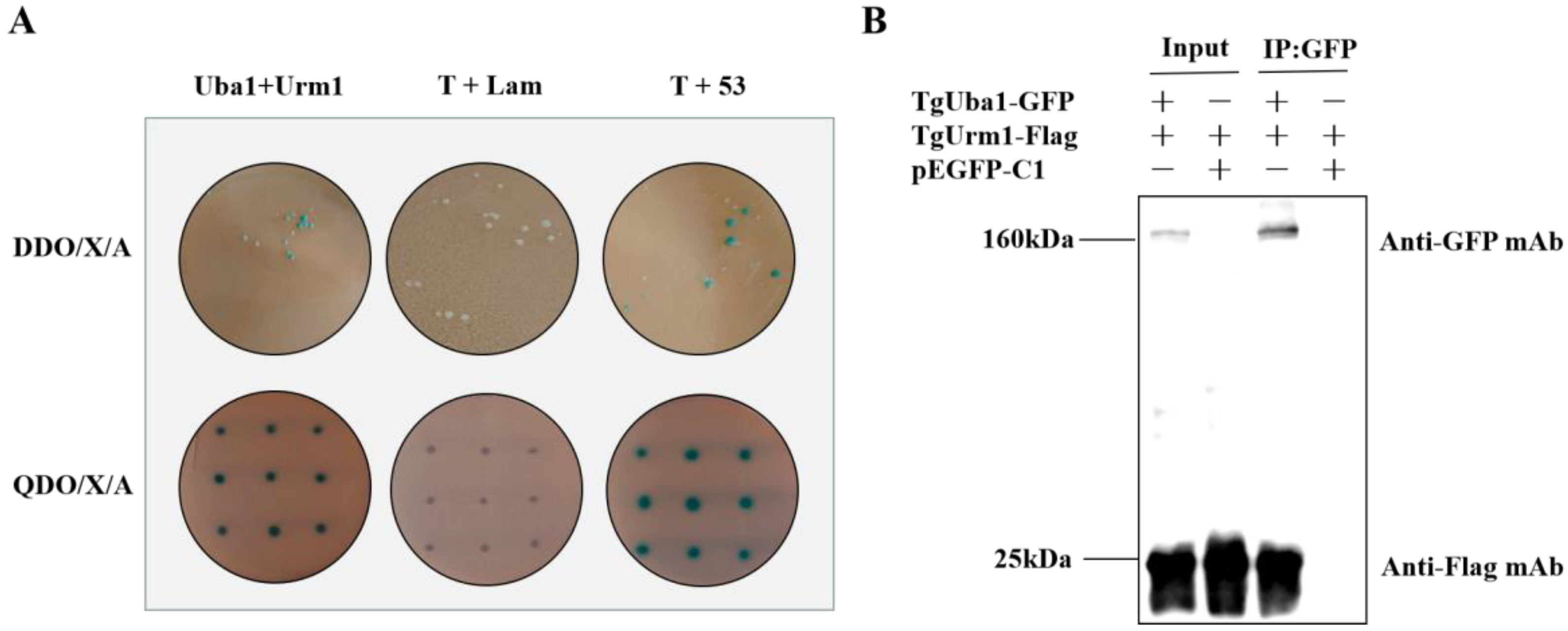
2.2. Confirmation of the Interaction of TgUba1-TgUrm1
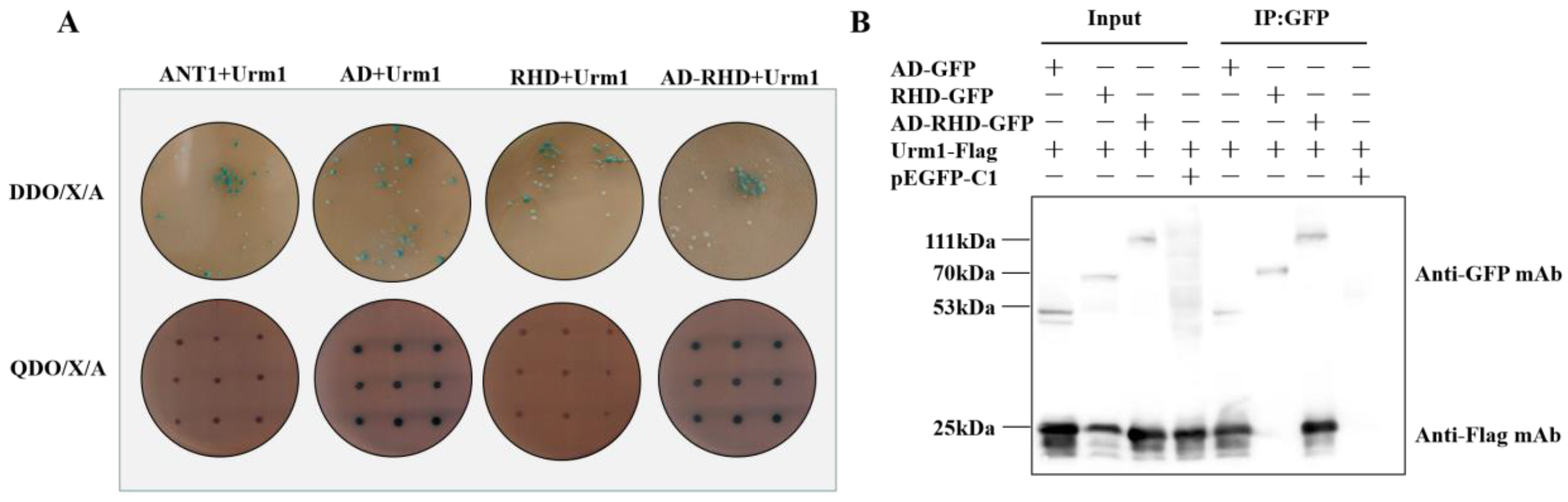
2.3. AD Domain as the Key Domain Targeting TgUrm1
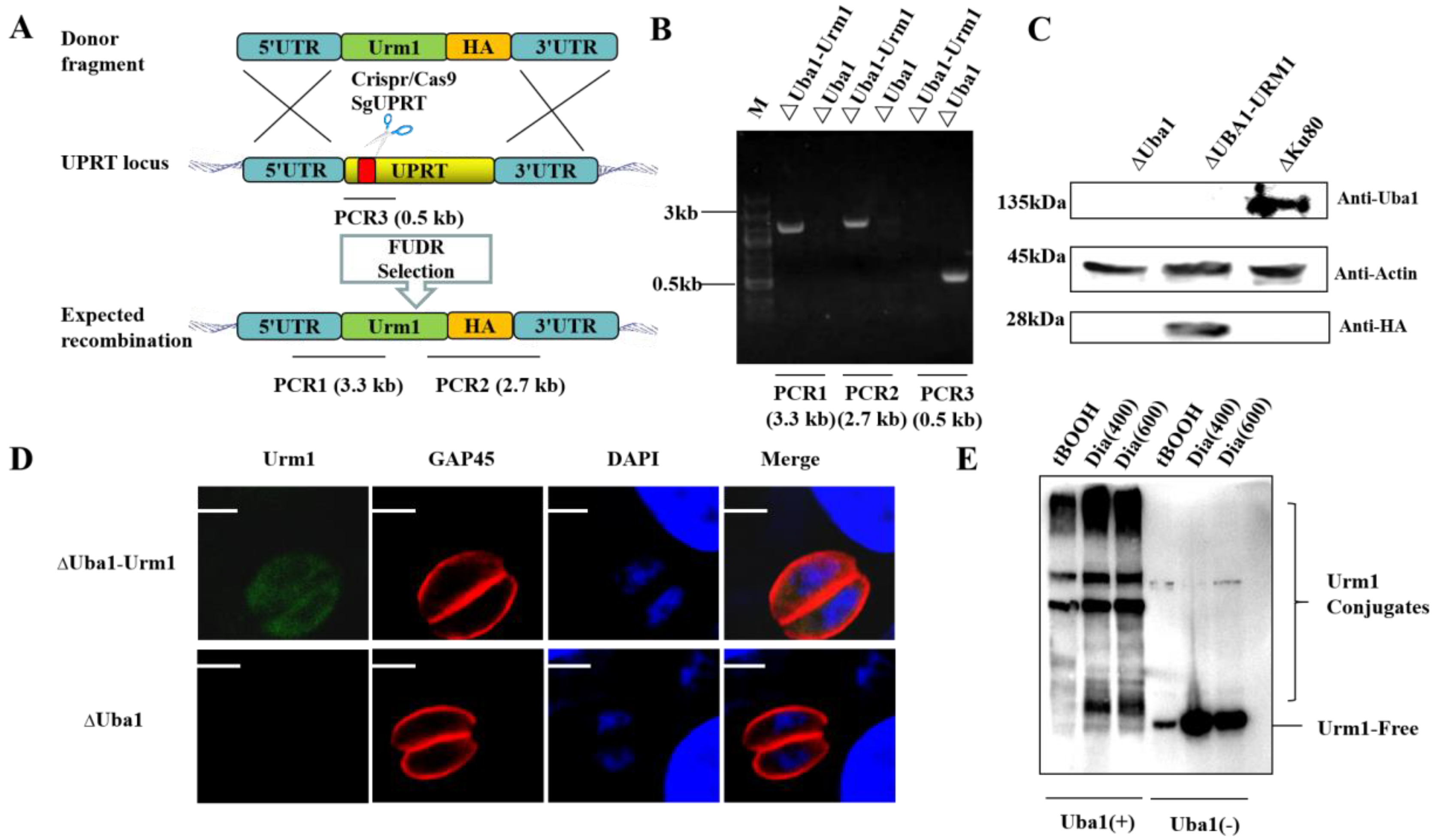
2.4. Construction of ΔUba1 and CM-Uba1 Strains by CRISPR/Cas9
2.5. Parasites Lacking TgUba1 Showed Significantly Reduced Proliferation and Virulence in Mice
2.6. TgUba1 Enhanced the Target Conjugation Ability of TgUrm1 upon Oxidative Stress
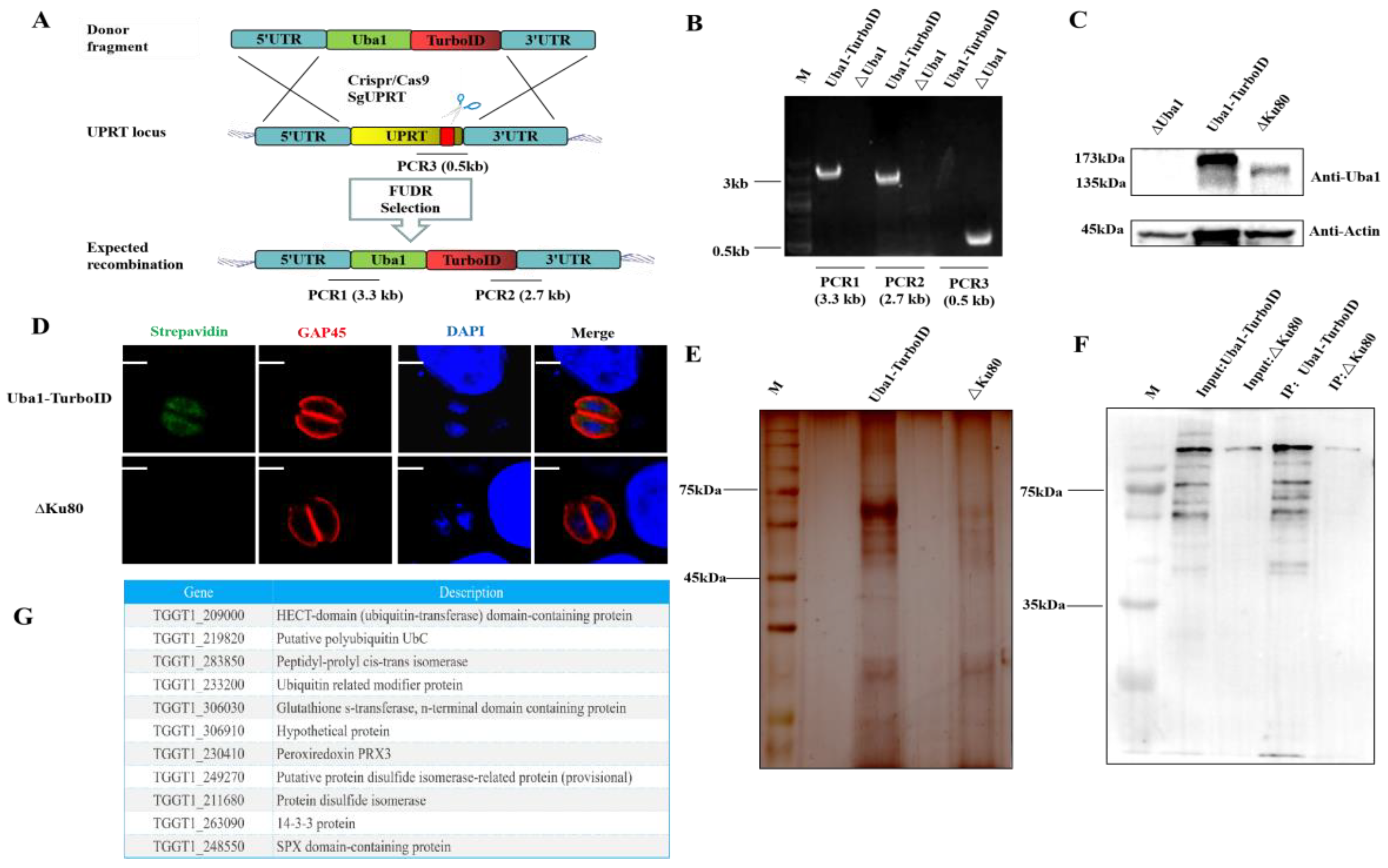
2.7. Screening for Other Interacting Proteins of TgUba1 by TurboID
3. Discussion
4. Materials and Methods
4.1. Cells and Parasites
4.2. Intracellular Invasion and Replication Assay
4.3. Egress Assay
4.4. Pathogenicity of T. gondii in Mice
4.5. Co-Immunoprecipitation Analysis
4.6. Yeast Two-Hybrid Screen
4.7. Enrichment and Identification of Biotinylated Proteins
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kochanowsky, J.A.; Koshy, A.A. Toxoplasma gondii. Curr. Biol. 2018, 28, R770–R771. [Google Scholar] [CrossRef]
- Harker, K.S.; Ueno, N.; Lodoen, M.B. Toxoplasma gondii dissemination: A parasite’s journey through the infected host. Parasite Immunol. 2015, 37, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Wang, F.; Liu, M.; Qiu, R.; Ji, C. The dual role of ubiquitin-like protein Urm1 as a protein modifier and sulfur carrier. Protein Cell 2011, 2, 612–619. [Google Scholar] [CrossRef]
- Petroski, M.D.; Salvesen, G.S.; Wolf, D.A. Urm1 couples sulfur transfer to ubiquitin-like protein function in oxidative stress. Proc. Natl. Acad. Sci. USA 2011, 108, 1749–1750. [Google Scholar] [CrossRef] [PubMed]
- Van der Veen, A.G.; Schorpp, K.; Schlieker, C.; Buti, L.; Damon, J.R.; Spooner, E.; Ploegh, H.L.; Jentsch, S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. USA 2011, 108, 1763–1770. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.L. The emerging roles of ubiquitin-like protein Urm1 in eukaryotes. Cell. Signal. 2021, 81, 109946. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Wang, J.; Chen, J.; Liu, X.; Chen, X.; Xiao, Q.; Li, J.; Li, H.; Zhao, X.; Zhang, X. Involvement of Urm1, a Ubiquitin-Like Protein, in the Regulation of Oxidative Stress Response of Toxoplasma gondii. Microbiol. Spectr. 2022, 10, e0239421. [Google Scholar] [CrossRef]
- Wang, J.; Tan, Q.; Chen, J.; Liu, X.; Di, Z.; Xiao, Q.; Li, J.; Zhao, X.; Zhang, X. Alkyl Hydroperoxide Reductase as a Determinant of Parasite Antiperoxide Response in Toxoplasma gondii. Oxidative Med. Cell. Longev. 2021, 2021, 1675652. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [Green Version]
- Taherbhoy, A.M.; Schulman, B.A.; Kaiser, S.E. Ubiquitin-like modifiers. Essays Biochem. 2012, 52, 51–63. [Google Scholar] [CrossRef]
- Cappadocia, L.; Lima, C.D. Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem. Rev. 2018, 118, 889–918. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Termathe, M.; Leidel, S.A. The Uba4 domain interplay is mediated via a thioester that is critical for tRNA thiolation through Urm1 thiocarboxylation. Nucleic Acids Res. 2018, 46, 5171–5181. [Google Scholar] [CrossRef]
- Jüdes, A.; Bruch, A.; Klassen, R.; Helm, M.; Schaffrath, R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb. Cell 2016, 3, 554–564. [Google Scholar] [CrossRef]
- Termathe, M.; Leidel, S.A. Urm1: A Non-Canonical UBL. Biomolecules 2021, 11, 139. [Google Scholar] [CrossRef]
- Schmitz, J.; Chowdhury, M.M.; Hänzelmann, P.; Nimtz, M.; Lee, E.Y.; Schindelin, H.; Leimkühler, S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 2008, 47, 6479–6489. [Google Scholar] [CrossRef]
- Furukawa, K.; Mizushima, N.; Noda, T.; Ohsumi, Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 2000, 275, 7462–7465. [Google Scholar] [CrossRef]
- Leimkühler, S.; Wuebbens, M.M.; Rajagopalan, K.V. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 2001, 276, 34695–34701. [Google Scholar] [CrossRef]
- Olsen, S.K.; Capili, A.D.; Lu, X.; Tan, D.S.; Lima, C.D. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 2010, 463, 906–912. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Yamamoto, H.; Sakamoto, H.; Oku, M.; Mutungi, J.K.; Sahani, M.H.; Kurikawa, Y.; Kita, K.; Noda, N.N.; Sakai, Y.; et al. Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat. Struct. Mol. Biol. 2019, 26, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Kayashima, Y.; Yamakawa-Kobayashi, K. Involvement of Prx3, a Drosophila ortholog of the thiol-dependent peroxidase PRDX3, in age-dependent oxidative stress resistance. Biomed. Res. 2012, 33, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef]
- Yamada, Y.; Suzuki, N.N.; Hanada, T.; Ichimura, Y.; Kumeta, H.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 2007, 282, 8036–8043. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef]
- Suzuki, T.; Sugiyama, M.; Wakazono, K.; Kaneko, Y.; Harashima, S. Lactic-acid stress causes vacuolar fragmentation and impairs intracellular amino-acid homeostasis in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2012, 113, 421–430. [Google Scholar] [CrossRef]
- Rubio-Texeira, M. Urmylation controls Nil1p and Gln3p-dependent expression of nitrogen-catabolite repressed genes in Saccharomyces cerevisiae. FEBS Lett. 2007, 581, 541–550. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Yao, Z.; Shi, Y.; Xiong, J.; Zhou, J.; Su, Z.; Huang, Y. 14-3-3/Tau Interaction and Tau Amyloidogenesis. J. Mol. Neurosci. 2019, 68, 620–630. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Q.; Li, J.; Chen, J.; Tan, Q.; Chen, X.; Li, H.; Zhao, X.; Zhang, X. Uba1: A Potential Ubiquitin-like Activator Protein of Urm1 in Toxoplasma gondii. Int. J. Mol. Sci. 2022, 23, 10298. https://doi.org/10.3390/ijms231810298
Xiao Q, Li J, Chen J, Tan Q, Chen X, Li H, Zhao X, Zhang X. Uba1: A Potential Ubiquitin-like Activator Protein of Urm1 in Toxoplasma gondii. International Journal of Molecular Sciences. 2022; 23(18):10298. https://doi.org/10.3390/ijms231810298
Chicago/Turabian StyleXiao, Qianqian, Jinxuan Li, Junpeng Chen, Qianqian Tan, Xiao Chen, Hongmei Li, Xiaomin Zhao, and Xiao Zhang. 2022. "Uba1: A Potential Ubiquitin-like Activator Protein of Urm1 in Toxoplasma gondii" International Journal of Molecular Sciences 23, no. 18: 10298. https://doi.org/10.3390/ijms231810298



