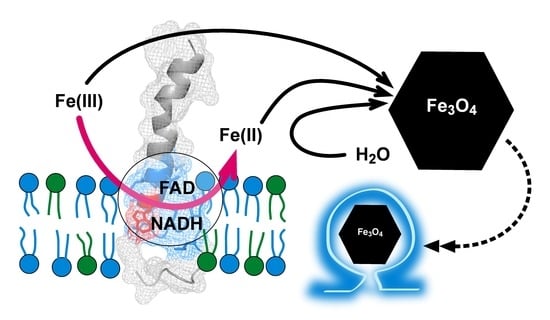
The Magnetosome Protein, Mms6 from Magnetospirillum magneticum Strain AMB-1, Is a Lipid-Activated Ferric Reductase
Abstract
:1. Introduction
2. Results
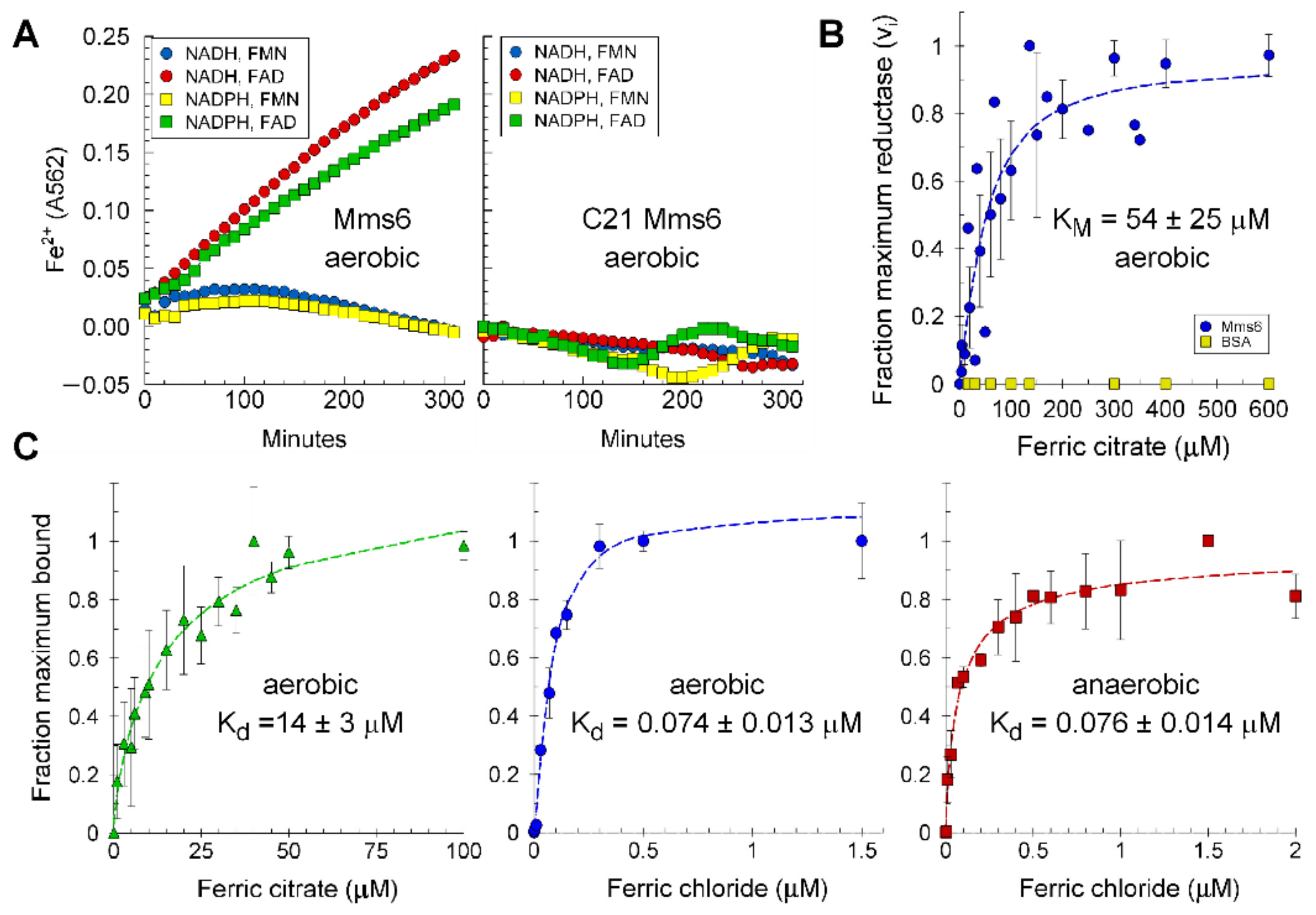
2.1. Mms6 Is a Ferric Reductase
2.2. Electron Donor and Cofactor Requirement
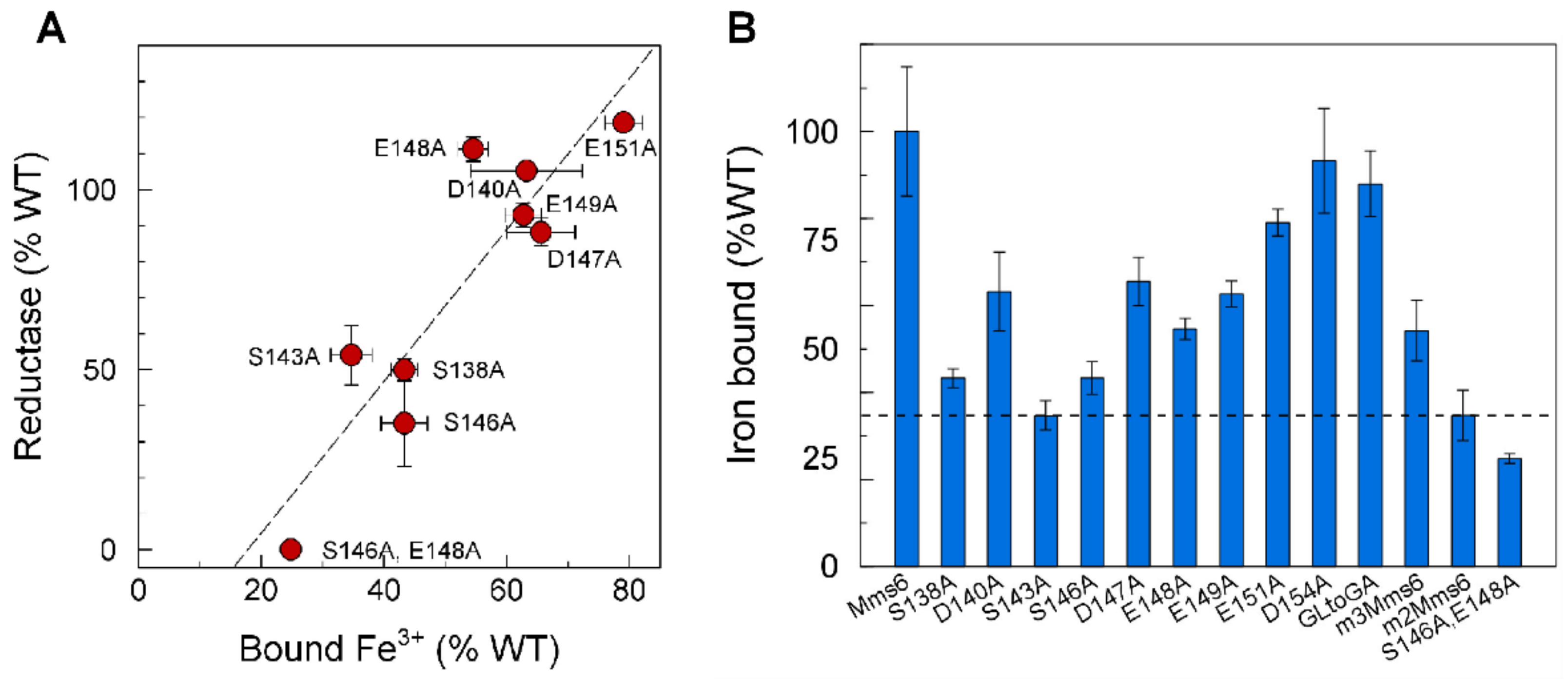
2.3. Iron Binding Residues in Mms6
2.4. Mms6 Primary Sequence and Tertiary Structure Predictions
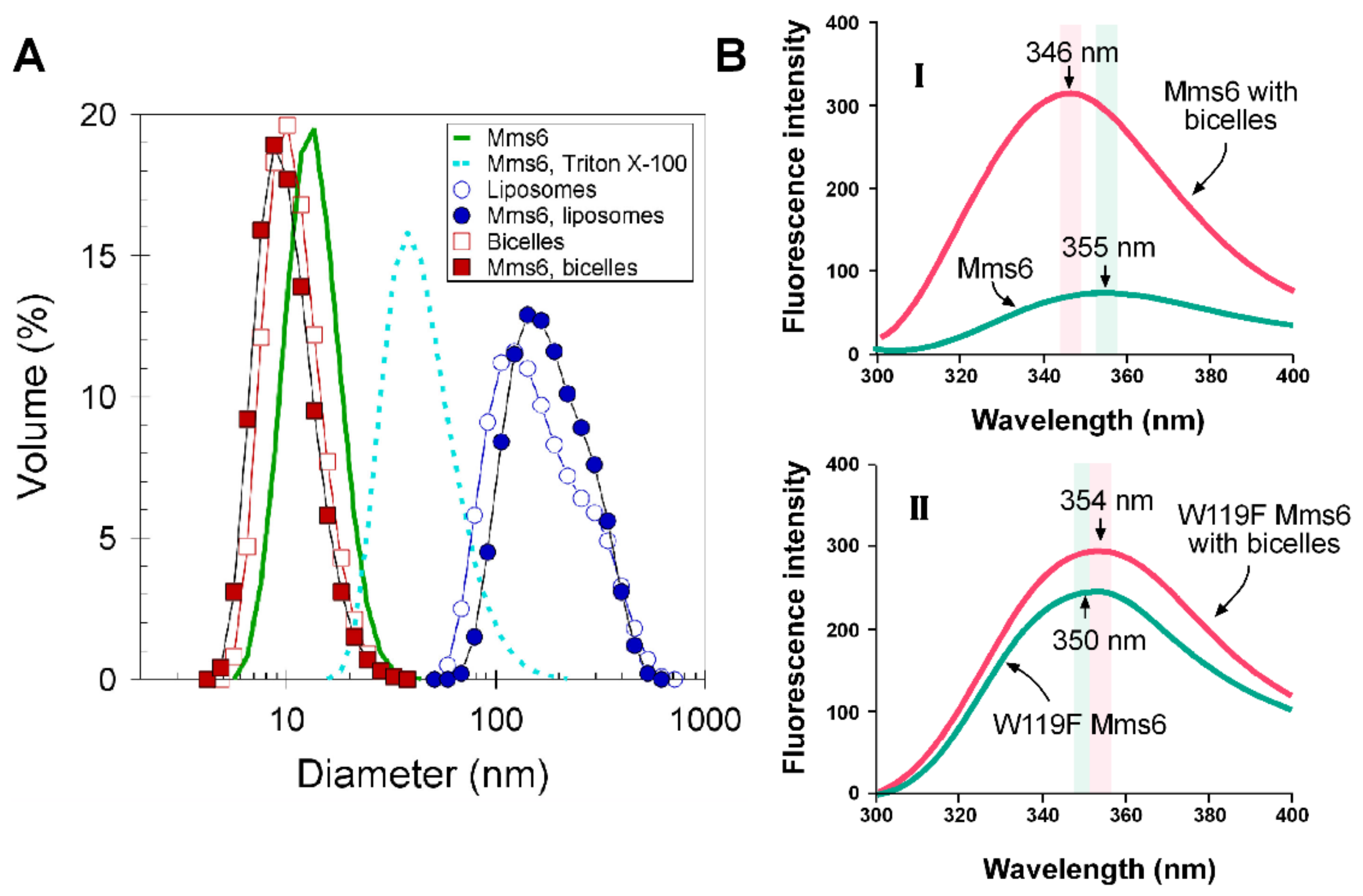
2.5. Mms6 Integrates into Lipid Membranes
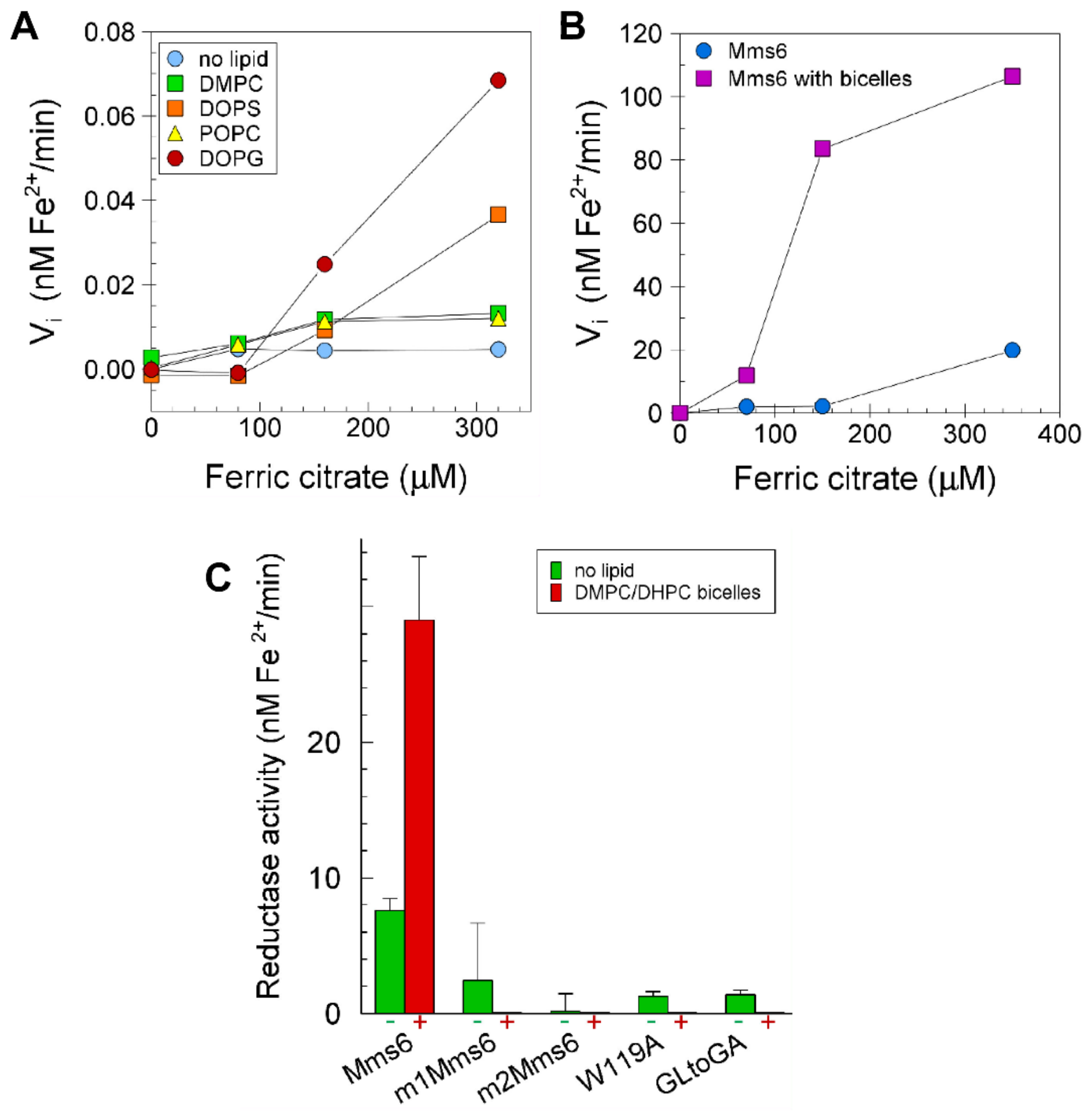
2.6. Lipids Promote Higher Ferric Reductase Activity of Mms6
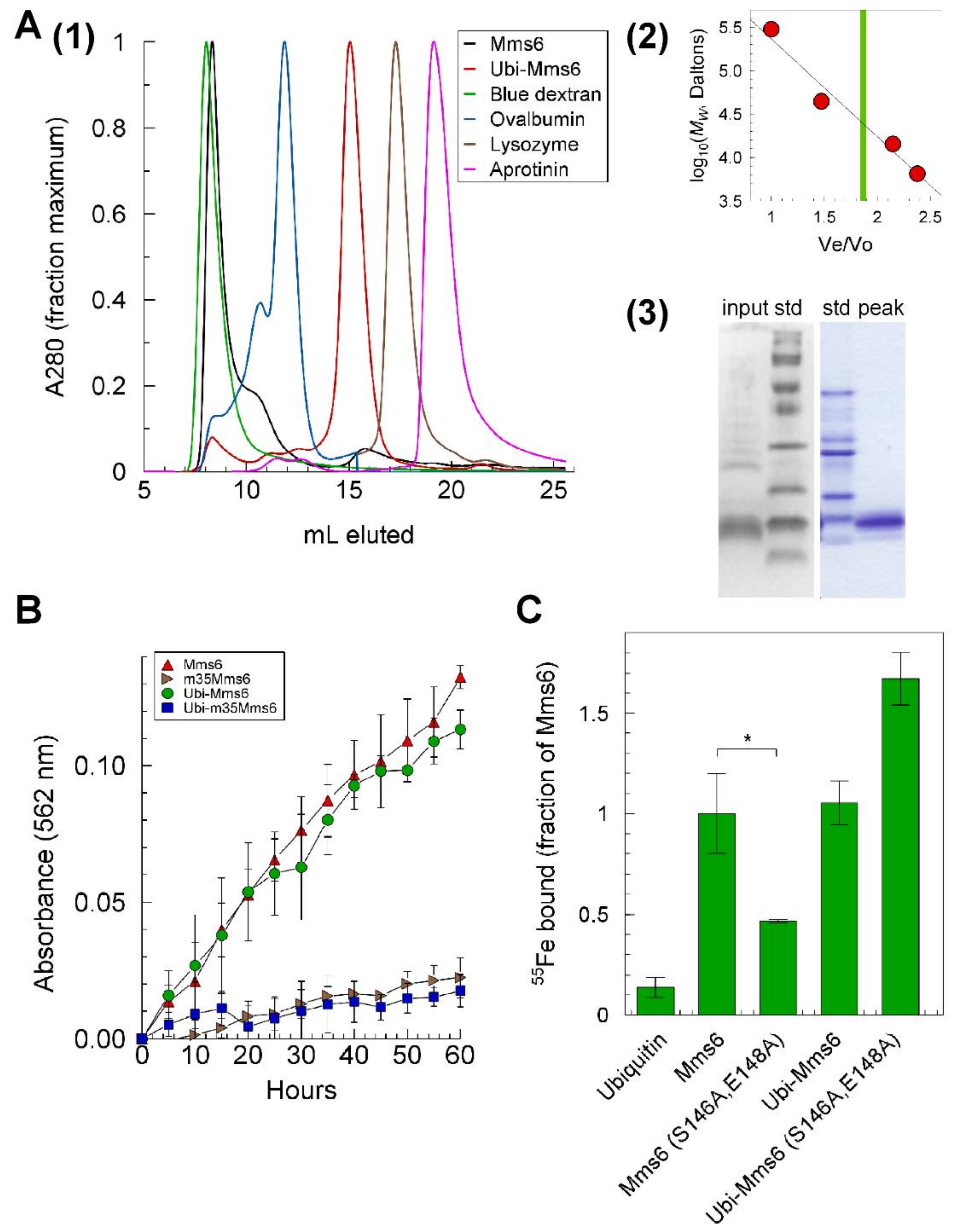
2.7. Reductase Activity of the Mms6 Ubiquitin Monomer
3. Discussion
4. Materials and Methods
4.1. Reagents, Proteins, and Preparation of Mutants and Sequence Variants
4.2. Bicelle Preparation and Integration of Mms6 into Bicelles
4.3. Liposome Preparation and Integration of Mms6 into Liposomes
4.4. Ferric Reductase Activity
4.5. Iron Binding by Filter Capture
4.6. Intrinsic Fluorescence Spectroscopy
4.7. Dynamic Light Scattering (DLS)
4.8. Size Exclusion Chromatography
4.9. Analysis of Binding Isotherms and Statistical Evaluations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blakemore, R. Magnetotactic bacteria. Science 1975, 190, 377–379. [Google Scholar] [CrossRef]
- Blakemore, R.P. Magnetotactic Bacteria. Annu. Rev. Microbiol. 1982, 36, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Schüler, D.; Monteil, C.L.; Lefevre, C.T. Magnetospirillum gryphiswaldense. Trends Microbiol. 2020, 28, 947–948. [Google Scholar] [CrossRef]
- Müller, F.D.; Schüler, D.; Pfeiffer, D. A Compass to Boost Navigation: Cell Biology of Bacterial Magnetotaxis. J. Bacteriol. 2020, 202, e00398-20. [Google Scholar] [CrossRef] [PubMed]
- Zwiener, T.; Dziuba, M.; Mickoleit, F.; Rückert, C.; Busche, T.; Kalinowski, J.; Uebe, R.; Schüler, D. Towards a ‘chassis’ for bacterial magnetosome biosynthesis: Genome streamlining of Magnetospirillum gryphiswaldense by multiple deletions. Microb. Cell Factories 2021, 20, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Zhang, Y.; Liu, J.; Wen, T.; Miao, T.; Basit, A.; Jiang, W. OxyR controls magnetosome formation by regulating magnetosome island (MAI) genes, iron metabolism, and redox state. Free Radic. Biol. Med. 2020, 161, 272–282. [Google Scholar] [CrossRef]
- Lohße, A.; Kolinko, I.; Raschdorf, O.; Uebe, R.; Borg, S.; Brachmann, A.; Plitzko, J.M.; Müller, R.; Zhang, Y.; Schüler, D. Overproduction of Magnetosomes by Genomic Amplification of Biosynthesis-Related Gene Clusters in a Magnetotactic Bacterium. Appl. Environ. Microbiol. 2016, 82, 3032–3041. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, A.; Webb, J.; Matsunaga, T. A Novel Protein Tightly Bound to Bacterial Magnetic Particles in Magnetospirillum magneticum Strain AMB-1. J. Biol. Chem. 2003, 278, 8745–8750. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Prozorov, T.; Palo, P.E.; Liu, X.; Vaknin, D.; Prozorov, R.; Mallapragada, S.; Nilsen-Hamilton, M. Self-Assembly and Biphasic Iron-Binding Characteristics of Mms6, A Bacterial Protein That Promotes the Formation of Superparamagnetic Magnetite Nanoparticles of Uniform Size and Shape. Biomacromolecules 2012, 13, 98–105. [Google Scholar] [CrossRef]
- Prozorov, T.; Mallapragada, S.K.; Narasimhan, B.; Wang, L.; Palo, P.; Nilsen-Hamilton, M.; Williams, T.J.; Bazylinski, D.A.; Prozorov, R.; Canfield, P.C. Protein-Mediated Synthesis of Uniform Superparamagnetic Magnetite Nanocrystals. Adv. Funct. Mater. 2007, 17, 951–957. [Google Scholar] [CrossRef]
- Tanaka, M.; Mazuyama, E.; Arakaki, A.; Matsunaga, T. MMS6 Protein Regulates Crystal Morphology during Nano-sized Magnetite Biomineralization In Vivo. J. Biol. Chem. 2011, 286, 6386–6392. [Google Scholar] [CrossRef]
- Mirabello, G.; Lenders, J.J.M.; Sommerdijk, N.A.J.M. Bioinspired synthesis of magnetite nanoparticles. Chem. Soc. Rev. 2016, 45, 5085–5106. [Google Scholar] [CrossRef]
- Bain, J.; Legge, C.J.; Beattie, D.L.; Sahota, A.; Dirks, C.; Lovett, J.R.; Staniland, S.S. A biomimetic magnetosome: Formation of iron oxide within carboxylic acid terminated polymersomes. Nanoscale 2019, 11, 11617–11625. [Google Scholar] [CrossRef]
- Bird, S.M.; Rawlings, A.E.; Galloway, J.M.; Staniland, S.S. Using a biomimetic membrane surface experiment to investigate the activity of the magnetite biomineralisation protein Mms6. RSC Adv. 2016, 6, 7356–7363. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.M.; Galloway, J.M.; Rawlings, A.E.; Bramble, J.P.; Staniland, S.S. Taking a hard line with biotemplating: Cobalt-doped magnetite magnetic nanoparticle arrays. Nanoscale 2015, 7, 7340–7351. [Google Scholar] [CrossRef] [PubMed]
- Ali, J.; Ali, N.; Wang, L.; Waseem, H.; Pan, G. Revisiting the mechanistic pathways for bacterial mediated synthesis of noble metal nanoparticles. J. Microbiol. Methods 2019, 159, 18–25. [Google Scholar] [CrossRef]
- Chen, H.; Sun, J.; Wang, Z.; Zhou, Y.; Lou, Z.; Chen, B.; Wang, P.; Guo, Z.; Tang, H.; Ma, J.; et al. Magnetic Cell–Scaffold Interface Constructed by Superparamagnetic IONP Enhanced Osteogenesis of Adipose-Derived Stem Cells. ACS Appl. Mater. Interfaces 2018, 10, 44279–44289. [Google Scholar] [CrossRef]
- Mori, J.F.; Lu, S.; Händel, M.; Totsche, K.U.; Neu, T.R.; Iancu, V.V.; Tarcea, N.; Popp, J.; Küsel, K. Schwertmannite formation at cell junctions by a new filament-forming Fe(II)-oxidizing isolate affiliated with the novel genus Acidithrix. Microbiology 2016, 162, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, L.; Palo, P.; Liu, X.; Mallapragada, S.K.; Nilsen-Hamilton, M. Integrated Self-Assembly of the Mms6 Magnetosome Protein to Form an Iron-Responsive Structure. Int. J. Mol. Sci. 2013, 14, 14594–14606. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Bu, W.; Wang, L.; Palo, P.E.; Mallapragada, S.; Nilsen-Hamilton, M.; Vaknin, D. Interfacial Properties and Iron Binding to Bacterial Proteins That Promote the Growth of Magnetite Nanocrystals: X-ray Reflectivity and Surface Spectroscopy Studies. Langmuir 2012, 28, 4274–4282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawlings, A.E.; Bramble, J.P.; Hounslow, A.M.; Williamson, M.P.; Monnington, A.E.; Cooke, D.J.; Staniland, S.S. Ferrous Iron Binding Key to Mms6 Magnetite Biomineralisation: A Mechanistic Study to Understand Magnetite Formation Using pH Titration and NMR Spectroscopy. Chem. Eur. J. 2016, 22, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Ofer, S.; Nowik, I.; Bauminger, E.; Papaefthymiou, G.; Frankel, R.; Blakemore, R. Magnetosome dynamics in magnetotactic bacteria. Biophys. J. 1984, 46, 57–64. [Google Scholar] [CrossRef]
- Baumgartner, J.; Dey, A.; Bomans, P.H.H.; Le Coadou, C.; Fratzl, P.; Sommerdijk, N.A.J.M.; Faivre, D. Nucleation and growth of magnetite from solution. Nat. Mater. 2013, 12, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Staniland, S.; Ward, B.; Harrison, A.; van der Laan, G.; Telling, N. Rapid magnetosome formation shown by real-time X-ray magnetic circular dichroism. Proc. Natl. Acad. Sci. USA 2007, 104, 19524–19528. [Google Scholar] [CrossRef] [PubMed]
- Frankel, R.B.; Blakemore, R.P. Precipitation of Fe3O4 in Magnetotactic Bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1984, 304, 567–574. [Google Scholar] [CrossRef]
- Frankel, R.B.; Papaefthymiou, G.C.; Blakemore, R.P.; O’Brien, W. Fe3O4 precipitation in magnetotactic bacteria. Biochim. Biophys. Acta 1983, 763, 147–159. [Google Scholar] [CrossRef]
- Fdez-Gubieda, M.L.; Muela, A.; Alonso, J.; García-Prieto, A.; Olivi, L.; Fernández-Pacheco, R.; Barandiarán, J.M. Magnetite Biomineralization in Magnetospirillum gryphiswaldense: Time-Resolved Magnetic and Structural Studies. ACS Nano 2013, 7, 3297–3305. [Google Scholar] [CrossRef]
- Arakaki, A.; Yamagishi, A.; Fukuyo, A.; Tanaka, M.; Matsunaga, T. Co-ordinated functions of Mms proteins define the surface structure of cubo-octahedral magnetite crystals in magnetotactic bacteria. Mol. Microbiol. 2014, 93, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-Align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Fischer, H.; Polikarpov, I.; Craievich, A.F. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004, 13, 2825–2828. [Google Scholar] [CrossRef] [PubMed]
- Beaugrand, M.; Arnold, A.; Hénin, J.; Warschawski, D.; Williamson, P.T.; Marcotte, I. Lipid Concentration and Molar Ratio Boundaries for the Use of Isotropic Bicelles. Langmuir 2014, 30, 6162–6170. [Google Scholar] [CrossRef]
- Amemiya, Y.; Arakaki, A.; Staniland, S.S.; Tanaka, T.; Matsunaga, T. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials 2007, 28, 5381–5389. [Google Scholar] [CrossRef]
- Lohße, A.; Borg, S.; Raschdorf, O.; Kolinko, I.; Tompa, É.; Pósfai, M.; Faivre, D.; Baumgartner, J.; Schüler, D. Genetic Dissection of the mamAB and mms6 Operons Reveals a Gene Set Essential for Magnetosome Biogenesis in Magnetospirillum gryphiswaldense. J. Bacteriol. 2014, 196, 2658–2669. [Google Scholar] [CrossRef]
- Zhang, X.; Krause, K.-H.; Xenarios, I.; Soldati, T.; Boeckmann, B. Evolution of the Ferric Reductase Domain (FRD) Superfamily: Modularity, Functional Diversification, and Signature Motifs. PLoS ONE 2013, 8, e58126. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, A.E.; Liravi, P.; Corbett, S.; Holehouse, A.S.; Staniland, S.S. Investigating the ferric ion binding site of magnetite biomineralisation protein Mms6. PLoS ONE 2020, 15, e0228708. [Google Scholar] [CrossRef] [Green Version]
- Hasan, N.; Nester, E.W. Purification and characterization of NADPH-dependent flavin reductase. An enzyme required for the activation of chorismate synthase in Bacillus subtilis. J. Biol. Chem. 1978, 253, 4987–4992. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Suzuki, E.; Oestreicher, Z.; Minamide, H.; Endoh, H.; Fukumori, Y.; Taoka, A. A protein-protein interaction in magnetosomes: TPR protein MamA interacts with an Mms6 protein. Biochem. Biophys. Rep. 2016, 7, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Peigneux, A.; Jabalera, Y.; Vivas, M.A.F.; Casares, S.; Azuaga, A.I.; Jimenez-Lopez, C. Tuning properties of biomimetic magnetic nanoparticles by combining magnetosome associated proteins. Sci. Rep. 2019, 9, 8804. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Browne, P.J.; Hershey, D.M.; Montabana, E.; Iavarone, A.T.; Downing, K.H.; Komeili, A. A protease-mediated switch regulates the growth of magnetosome organelles in Magnetospirillum magneticum. Proc. Natl. Acad. Sci. USA 2022, 119, e2111745119. [Google Scholar] [CrossRef] [PubMed]
- Cain, T.J.; Smith, A.T. Ferric iron reductases and their contribution to unicellular ferrous iron uptake. J. Inorg. Biochem. 2021, 218, 111407. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Chen, X.; Huang, Y.; Wang, Z. Selection and Identification of a DNA Aptamer Targeted to Vibrio parahemolyticus. J. Agric. Food Chem. 2012, 60, 4034–4038. [Google Scholar] [CrossRef] [PubMed]
- Poget, S.F.; Girvin, M.E. Solution NMR of membrane proteins in bilayer mimics: Small is beautiful, but sometimes bigger is better. Biochim. Biophys. Acta (BBA) Biomembr. 2007, 1768, 3098–3106. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, Y.; Shin, Y.-K. Supramolecular SNARE assembly precedes hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Riemer, J.; Hoepken, H.H.; Czerwinska, H.; Robinson, S.R.; Dringen, R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal. Biochem. 2004, 331, 370–375. [Google Scholar] [CrossRef]
- Stookey, L.L. Ferrozine—A new spectrophotometric reagent for iron. Anal. Chem. 1970, 42, 779–781. [Google Scholar] [CrossRef] [Green Version]
- Williams, H.D.; Poole, R.K. Reduction of iron(III) byEscherichia coli K12: Lack of involvement of the respiratory chains. Curr. Microbiol. 1987, 15, 319–324. [Google Scholar] [CrossRef]
- Coves, J.; Fontecave, M. Reduction and mobilization of iron by a NAD(P)H: Flavin oxidoreductase from Escherichia coli. JBIC J. Biol. Inorg. Chem. 1993, 211, 635–641. [Google Scholar] [CrossRef]
- Fischer, E.; Strehlow, B.; Hartz, D.; Braun, V. Soluble and membrane-bound ferrisiderophore reductases of Escherichia coli K-12. Arch. Microbiol. 1990, 153, 329–336. [Google Scholar] [CrossRef]
- Shen, C.; Menon, R.; Das, D.; Bansal, N.; Nahar, N.; Guduru, N.; Jaegle, S.; Peckham, J.; Reshetnyak, Y.K. The protein fluorescence and structural toolkit: Database and programs for the analysis of protein fluorescence and structural data. Proteins Struct. Funct. Bioinform. 2008, 71, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Suslov, N.B.; Li, N.-S.; Shelke, S.; Evans, M.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef]
- Vadas, A.; Monbouquette, H.G.; Johnson, E.; Schröder, I. Identification and characterization of a novel ferric reductase from the hyperthermophilic Archaeon Archaeoglobus fulgidus. J. Biol.Chem. 1999, 274, 36715–36721. [Google Scholar] [CrossRef]
- Huyer, M.; Page, W.J. Ferric reductase activity in Azotobacter vinelandii and its inhibition by Zn2+. J. Bacteriol. 1989, 171, 4031–4037. [Google Scholar] [CrossRef]
- Gaines, C.G.; Lodge, J.S.; Arceneaux, J.E.; Byers, B.R. Ferrisiderophore reductase activity associated with an aromatic biosynthetic enzyme complex in Bacillus subtilis. J. Bacteriol. 1981, 148, 527–533. [Google Scholar] [CrossRef]
- Poch, M.T.; Johnson, W. Ferric reductases of Legionella pneumophila. Biometals 1993, 6, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, Y.; Fujiwara, T.; Yoshimatsu, K.; Fukumori, Y. Iron reductase for magnetite synthesis in the magnetotactic bacterium Magnetospirillum magnetotacticum. J. Bacteriol. 1999, 181, 2142–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, M.D.; Dailey, H.A. Ferric iron reductase of Rhodopseudomonas sphaeroides. J. Bacteriol. 1985, 163, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Hongyu, M.; Chen, Q.; Du, J.; Tang, L.; Qin, F.; Miao, B.; Wu, X.; Zeng, J. Ferric Reductase Activity of the ArsH Protein from Acidithiobacillus ferrooxidans. J. Microbiol. Biotechnol. 2011, 21, 464–469. [Google Scholar] [CrossRef]
- Mazoch, J.; Tesařík, R.; Sedláček, V.; Kučera, I.; Turánek, J. Isolation and biochemical characterization of two soluble iron(III) reductases from Paracoccus denitrificans. Eur. J. Biochem. 2004, 271, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Yeom, J.; Jeon, C.O.; Madsen, E.L.; Park, W. Ferredoxin-NADP+ reductase from Pseudomonas putida functions as a ferric reductase. J. Bacteriol. 2009, 191, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Wei, J.; Lei, Y.; Ying, L. A novel ferric reductase purified from Magnetospirillum gryphiswaldense MSR-1. Curr. Microbiol. 2007, 55, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, F.; Lovley, D.R. Isolation and characterization of a soluble NADPH-dependent Fe(III) reductase from Geobacter sulfurreducens. J. Bacteriol. 2001, 183, 4468–4476. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singappuli-Arachchige, D.; Feng, S.; Wang, L.; Palo, P.E.; Shobade, S.O.; Thomas, M.; Nilsen-Hamilton, M. The Magnetosome Protein, Mms6 from Magnetospirillum magneticum Strain AMB-1, Is a Lipid-Activated Ferric Reductase. Int. J. Mol. Sci. 2022, 23, 10305. https://doi.org/10.3390/ijms231810305
Singappuli-Arachchige D, Feng S, Wang L, Palo PE, Shobade SO, Thomas M, Nilsen-Hamilton M. The Magnetosome Protein, Mms6 from Magnetospirillum magneticum Strain AMB-1, Is a Lipid-Activated Ferric Reductase. International Journal of Molecular Sciences. 2022; 23(18):10305. https://doi.org/10.3390/ijms231810305
Chicago/Turabian StyleSingappuli-Arachchige, Dilini, Shuren Feng, Lijun Wang, Pierre E. Palo, Samuel O. Shobade, Michelle Thomas, and Marit Nilsen-Hamilton. 2022. "The Magnetosome Protein, Mms6 from Magnetospirillum magneticum Strain AMB-1, Is a Lipid-Activated Ferric Reductase" International Journal of Molecular Sciences 23, no. 18: 10305. https://doi.org/10.3390/ijms231810305
APA StyleSingappuli-Arachchige, D., Feng, S., Wang, L., Palo, P. E., Shobade, S. O., Thomas, M., & Nilsen-Hamilton, M. (2022). The Magnetosome Protein, Mms6 from Magnetospirillum magneticum Strain AMB-1, Is a Lipid-Activated Ferric Reductase. International Journal of Molecular Sciences, 23(18), 10305. https://doi.org/10.3390/ijms231810305