Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma
Abstract
:1. Introduction
2. Results
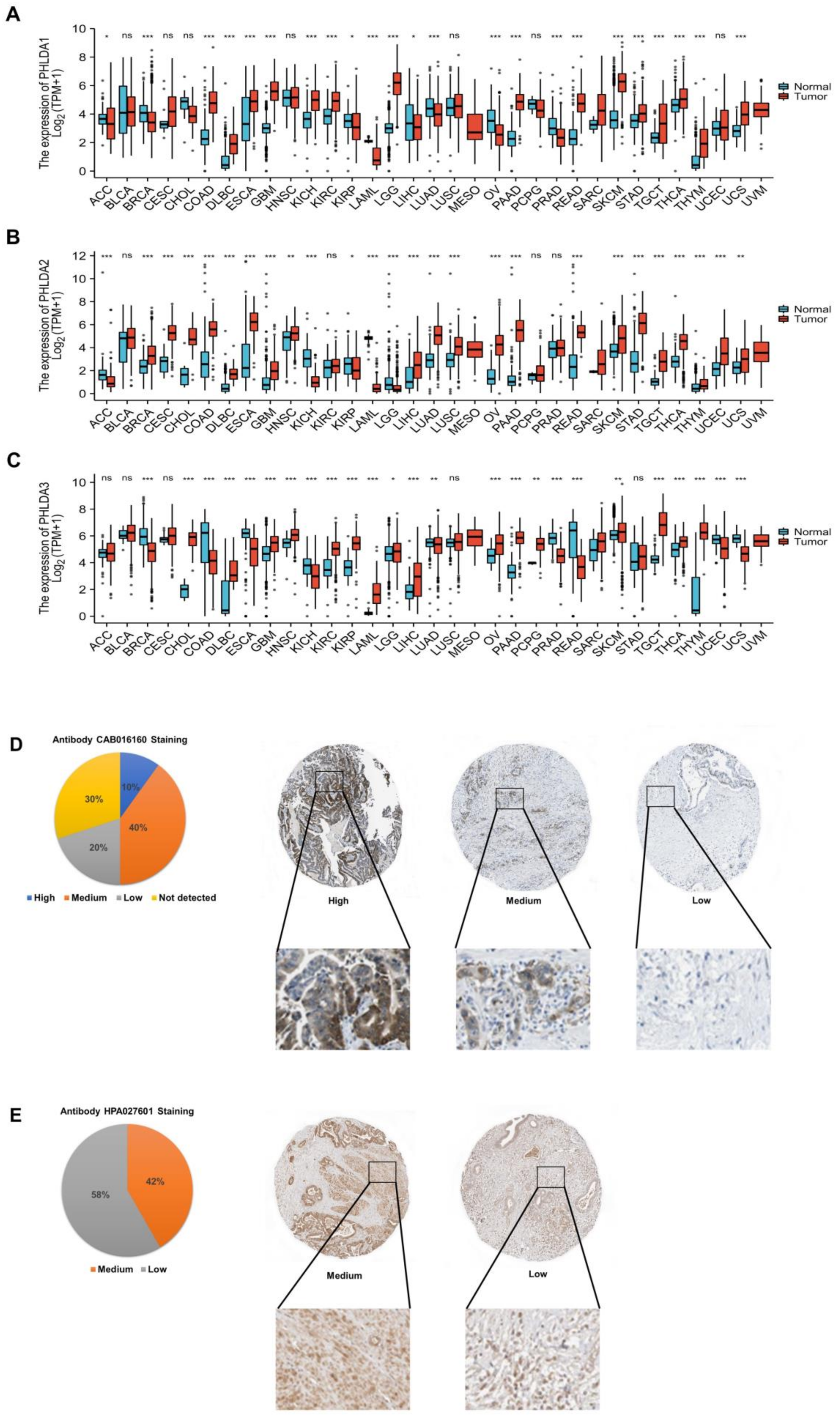
2.1. Aberrant Expression of the PHLDA Family Members across Cancers
2.2. Abnormal Expression of PHLDA1/3 Proteins in PAAD Tissues
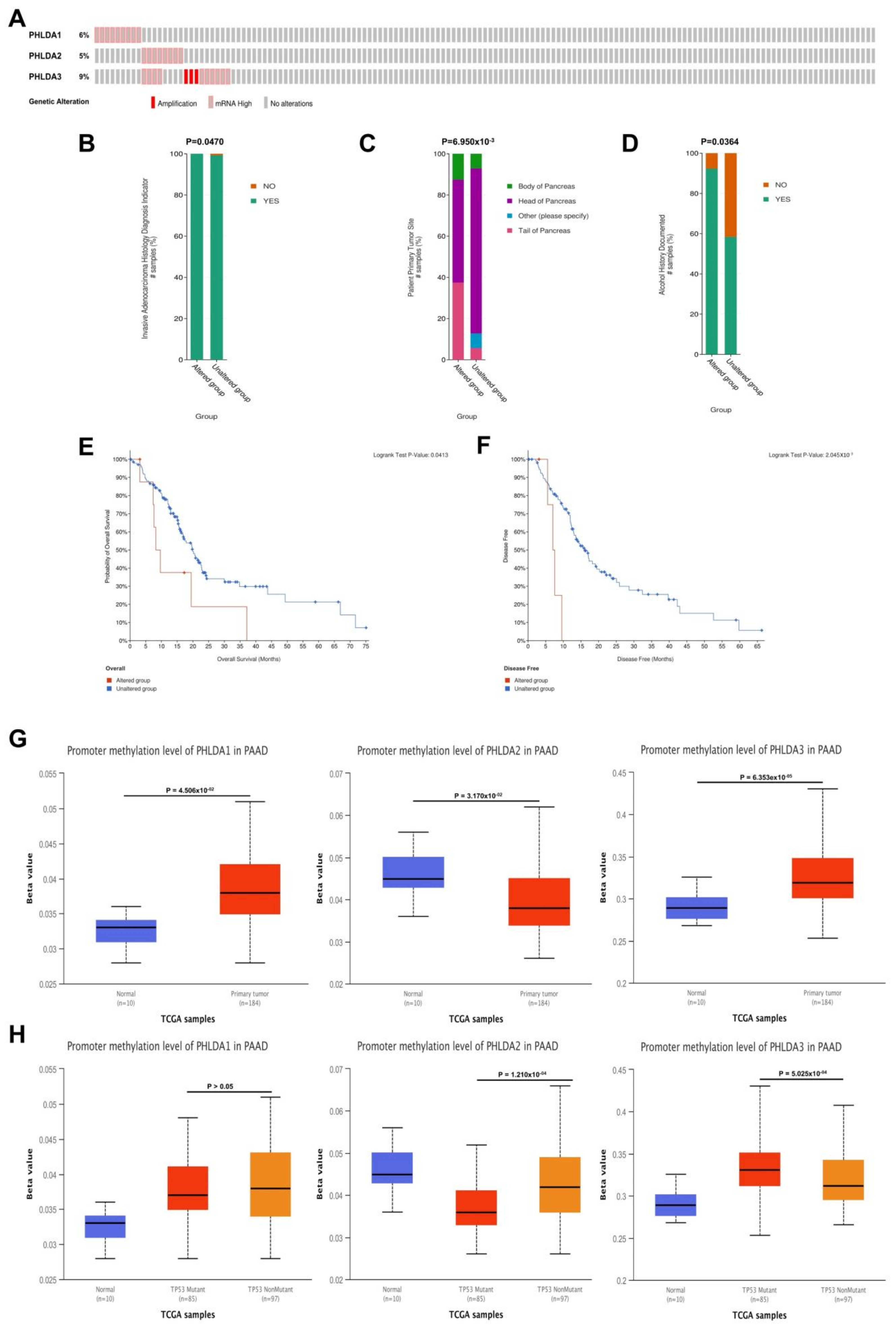
2.3. The Relationship between PHLDA Family Member Expression and the Clinicopathological Characteristics and Prognosis of PAAD Patients
2.4. Mutations in the PHLDA Family Member Genes Were Associated with Worse Clinicopathological Characteristics and a Worse Prognosis for PAAD Patients
2.5. Correlations between the Methylation Levels of the PHLDA Family Member Promoters and the Clinicopathological Characteristics of PAAD Patients
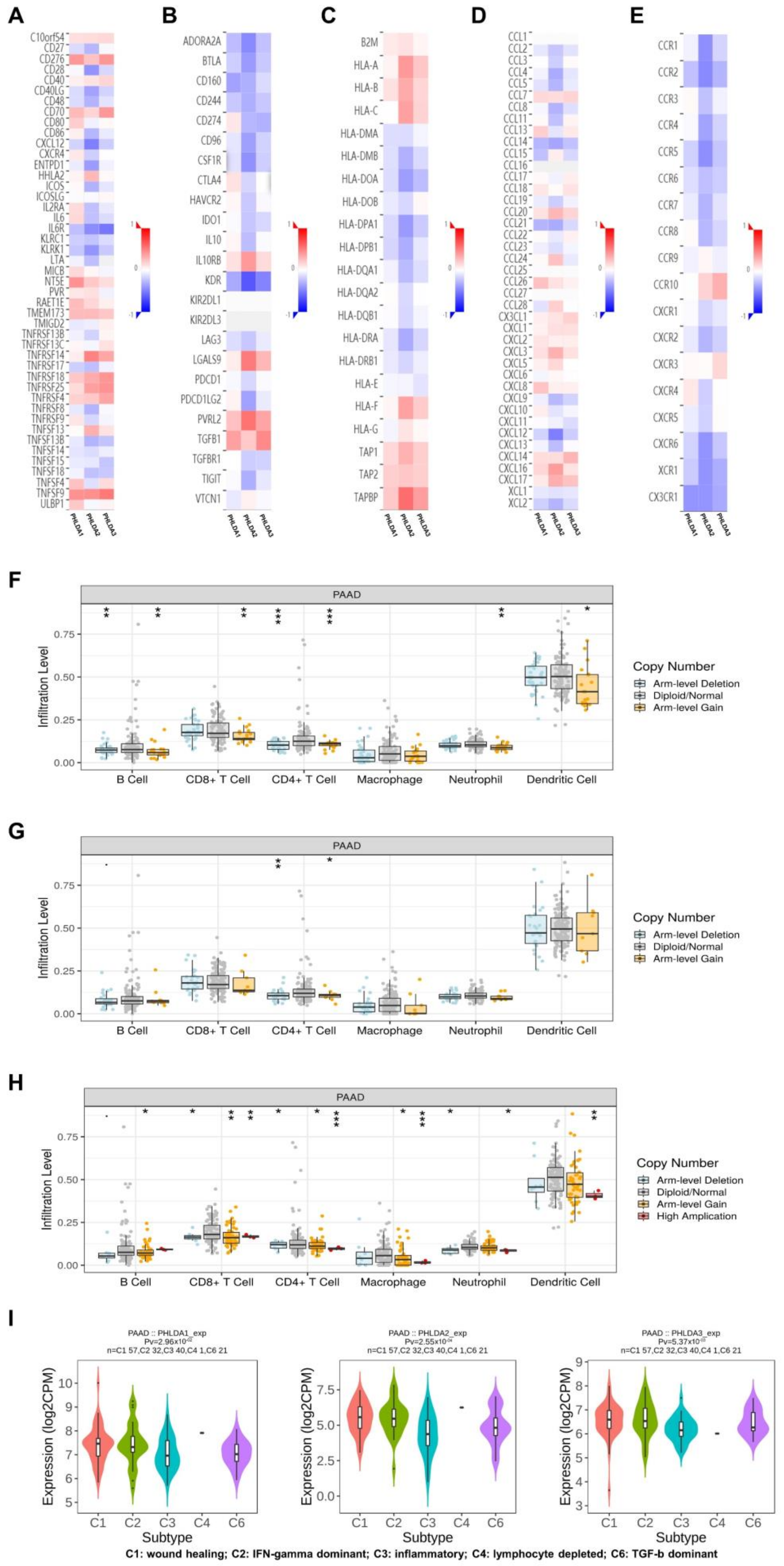
2.6. The Immune Landscape of PHLDA Family Member Expression and Variations in PAAD Patients
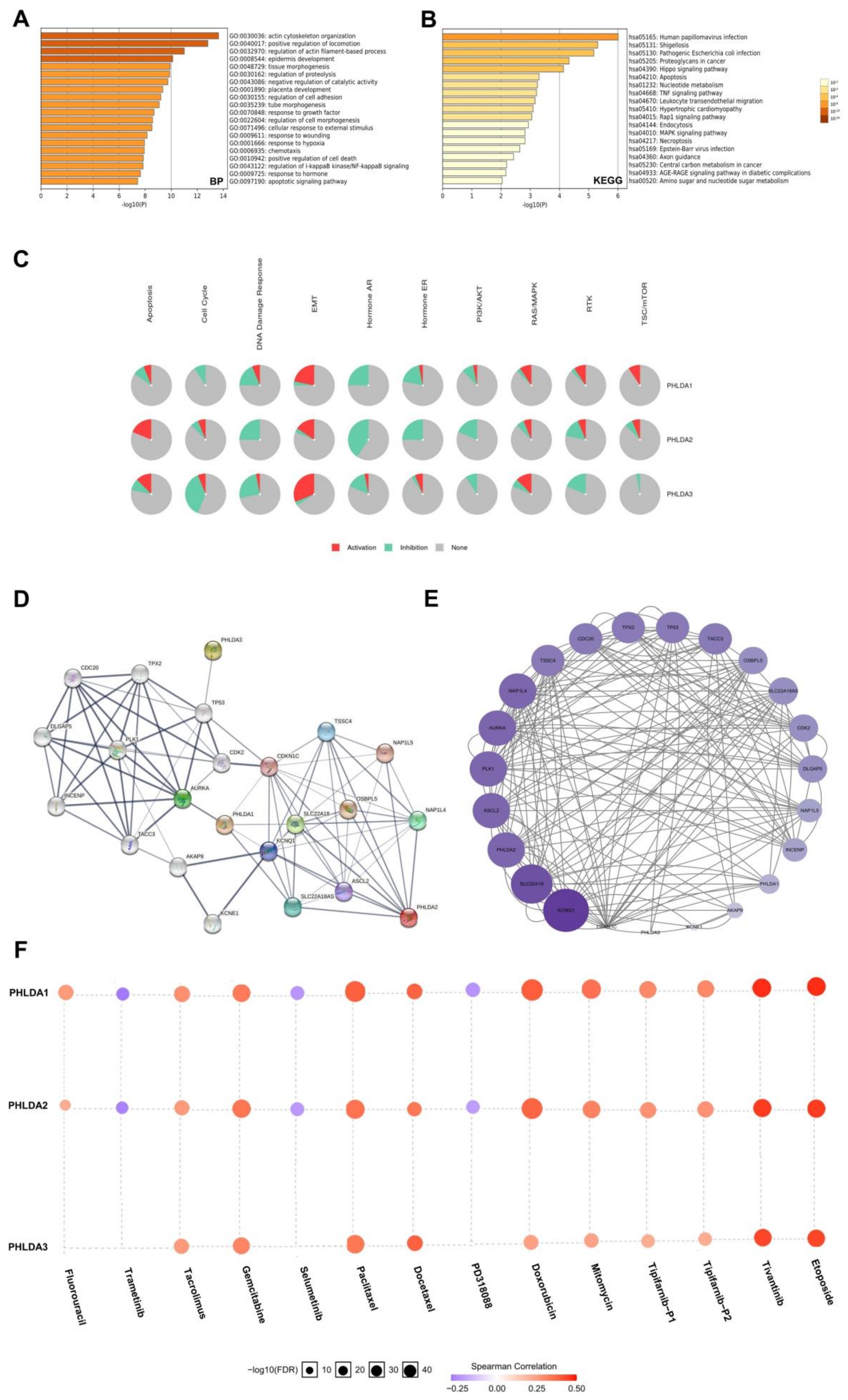
2.7. Enrichment Analysis of the PHLDA Family Members and the 600 Co-Expressed Genes
2.8. Construction and Analysis of the PPI Network Associated with the PHLDA Family Members
2.9. The Expression Levels of the PHLDA Family Members Affect the Treatment Sensitivity of Multiple Drugs
3. Discussion
4. Methods and Materials
4.1. Expression Analysis
4.2. HPA Analysis
4.3. Survival Analysis
4.4. Gene Variation Analysis
4.5. Methylation Analysis
4.6. Immune Infiltration Analysis
4.7. Gene Enrichment Analysis
4.8. Construction of the Functional PPI Network
4.9. Drug Sensitivity Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.; Donahue, T. Pancreatic Cancer. JAMA 2019, 322, 1426. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global surveillance of trends in cancer survival 2000–2014 (CONCORD-3): Analysis of individual records for 37,513,025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Adamek, H.E.; Albert, J.; Breer, H.; Weitz, M.; Schilling, D.; Riemann, J.F. Pancreatic cancer detection with magnetic resonance cholangiopancreatography and endoscopic retrograde cholangiopancreatography: A prospective controlled study. Lancet 2000, 356, 190–193. [Google Scholar] [CrossRef]
- Ringel, J.; Löhr, M. The MUC gene family: Their role in diagnosis and early detection of pancreatic cancer. Mol. Cancer. 2003, 2, 9. [Google Scholar] [CrossRef]
- Cui, X.H.; Hu, S.Y.; Zhu, C.F.; Qin, X.H. Expression and prognostic analyses of the insulin-like growth factor 2 mRNA binding protein family in human pancreatic cancer. BMC Cancer 2020, 20, 1160. [Google Scholar] [CrossRef]
- Haslam, R.J.; Koide, H.B.; Hemmings, B.A. Pleckstrin domain homology. Nature 1993, 363, 309–310. [Google Scholar] [CrossRef]
- Fuselier, T.T.; Lu, H. PHLD Class Proteins: A Family of New Players in the p53 Network. Int. J. Mol. Sci. 2020, 21, 3543. [Google Scholar] [CrossRef] [PubMed]
- Park, C.G.; Lee, S.Y.; Kandala, G.; Lee, S.Y.; Choi, Y. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity 1996, 4, 583–591. [Google Scholar] [CrossRef] [Green Version]
- Bonatto, N.; Carlini, M.J.; de Bessa Garcia, S.A.; Nagai, M.A. PHLDA1 (pleckstrin homology-like domain, family A, member 1) knockdown promotes migration and invasion of MCF10A breast epithelial cells. Cell Adhes. Migr. 2018, 12, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Takikawa, M.; Tsutsumi, S.; Yamaguchi, Y.; Okabe, A.; Shimada, M.; Kawase, T.; Sada, A.; Ezawa, I.; Takano, Y.; et al. PHLDA1, another PHLDA family protein that inhibits Akt. Cancer Sci. 2018, 109, 3532–3542. [Google Scholar] [CrossRef] [PubMed]
- Sakthianandeswaren, A.; Christie, M.; D’Andreti, C.; Tsui, C.; Jorissen, R.N.; Li, S.; Fleming, N.I.; Gibbs, P.; Lipton, L.; Malaterre, J.; et al. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 2011, 71, 3709–3719. [Google Scholar] [CrossRef]
- Nagai, M.A. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed. Rep. 2016, 4, 275–281. [Google Scholar] [CrossRef]
- Müller, S.; van den Boom, D.; Zirkel, D.; Köster, H.; Berthold, F.; Schwab, M.; Westphal, M.; Zumkeller, W. Retention of imprinting of the human apoptosis-related gene TSSC3 in human brain tumors. Hum. Mol. Genet. 2000, 9, 757–763. [Google Scholar] [CrossRef]
- McMinn, J.; Wei, M.; Schupf, N.; Cusmai, J.; Johnson, E.B.; Smith, A.C.; Weksberg, R.; Thaker, H.M.; Tycko, B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 2006, 27, 540–549. [Google Scholar] [CrossRef]
- Anderson, J.; Gordon, A.; McManus, A.; Shipley, J.; Pritchard-Jones, K. Disruption of imprinted genes at chromosome region 11p15.5 in paediatric rhabdomyosarcoma. Neoplasia 1999, 1, 340–348. [Google Scholar] [CrossRef]
- Xu, X.L.; Wu, L.C.; Du, F.; Davis, A.; Peyton, M.; Tomizawa, Y.; Maitra, A.; Tomlinson, G.; Gazdar, A.F.; Weissman, B.E.; et al. Inactivation of human SRBC, located within the 11p15.5-p15.4 tumor suppressor region, in breast and lung cancers. Cancer Res. 2001, 61, 7943–7949. [Google Scholar] [PubMed]
- Moon, H.G.; Oh, K.; Lee, J.; Lee, M.; Kim, J.Y.; Yoo, T.K.; Seo, M.W.; Park, A.K.; Ryu, H.S.; Jung, E.J.; et al. Prognostic and functional importance of the engraftment-associated genes in the patient-derived xenograft models of triple-negative breast cancers. Breast Cancer Res. Treat. 2015, 154, 13–22. [Google Scholar] [CrossRef]
- Kawase, T.; Ohki, R.; Shibata, T.; Tsutsumi, S.; Kamimura, N.; Inazawa, J.; Ohta, T.; Ichikawa, H.; Aburatani, H.; Tashiro, F.; et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 2009, 136, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Lei, L.; Wang, Y.; Li, Z.H.; Fei, L.R.; Huang, W.J.; Zheng, Y.W.; Liu, C.C.; Yang, M.Q.; Wang, Z.; Zou, Z.F.; et al. PHLDA3 promotes lung adenocarcinoma cell proliferation and invasion via activation of the Wnt signaling pathway. Lab. Investig. 2021, 101, 1130–1141. [Google Scholar] [CrossRef]
- Mangone, F.R.; Valoyes, M.A.; do Nascimento, R.G.; Conceição, M.P.F.; Bastos, D.R.; Pavanelli, A.C.; Soares, I.C.; de Mello, E.S.; Nonogaki, S.; de T Osório, C.A.B.; et al. Prognostic and predictive value of Pleckstrin homology-like domain, family A family members in breast cancer. Biomark. Med. 2020, 14, 1537–1552. [Google Scholar] [CrossRef] [PubMed]
- Indarte, M.; Puentes, R.; Maruggi, M.; Ihle, N.T.; Grandjean, G.; Scott, M.; Ahmed, Z.; Meuillet, E.J.; Zang, S.; Lemos, R.; et al. An Inhibitor of the Pleckstrin Homology Domain of CNK1 Selectively Blocks the Growth of Mutant KRAS Cells and Tumors. Cancer Res. 2019, 79, 3100–3111, Erratum in Cancer Res. 2019, 79, 5457. [Google Scholar] [CrossRef]
- Nowak, E.; Bednarek, I. Aspects of the Epigenetic Regulation of EMT Related to Cancer Metastasis. Cells 2021, 10, 3435. [Google Scholar] [CrossRef]
- Peng, X.; Yang, R.; Song, J.; Wang, X.; Dong, W. Calpain2 Upregulation Regulates EMT-Mediated Pancreatic Cancer Metastasis via the Wnt/β-Catenin Signaling Pathway. Front. Med. 2022, 9, 783592. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, C.; Zhang, M.; Shi, L.; Wang, J.; Zhang, H.; Ma, P.; Li, S. Ephrin-A2 promotes prostate cancer metastasis by enhancing angiogenesis and promoting EMT. J. Cancer Res. Clin. Oncol. 2021, 147, 2013–2023. [Google Scholar] [CrossRef]
- Kong, D.; Zhou, H.; Neelakantan, D.; Hughes, C.J.; Hsu, J.Y.; Srinivasan, R.R.; Lewis, M.T.; Ford, H.L. VEGF-C mediates tumor growth and metastasis through promoting EMT-epithelial breast cancer cell crosstalk. Oncogene 2021, 40, 964–979. [Google Scholar] [CrossRef]
- Rezatabar, S.; Karimian, A.; Rameshknia, V.; Parsian, H.; Majidinia, M.; Kopi, T.A.; Bishayee, A.; Sadeghinia, A.; Yousefi, M.; Monirialamdari, M.; et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J. Cell Physiol 2019. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Ledda, F.; Paratcha, G. Negative Regulation of Receptor Tyrosine Kinase (RTK) Signaling: A Developing Field. Biomark. Insights 2007, 2, 45–58. [Google Scholar] [CrossRef]
- Jham, B.C.; Ma, T.; Hu, J.; Chaisuparat, R.; Friedman, E.R.; Pandolfi, P.P.; Schneider, A.; Sodhi, A.; Montaner, S. Amplification of the angiogenic signal through the activation of the TSC/mTOR/HIF axis by the KSHV vGPCR in Kaposi’s sarcoma. PLoS ONE 2011, 6, e19103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monti, P.; Menichini, P.; Speciale, A.; Cutrona, G.; Fais, F.; Taiana, E.; Neri, A.; Bomben, R.; Gentile, M.; Gattei, V.; et al. Heterogeneity of TP53 Mutations and P53 Protein Residual Function in Cancer: Does It Matter? Front. Oncol. 2020, 10, 593383. [Google Scholar] [CrossRef] [PubMed]
- Marbun, V.M.G.; Erlina, L.; Lalisang, T.J.M. Genomic landscape of pathogenic mutation of APC, KRAS, TP53, PIK3CA, and MLH1 in Indonesian colorectal cancer. PLoS ONE 2022, 17, e0267090. [Google Scholar] [CrossRef]
- Selvaraj, J.; Yasothkumar, D.; Vishnu Priya, V.; Raj, A.T.; Babu, S.D.; Patil, S. Development and tumorigenic potential of TP53: A therapeutic target for head and neck squamous cell carcinoma. Oral Oncol. 2022, 130, 105922. [Google Scholar] [CrossRef]
- Vokes, N.I.; Chambers, E.; Nguyen, T.; Coolidge, A.; Lydon, C.A.; Le, X.; Sholl, L.; Heymach, J.V.; Nishino, M.; Van Allen, E.M.; et al. Concurrent TP53 Mutations Facilitate Resistance Evolution in EGFR-Mutant Lung Adenocarcinoma. J. Thorac. Oncol. 2022, 17, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Ceccarelli, V.; Racanicchi, S.; Martelli, M.P.; Nocentini, G.; Fettucciari, K.; Riccardi, C.; Marconi, P.; Di Nardo, P.; Grignani, F.; Binaglia, L.; et al. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J. Biol. Chem. 2011, 286, 27092–27102. [Google Scholar] [CrossRef]
- Barcena-Varela, M.; Colyn, L.; Fernandez-Barrena, M.G. Epigenetic Mechanisms in Hepatic Stellate Cell Activation During Liver Fibrosis and Carcinogenesis. Int. J. Mol. Sci. 2019, 20, 2507. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, C.; Li, S.; Qu, Y.; Xue, P.; Ma, Z.; Zhang, X.; Bai, H.; Wang, J. ERO1L Is a Novel and Potential Biomarker in Lung Adenocarcinoma and Shapes the Immune-Suppressive Tumor Microenvironment. Front. Immunol. 2021, 12, 677169. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef]
- Bolandi, N.; Derakhshani, A.; Hemmat, N.; Baghbanzadeh, A.; Asadzadeh, Z.; Afrashteh Nour, M.; Brunetti, O.; Bernardini, R.; Silvestris, N.; Baradaran, B. The Positive and Negative Immunoregulatory Role of B7 Family: Promising Novel Targets in Gastric Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 10719. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, L.; Wei, Y.; Wei, K.; Song, T.; Du, Z.; Feng, Z. KIF22 Promotes Development of Pancreatic Cancer by Regulating the MEK/ERK/P21 Signaling Axis. Biomed. Res. Int. 2022, 2022, 6000925. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Courau, T.; Borison, J.; Ritchie, A.J.; Mayer, A.T.; Krummel, M.F.; Collisson, E.A. Activating Immune Recognition in Pancreatic Ductal Adenocarcinoma via Autophagy Inhibition, MEK Blockade, and CD40 Agonism. Gastroenterology 2022, 162, 590–603.e14, Erratum in Gastroenterology 2022, 162, 1785. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Igarashi, K.; Miyake, K.; Lwin, T.M.; Miyake, M.; Kiyuna, T.; Hwang, H.K.; Murakami, T.; Delong, J.C.; Singh, S.R.; et al. MEK inhibitor trametinib in combination with gemcitabine regresses a patient-derived orthotopic xenograft (PDOX) pancreatic cancer nude mouse model. Tissue Cell 2018, 52, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Munkácsy, G.; Győrffy, B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021, 11, 6047. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404, Erratum in Cancer Discov. 2012, 2, 960. [Google Scholar] [CrossRef] [Green Version]
- Men, C.; Chai, H.; Song, X.; Li, Y.; Du, H.; Ren, Q. Identification of DNA methylation associated gene signatures in endometrial cancer via integrated analysis of DNA methylation and gene expression systematically. J. Gynecol. Oncol. 2017, 28, e83. [Google Scholar] [CrossRef]
- Shinawi, T.; Hill, V.K.; Krex, D.; Schackert, G.; Gentle, D.; Morris, M.R.; Wei, W.; Cruickshank, G.; Maher, E.R.; Latif, F. DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics 2013, 8, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Liu, C.J.; Hu, F.F.; Xia, M.X.; Han, L.; Zhang, Q.; Guo, A.Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef] [PubMed]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]

| Description | PHLDA1 | PHLDA2 | PHLDA3 | |||
|---|---|---|---|---|---|---|
| Purity | Purity | Purity | ||||
| Cor | p | Cor | p | Cor | p | |
| Act CD8 | −0.073 | 0.334 | −0.363 | *** | −0.013 | 0.858 |
| Tcm CD8 | 0.146 | 0.0514 | 0.008 | 0.911 | 0.061 | 0.415 |
| Tem CD8 | 0.021 | 0.776 | −0.244 | ** | −0.028 | 0.715 |
| Act CD4 | 0.249 | *** | −0.042 | 0.576 | −0.087 | 0.247 |
| Tcm CD4 | 0.341 | *** | 0.209 | ** | 0.298 | *** |
| Tem CD4 | −0.106 | 0.158 | −0.546 | *** | −0.313 | *** |
| Tfh | −0.05 | 0.509 | −0.294 | *** | −0.077 | 0.305 |
| Tgd | 0.073 | 0.329 | −0.141 | 0.0589 | 0.135 | 0.0707 |
| Th1 | −0.001 | 0.993 | −0.251 | *** | −0.024 | 0.755 |
| Th17 | 0.027 | 0.722 | 0.363 | *** | −0.021 | 0.782 |
| Th2 | 0.238 | ** | −0.157 | * | −0.174 | * |
| Treg | 0.036 | 0.627 | −0.304 | *** | −0.051 | 0.495 |
| Act B | −0.218 | ** | −0.363 | *** | −0.142 | 0.0587 |
| Imm B | −0.088 | 0.242 | −0.326 | *** | −0.071 | 0.345 |
| Mem B | −0.042 | 0.578 | −0.429 | *** | −0.198 | ** |
| NK | 0.044 | 0.557 | −0.272 | *** | −0.041 | 0.587 |
| CD56bright | 0.279 | *** | 0.328 | *** | 0.405 | *** |
| CD56dim | 0.34 | *** | 0.459 | *** | 0.412 | *** |
| MDSC | 0.018 | 0.811 | −0.171 | * | 0.034 | 0.655 |
| NKT | 0.051 | 0.499 | −0.243 | ** | −0.087 | 0.248 |
| Act DC | 0.199 | ** | 0.136 | 0.0701 | 0.178 | * |
| pDC | 0.257 | *** | −0.053 | 0.481 | 0.049 | 0.51 |
| iDC | −0.13 | 0.084 | −0.192 | * | −0.194 | *** |
| Macrophage | 0.006 | 0.932 | −0.251 | *** | 0.029 | 0.696 |
| Eosinophil | −0.106 | 0.159 | −0.379 | *** | −0.253 | *** |
| Mast | 0.018 | 0.812 | −0.391 | *** | −0.119 | 0.111 |
| Monocyte | 0.074 | 0.325 | 0.163 | * | 0.007 | 0.925 |
| Neutrophil | 0.032 | 0.668 | −0.078 | 0.299 | −0.097 | 0.196 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Du, Y.; Gu, Z.; Zheng, X.; Wang, C. Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma. Int. J. Mol. Sci. 2022, 23, 10316. https://doi.org/10.3390/ijms231810316
Duan Y, Du Y, Gu Z, Zheng X, Wang C. Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma. International Journal of Molecular Sciences. 2022; 23(18):10316. https://doi.org/10.3390/ijms231810316
Chicago/Turabian StyleDuan, Yunjie, Yongxing Du, Zongting Gu, Xiaohao Zheng, and Chengfeng Wang. 2022. "Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma" International Journal of Molecular Sciences 23, no. 18: 10316. https://doi.org/10.3390/ijms231810316
APA StyleDuan, Y., Du, Y., Gu, Z., Zheng, X., & Wang, C. (2022). Prognostic Value, Immune Signature, and Molecular Mechanisms of the PHLDA Family in Pancreatic Adenocarcinoma. International Journal of Molecular Sciences, 23(18), 10316. https://doi.org/10.3390/ijms231810316




