Salt Tolerance of Rice Is Enhanced by the SS3 Gene, Which Regulates Ascorbic Acid Synthesis and ROS Scavenging
Abstract
:1. Introduction
2. Results
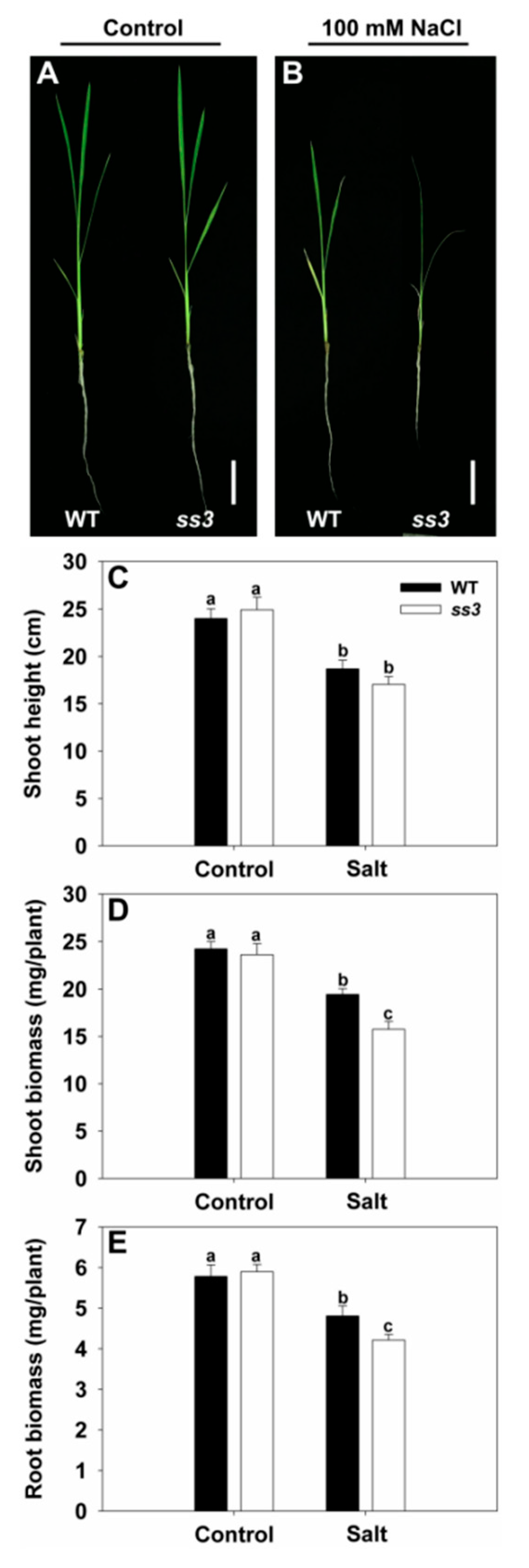
2.1. Isolation of a Rice Mutant Sensitive to Salt Stress
2.2. Map-Based Cloning of SS3
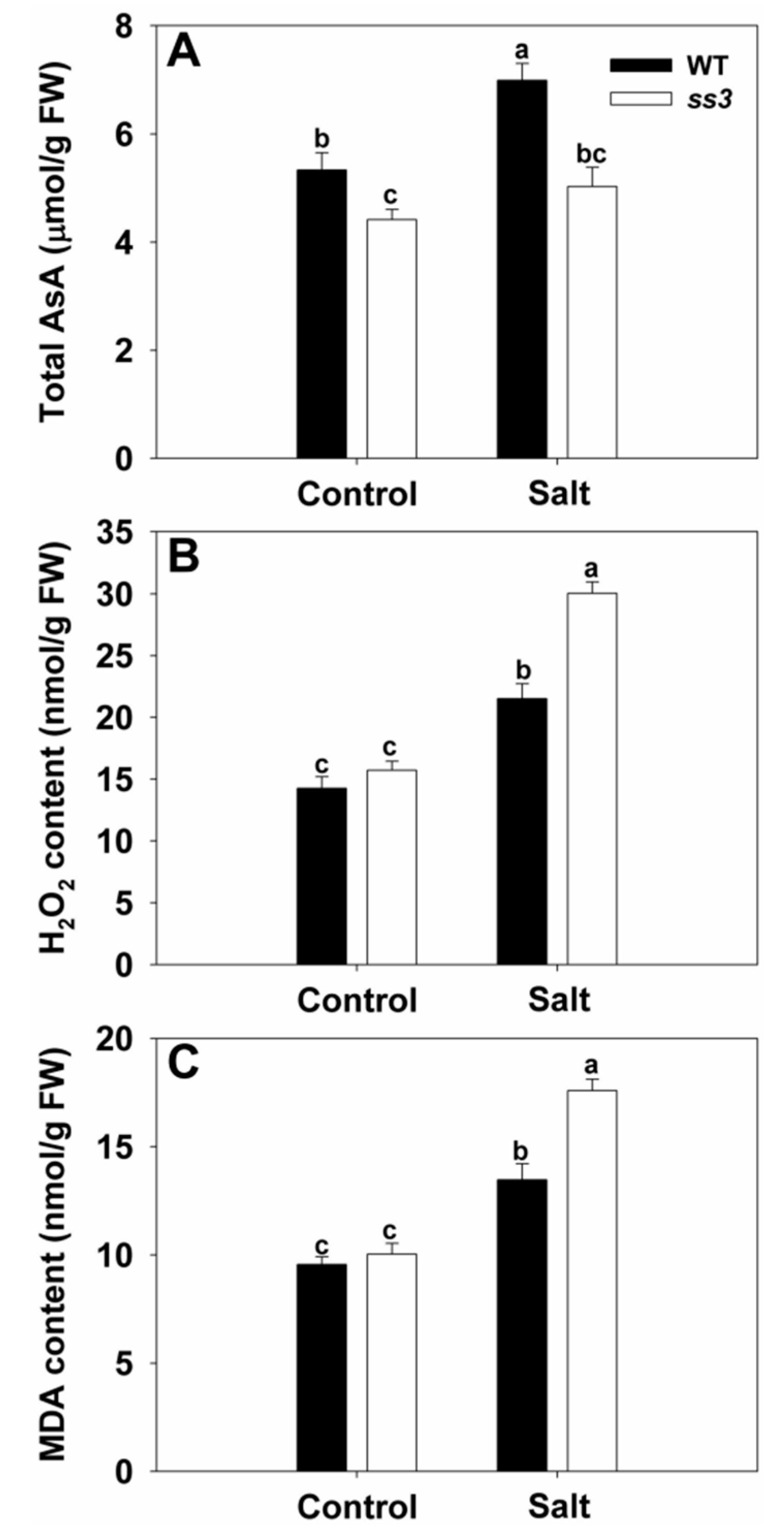
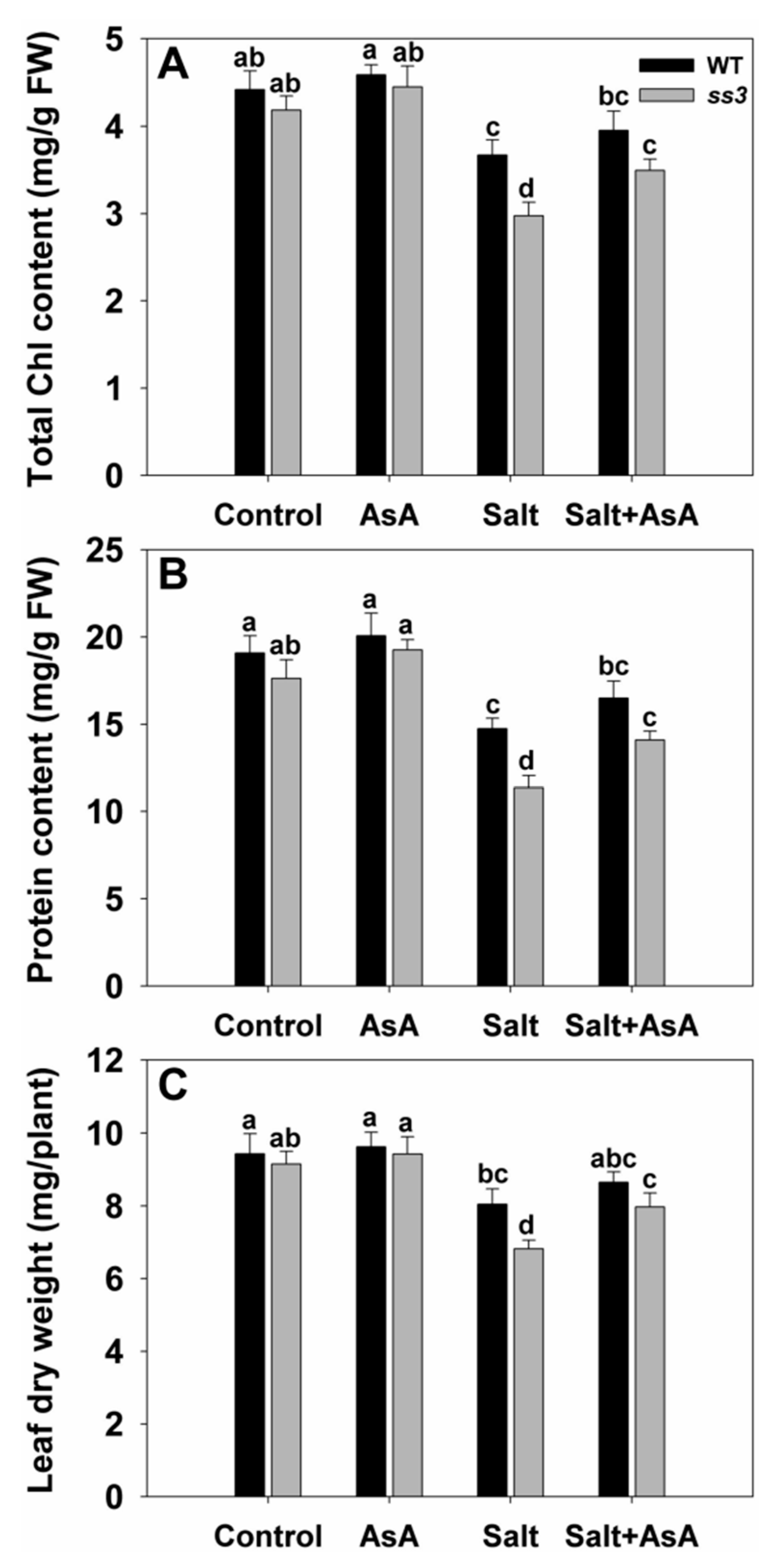
2.3. SS3 Is Involved in AsA Production and ROS Scavenging in Rice
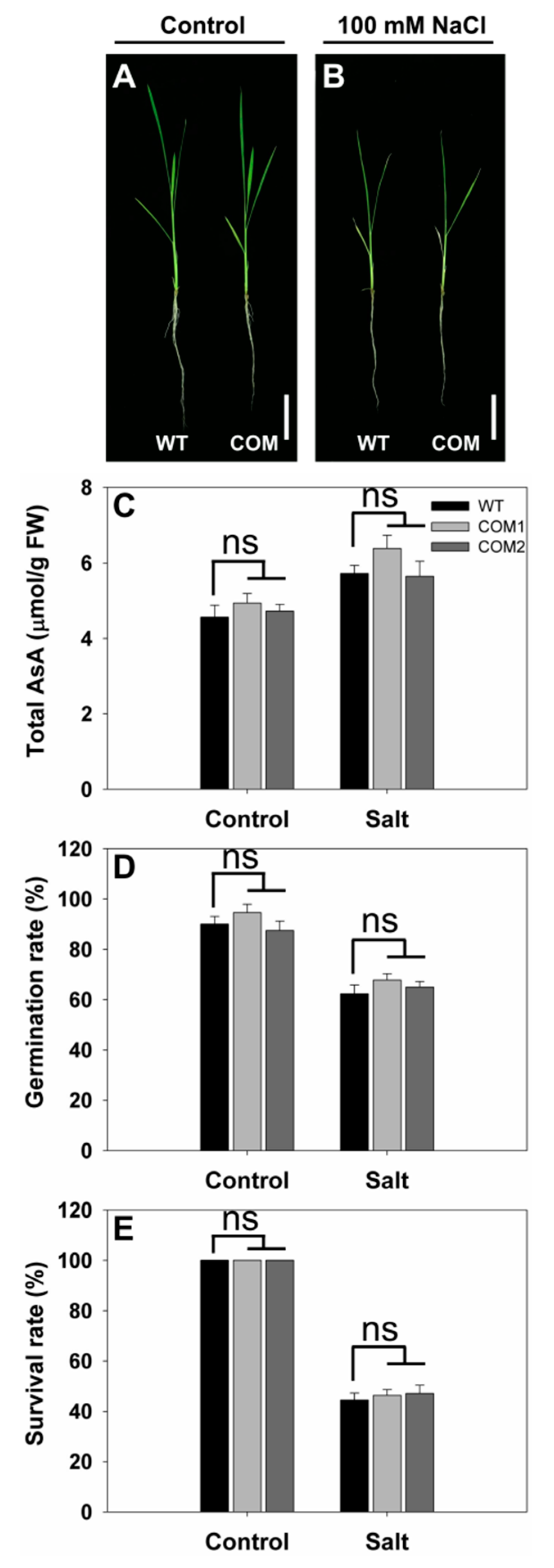
2.4. Generation of Complementation Lines for SS3
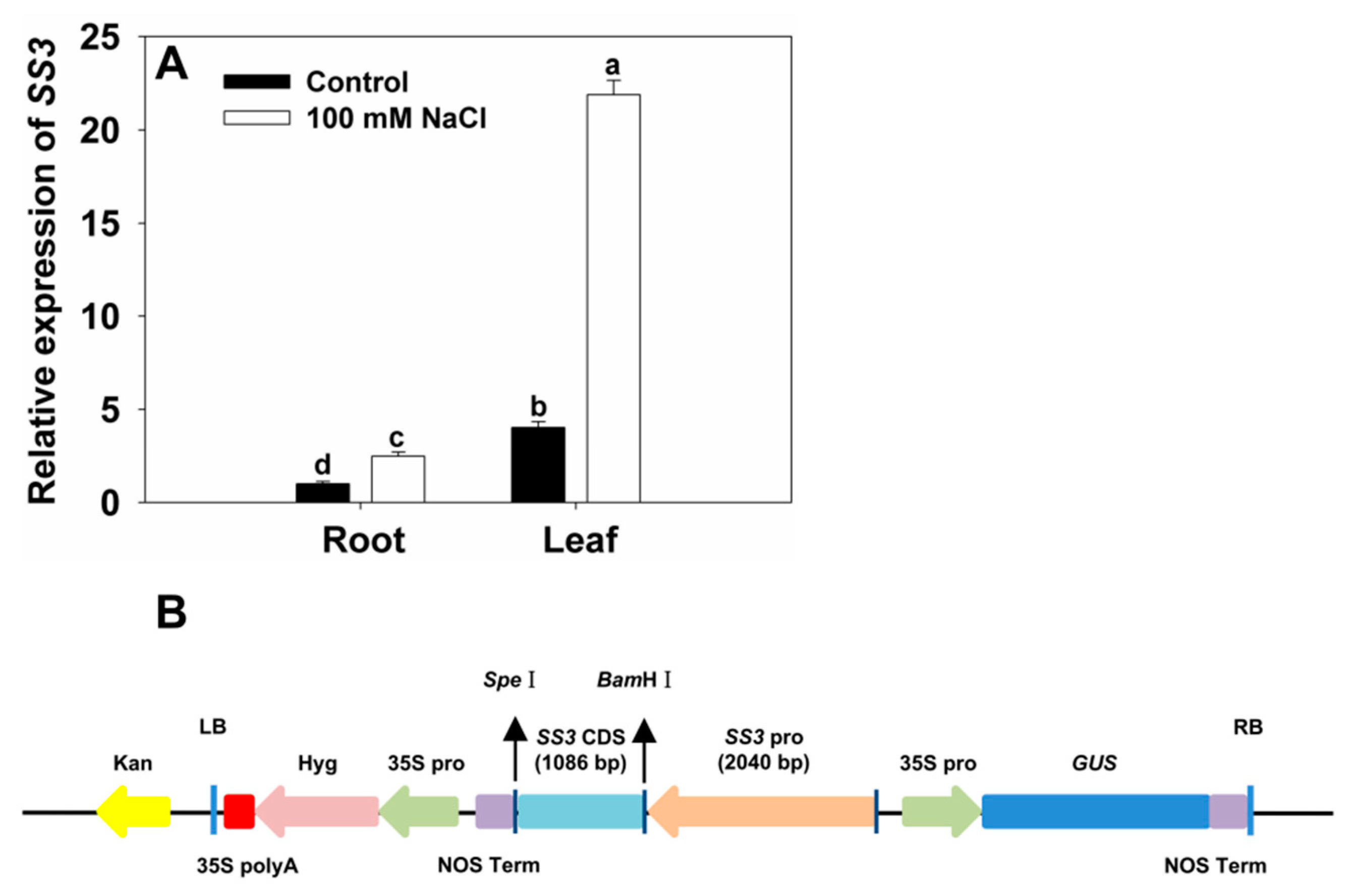
2.5. SS3 Is Induced by Salinity Stress
2.6. Generation of Transgenic Rice Expressing SS3p:SS3
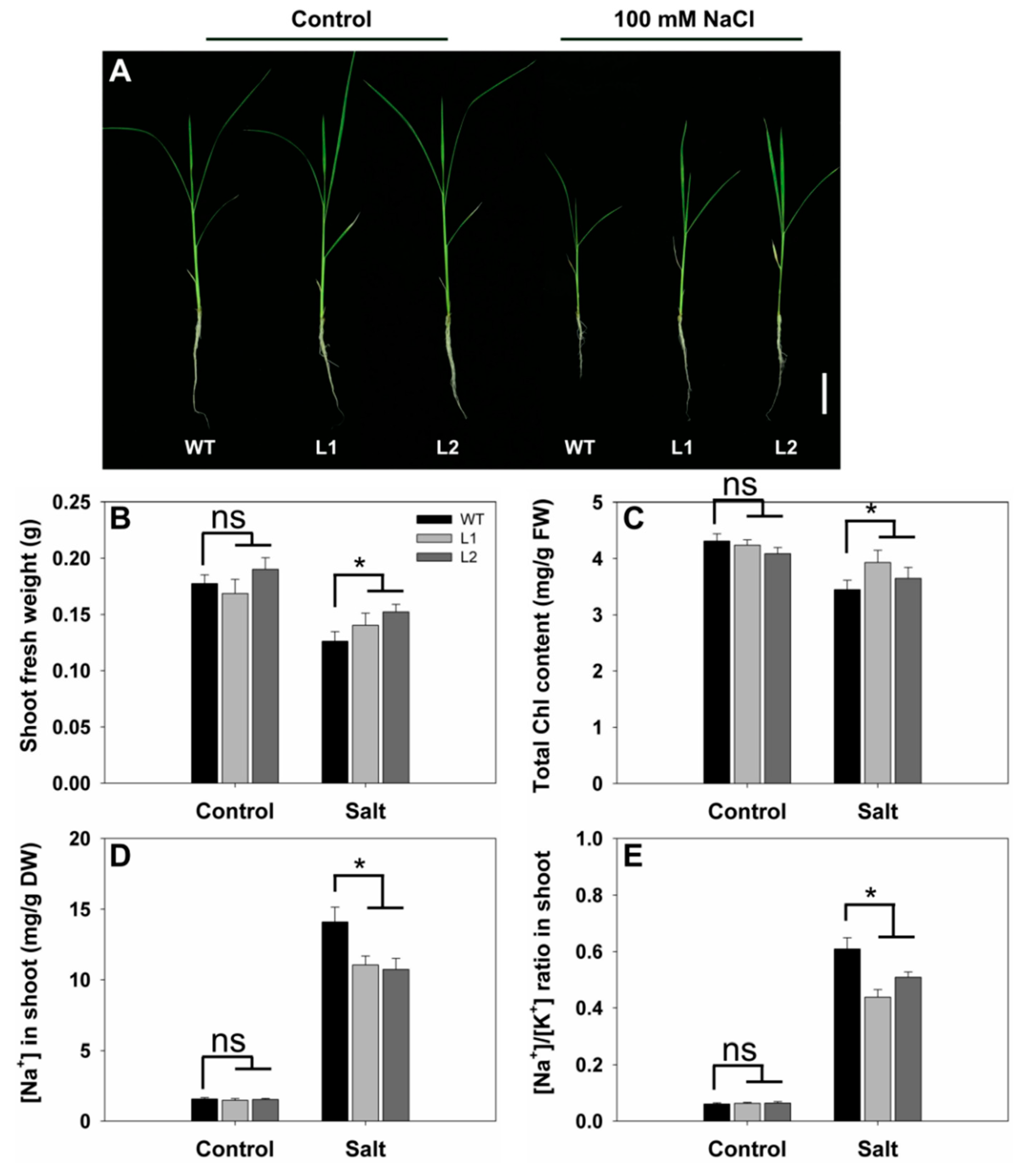
2.7. Effects of SS3p:SS3 Expression on Rice Growth and Na+/K+ Homeostasis under Salt Stress
2.8. SS3p:SS3 Transgenic Plants under Salt Stress Present Altered Expression of Senescence- and Autophagy-Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Map-Based Cloning
4.3. Generation of Transgenic Lines
4.4. Determination of AsA Content
4.5. Determination of H2O2 and MDA Content
4.6. Measurement of Chlorophyll and Soluble Protein Content
4.7. Determination of Sodium and Potassium Ions
4.8. Quantitative Real Time PCR (qRT-PCR)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules 2019, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Kaur, N.; Pati, P.K. Reactive oxygen species dynamics in roots of salt sensitive and salt tolerant cultivars of rice. Anal. Biochem. 2018, 550, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Shannon, M.C.; Lesch, S.M. Timing of salinity stress afects rice growth and yield components. Agric. Water Manag. 2001, 48, 191–206. [Google Scholar] [CrossRef]
- Chen, G.; Zheng, D.; Feng, N.; Zhou, H.; Mu, D.; Zhao, L.; Shen, X.; Rao, G.; Meng, F.; Huang, A. Physiological mechanisms of ABA-induced salinity tolerance in leaves and roots of rice. Sci. Rep. 2022, 12, 8228. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef] [PubMed]
- De Pinto, M.C.; De Gara, L. Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell diferentiation. J. Exp. Bot. 2004, 55, 2559–2569. [Google Scholar] [CrossRef] [PubMed]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-a potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Shalata, A.; Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: The root antioxidative system. Physiol. Plant 2001, 112, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.D.; Viola, R. Biosynthesis and catabolism of L-ascorbic acid in plants. Crit. Rev. Plant Sci. 2005, 24, 167–188. [Google Scholar] [CrossRef]
- Viviani, A.; Verma, B.C.; Giordani, T.; Fambrini, M. L-Ascorbic acid in plants: From biosynthesis to its role in plant development and stress response. Agrochim. Int. J. Plant Chem. Soil Sci. Plant Nutr. Univ. Pisa 2021, 65, 151–171. [Google Scholar] [CrossRef]
- Conklin, P.; Saracco, S.; Norris, S.; Last, R. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 2000, 154, 847–856. [Google Scholar] [CrossRef]
- Lamanchai, K.; Salmon, D.L.; Smirnoff, N.; Sutthinon, P.; Roytrakul, S.; Leetanasaksakul, K.; Kittisenachai, S.; Jantasuriyarat, C. OsVTC1-1 RNAi mutant with reduction of ascorbic acid synthesis alters cell wall sugar composition and cell wall-associated proteins. Agronomy 2022, 12, 1272. [Google Scholar] [CrossRef]
- Conklin, P.; Norris, S.; Wheeler, G.; Williams, E.; Smirnoff, N.; Last, R. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin c) biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4198–4203. [Google Scholar] [CrossRef]
- Qin, H.; Deng, Z.; Zhang, C.; Wang, Y.; Wang, J.; Liu, H.; Zhang, Z.; Huang, R.; Zhang, Z. Rice GDP-mannose pyrophosphorylase OsVTC1-1 and OsVTC1-3 play different roles in ascorbic acid synthesis. Plant Mol. Biol. 2016, 90, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, Y.; Wang, J.; Liu, H.; Zhao, H.; Deng, Z.; Zhang, Z.; Huang, R.; Zhang, Z. Knocking down the expression of GMPase gene OsVTC1-1 decreases salt tolerance of rice at seedling and reproductive stages. PLoS ONE 2016, 11, e0168650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, H.; Qin, H.; Li, Z.; Liu, H.; Wang, J.; Zhang, H.; Quan, R.; Huang, R.; Zhang, Z. The synthesis of ascorbic acid in rice roots plays an important role in the salt tolerance of rice by scavenging ROS. Int. J. Mol. Sci. 2018, 19, 3347. [Google Scholar] [CrossRef] [PubMed]
- Kakan, X.; Yu, Y.; Li, S.; Li, X.; Huang, R.; Wang, J. Ascorbic acid modulation by ABI4 transcriptional repression of VTC2 in the salt tolerance of Arabidopsis. BMC Plant Biol. 2021, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.S.; Yu, C.; Zhu, Z.J.; Yu, X.C. Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep. 2011, 30, 1029–1040. [Google Scholar] [CrossRef]
- Conklin, P.L.; Williams, E.H.; Last, R.L. Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc. Natl. Acad. Sci. USA 1996, 93, 9970–9974. [Google Scholar] [CrossRef]
- Huang, C.; He, W.; Guo, J.; Chang, X.; Su, P.; Zhang, L. Increased sensitivity to salt stress in an ascorbate deficient Arabidopsis mutant. J. Exp. Bot. 2005, 56, 3041–3049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. Ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Kumar, R.; Mustafiz, A.; Sahoo, K.K.; Sharma, V.; Samanta, S.; Sopory, S.K.; Pareek, A.; Singla-Pareek, S.L. Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 2012, 79, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, C.; He, L.; Qiu, Z.; Zhang, S.; Zhang, Y.; Guo, L.; Zeng, D.; Hu, J.; Ren, D.; et al. Knocking out the gene RLS1 induces hypersensitivity to oxidative stress and premature leaf senescence in rice. Int. J. Mol. Sci. 2018, 19, 2853. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Song, W.; Wang, F.; Chen, L.; Ma, R.; Zuo, X.; Cao, A.; Xie, S.; Chen, X.; Jin, X.; Li, H. GhVTC1, the key gene for ascorbate biosynthesis in Gossypium hirsutum, involves in cell elongation under control of ethylene. Cells 2019, 8, 1039. [Google Scholar] [CrossRef]
- Wang, J.; Yu, Y.; Zhang, Z.; Quan, R.; Zhang, H.; Ma, L.; Deng, X.; Huang, R. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 2013, 25, 625–636. [Google Scholar] [CrossRef]
- Li, Y.; Chu, Z.; Luo, J.; Zhou, Y.; Cai, Y.; Lu, Y.; Xia, J.; Kuang, H.; Ye, Z.; Ouyang, B. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnol. J. 2018, 16, 1201–1213. [Google Scholar] [CrossRef]
- Inzé, D.; Van Montague, M. Oxidative stress in plants. Curr. Opin. Biotechnol. 1995, 6, 153–158. [Google Scholar] [CrossRef]
- Chen, L.; Wuriyanghan, H.; Zhang, Y.; Duan, K.; Chen, H.; Li, Q.; Lu, X.; He, S.; Ma, B.; Zhang, W. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, Y.; Hong, X.; Hu, D.; Liu, C.; Yang, J.; Li, Y.; Huang, Y.; Feng, Y.; Gong, H. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 2015, 66, 973–987. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, Y.; Sinumporn, S.; Yu, N.; Zhan, X.; Shen, X.; Chen, D.; Yu, P.; Wu, W.; Liu, Q.; et al. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018, 95, 877–891. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, T.; Peng, H.; Luo, S.; Tan, J.; Jiang, K.; Heng, Y.; Zhang, X.; Guo, X.; Zheng, J.; et al. Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), is involved in leaf senescence. Front. Plant Sci. 2018, 9, 560. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yu, Q.; Wang, Z.; Pan, Y.; Lv, W.; Zhu, L.; Chen, R.; He, G. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ. 2013, 36, 1476–1489. [Google Scholar] [CrossRef] [PubMed]
- Akhter, D.; Qin, R.; Nath, U.K.; Alamin, M.; Jin, X.; Shi, C. The brown midrib leaf (bml) mutation in rice (Oryza sativa L.) causes premature leaf senescence and the induction of defense responses. Genes 2018, 9, 203. [Google Scholar] [CrossRef]
- Wang, M.; Guo, W.; Li, J.; Pan, X.; Pan, L.; Zhao, J.; Zhang, Y.; Cai, S.; Huang, X.; Wang, A.; et al. The miR528-AO module confers enhanced salt tolerance in rice by modulating the ascorbic acid and abscisic acid metabolism and ROS scavenging. J. Agric. Food Chem. 2021, 69, 8634–8648. [Google Scholar] [CrossRef]
- Chen, G.; Feng, H.; Hu, Q.; Qu, H.; Chen, A.; Yu, L.; Xu, G. Improving rice tolerance to potassium deficiency by enhancing OsHAK16p:WOX11-controlled root development. Plant Biotechnol. J. 2015, 13, 833–848. [Google Scholar] [CrossRef]
- Wu, J.; Yu, C.; Huang, L.; Gan, Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021, 173, 1120–1135. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, J.; Huang, L.; Leng, Y.; Dai, L.; Rao, Y.; Chen, L.; Wang, Y.; Tu, Z.; Hu, J.; et al. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 2016, 67, 1297–1310. [Google Scholar] [CrossRef]
- Chen, G.; Hu, J.; Lian, J.; Zhang, Y.; Zhu, L.; Zeng, D.; Guo, L.; Yu, L.; Xu, G.; Qian, Q. Functional characterization of OsHAK1 promoter in response to osmotic/drought stress by deletion analysis in transgenic rice. Plant Growth Regul. 2019, 88, 241–251. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Zhang, A.; Zhu, L.; Hu, J.; Ren, D.; Yu, L.; Xu, G.; et al. Variation in the abundance of OsHAK1 transcript underlies the differential salinity tolerance of an indica and a japonica rice cultivar. Front. Plant Sci. 2018, 8, 2216. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Jiang, H.; Zhu, L.; Ren, D.; Yu, L.; Xu, G.; Qian, Q. OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 2017, 8, 1885. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Zhu, L.; Hu, J.; Ren, D.; Xu, G.; Qian, Q. Driving the expression of RAA1 with a drought-responsive promoter enhances root growth in rice, its accumulation of potassium and its tolerance to moisture stress. Environ. Exp. Bot. 2018, 147, 147–156. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, Y.; Ruan, B.; Guo, L.; Zeng, D.; Gao, Z.; Zhu, L.; Hu, J.; Ren, D.; Yu, L.; et al. OsHAK1 controls the vegetative growth and panicle fertility of rice by its effect on potassium-mediated sugar metabolism. Plant Sci. 2018, 274, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, Q.; Luo, L.; Yang, T.; Zhang, S.; Hu, Y.; Yu, L.; Xu, G. Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 2015, 38, 2747–2765. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Han, H.; Yang, X.; Du, R.; Wang, X. Salt Tolerance of Rice Is Enhanced by the SS3 Gene, Which Regulates Ascorbic Acid Synthesis and ROS Scavenging. Int. J. Mol. Sci. 2022, 23, 10338. https://doi.org/10.3390/ijms231810338
Chen G, Han H, Yang X, Du R, Wang X. Salt Tolerance of Rice Is Enhanced by the SS3 Gene, Which Regulates Ascorbic Acid Synthesis and ROS Scavenging. International Journal of Molecular Sciences. 2022; 23(18):10338. https://doi.org/10.3390/ijms231810338
Chicago/Turabian StyleChen, Guang, Huimin Han, Xiuli Yang, Ruiying Du, and Xu Wang. 2022. "Salt Tolerance of Rice Is Enhanced by the SS3 Gene, Which Regulates Ascorbic Acid Synthesis and ROS Scavenging" International Journal of Molecular Sciences 23, no. 18: 10338. https://doi.org/10.3390/ijms231810338
APA StyleChen, G., Han, H., Yang, X., Du, R., & Wang, X. (2022). Salt Tolerance of Rice Is Enhanced by the SS3 Gene, Which Regulates Ascorbic Acid Synthesis and ROS Scavenging. International Journal of Molecular Sciences, 23(18), 10338. https://doi.org/10.3390/ijms231810338






