Colitis Induces Sex-Specific Intestinal Transcriptomic Responses in Mice
Abstract
:1. Introduction
2. Results
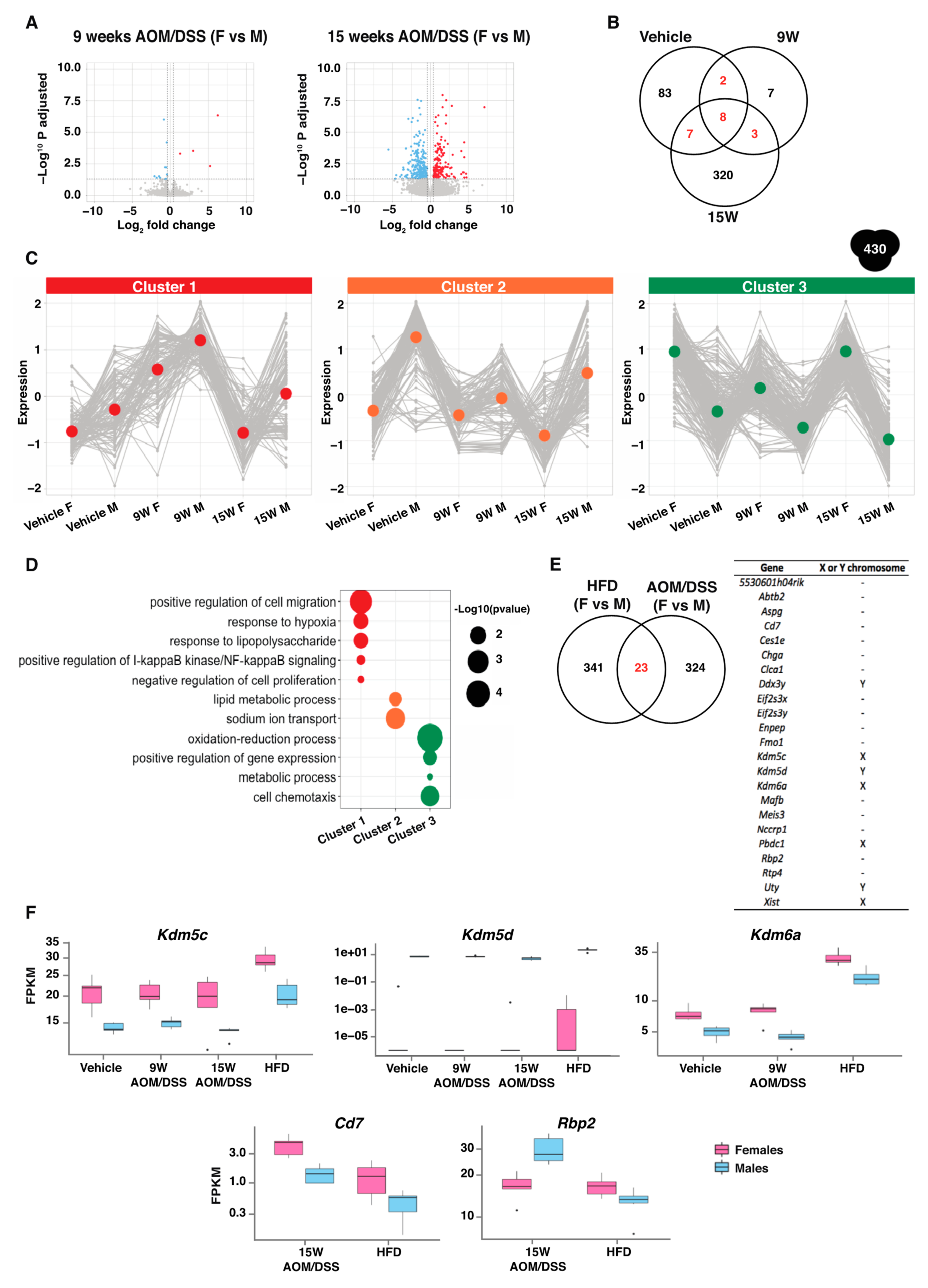
2.1. Sex Differences Are Apparent in the Colon Transcriptome
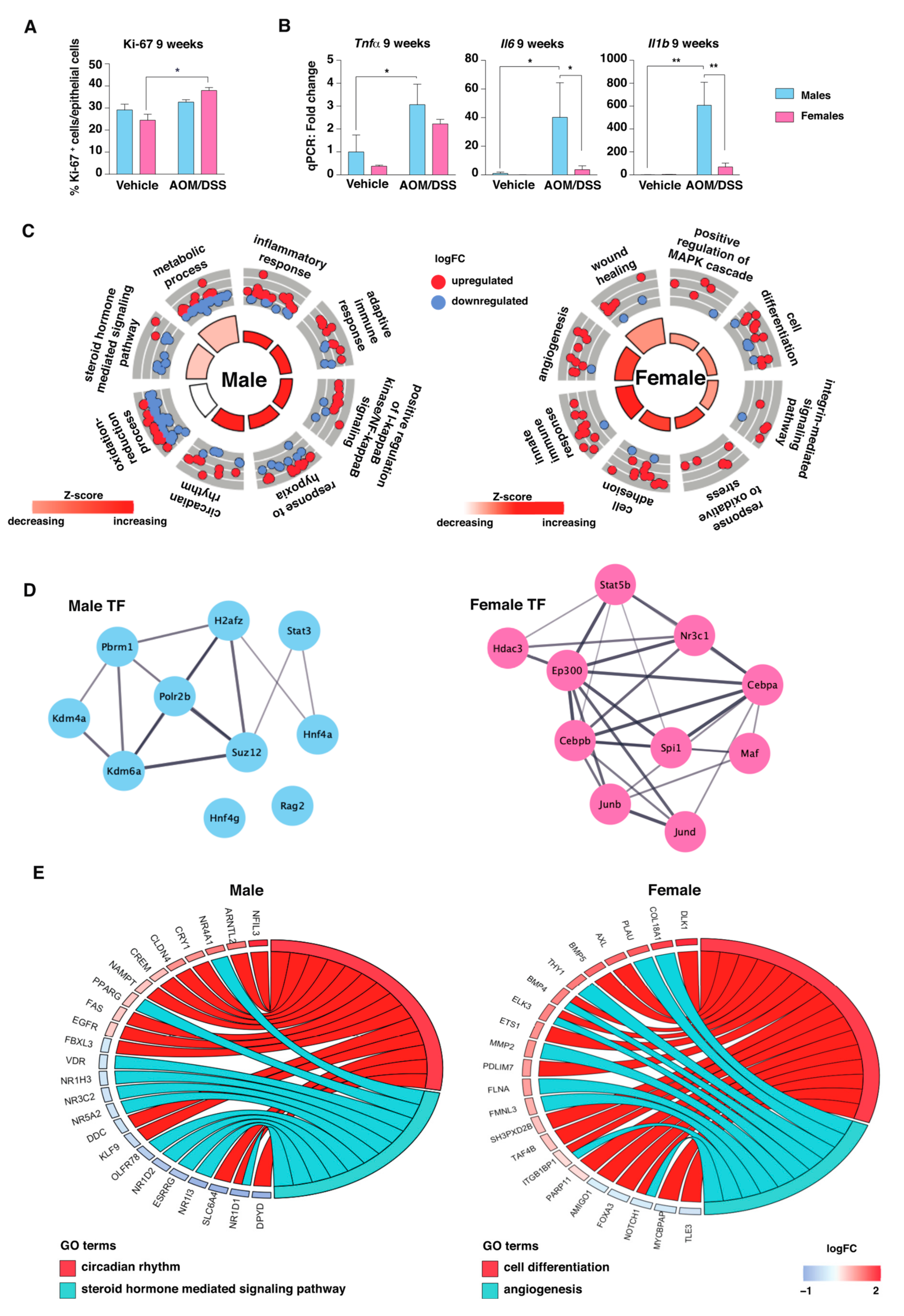
2.2. AOM/DSS Treatment Induces an Immune and Inflammatory Response in Both Sexes
2.3. The Transcriptomic Response to Colitis Differs between Males and Females
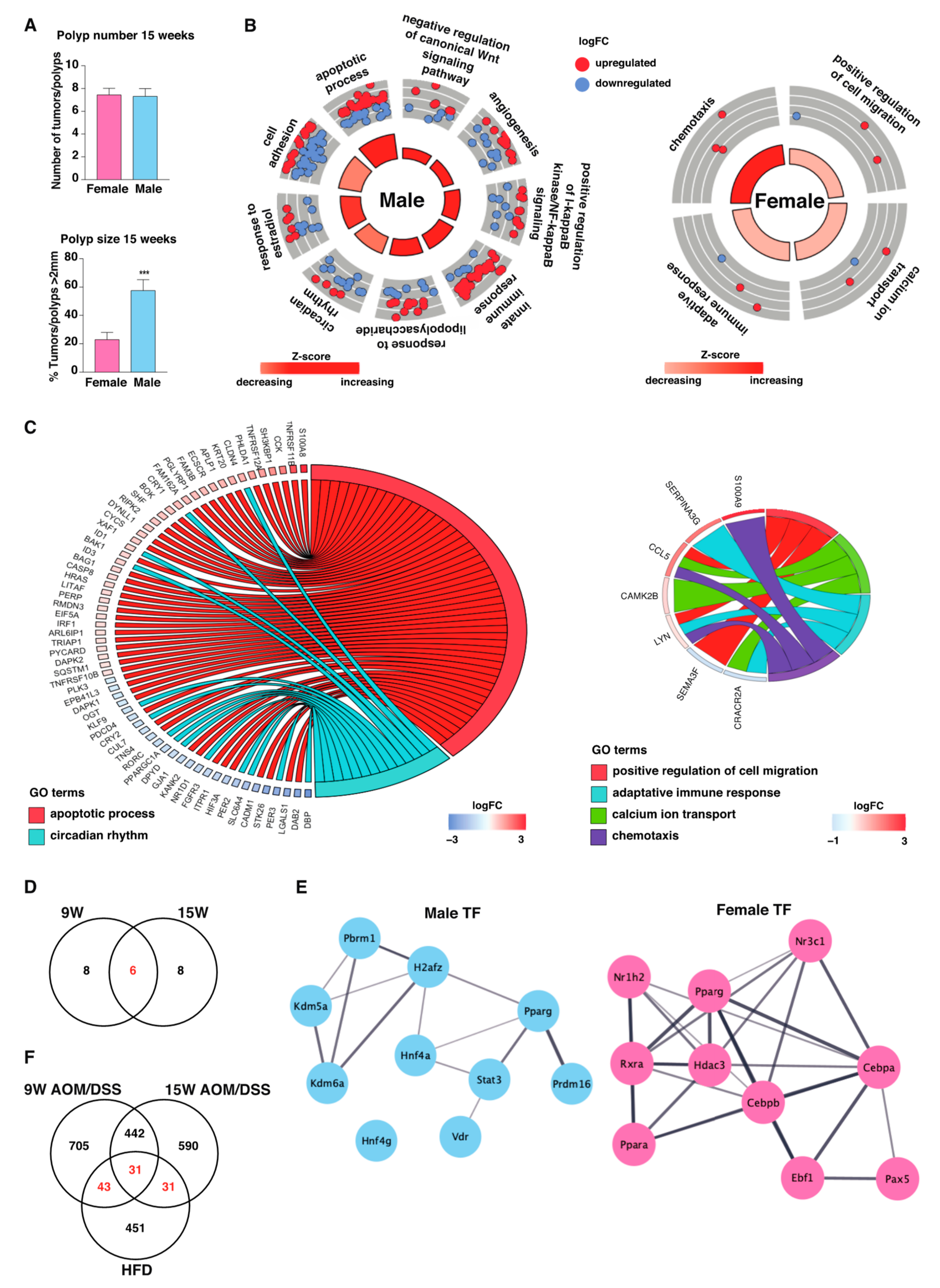
2.4. Males Exhibit Stronger Long-Term Inflammatory Response during Tumor Development
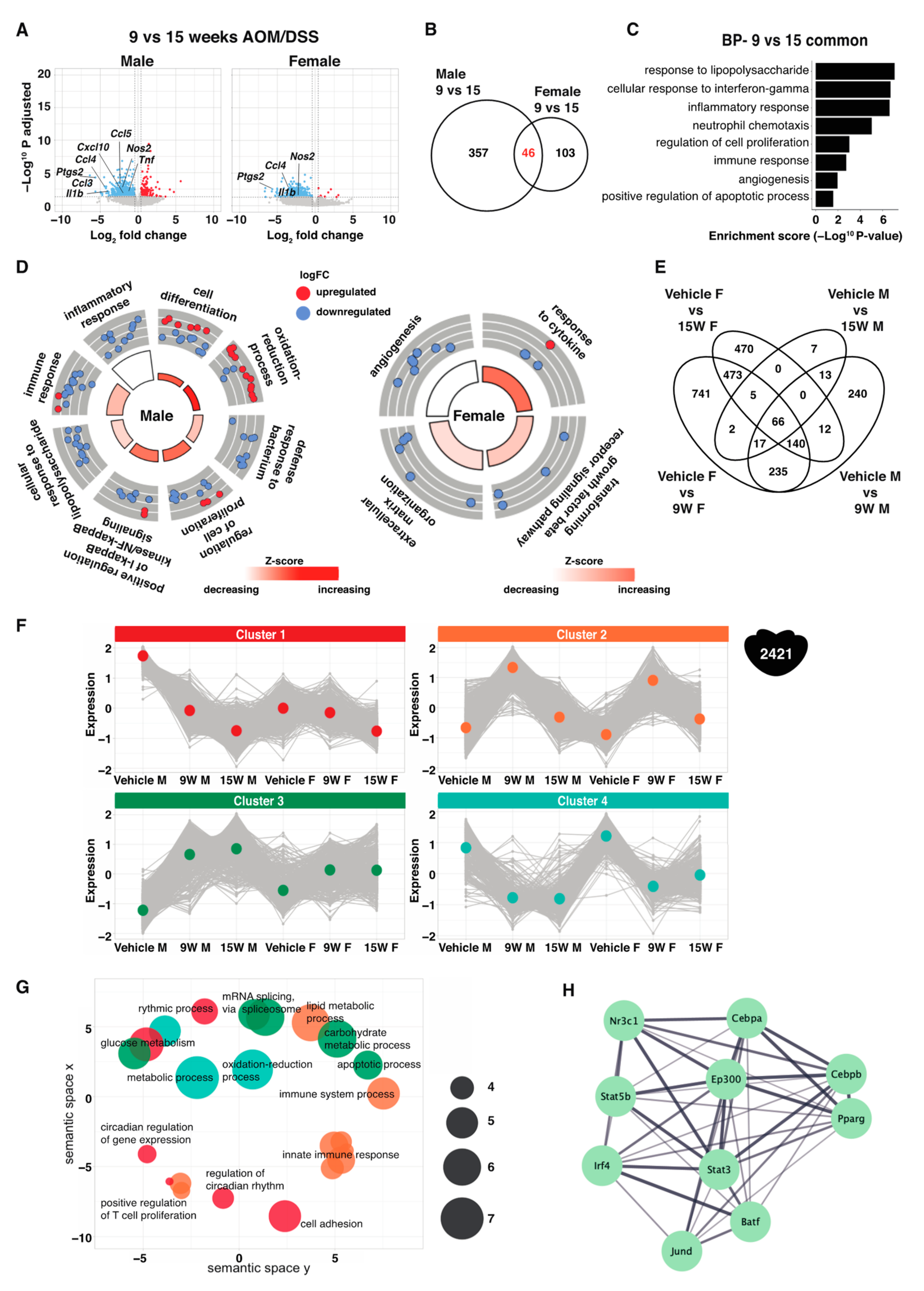
2.5. Consistent Sex Differences Were Most Apparent at Chronic State of Inflammation
2.6. Sex Differences over the Course of Acute-to-Chronic Inflammation
3. Discussion
4. Materials and Methods
4.1. Animal Experiment
4.2. RNA Isolation
4.3. RNA-Seq and Bioinformatic Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Terzic, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Shivaji, U.N.; Kasturi, R.; Sigamani, A.; Ghosh, S.; Iacucci, M. Inflammatory bowel disease-related colorectal cancer: Past, present and future perspectives. World J. Gastrointest. Oncol. 2022, 14, 547–567. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, S.; Granath, F.; Brostrom, O.; Karlen, P.; Lofberg, R.; Ekbom, A.; Askling, J. Inflammatory bowel disease confers a lower risk of colorectal cancer to females than to males. Gastroenterology 2010, 138, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Kim, S.E.; Paik, H.Y.; Yoon, H.; Lee, J.E.; Kim, N.; Sung, M.K. Sex- and gender-specific disparities in colorectal cancer risk. World J. Gastroenterol. 2015, 21, 5167–5175. [Google Scholar] [CrossRef]
- Zheng, D.; Williams, C.; Vold, J.A.; Nguyen, J.H.; Harnois, D.M.; Bagaria, S.P.; McLaughlin, S.A.; Li, Z. Regulation of sex hormone receptors in sexual dimorphism of human cancers. Cancer Lett. 2018, 438, 24–31. [Google Scholar] [CrossRef]
- Brozek, W.; Kriwanek, S.; Bonner, E.; Peterlik, M.; Cross, H.S. Mutual associations between malignancy, age, gender, and subsite incidence of colorectal cancer. Anticancer. Res. 2009, 29, 3721–3726. [Google Scholar]
- Tsai, Y.J.; Huang, S.C.; Lin, H.H.; Lin, C.C.; Lan, Y.T.; Wang, H.S.; Yang, S.H.; Jiang, J.K.; Chen, W.S.; Lin, T.C.; et al. Differences in gene mutations according to gender among patients with colorectal cancer. World J. Surg. Oncol. 2018, 16, 128. [Google Scholar] [CrossRef]
- Hases, L.; Ibrahim, A.; Chen, X.; Liu, Y.; Hartman, J.; Williams, C. The Importance of Sex in the Discovery of Colorectal Cancer Prognostic Biomarkers. Int. J. Mol. Sci. 2021, 22, 1354. [Google Scholar] [CrossRef]
- Hases, L.; Archer, A.; Indukuri, R.; Birgersson, M.; Savva, C.; Korach-André, M.; Williams, C. High-fat diet and estrogen impacts the colon and its transcriptome in a sex-dependent manner. Sci. Rep. 2020, 10, 16160. [Google Scholar] [CrossRef]
- Shah, S.C.; Itzkowitz, S.H. Colorectal Cancer in Inflammatory Bowel Disease: Mechanisms and Management. Gastroenterology 2022, 162, 715–730.e3. [Google Scholar] [CrossRef]
- Hases, L.; Indukuri, R.; Birgersson, M.; Nguyen-Vu, T.; Lozano, R.; Saxena, A.; Hartman, J.; Frasor, J.; Gustafsson, J.; Katajisto, P.; et al. Intestinal estrogen receptor beta suppresses colon inflammation and tumorigenesis in both sexes. Cancer Lett. 2020, 492, 54–62. [Google Scholar] [CrossRef]
- Mank, J.E.; Rideout, E.J. Developmental mechanisms of sex differences: From cells to organisms. Development 2021, 148, dev199750. [Google Scholar] [CrossRef] [PubMed]
- Hudry, B.; Khadayate, S.; Miguel-Aliaga, I. The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature 2016, 530, 344–348. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, N.; Son, H.J.; Park, J.H.; Nam, R.H.; Ham, M.H.; Choi, D.; Sohn, S.H.; Shin, E.; Hwang, Y.J.; et al. The Effect of Sex on the Azoxymethane/Dextran Sulfate Sodium-treated Mice Model of Colon Cancer. J. Cancer Prev. 2016, 21, 271–278. [Google Scholar] [CrossRef]
- Tang, A.; Li, N.; Li, X.; Yang, H.; Wang, W.; Zhang, L.; Li, G.; Xiong, W.; Ma, J.; Shen, S. Dynamic activation of the key pathways: Linking colitis to colorectal cancer in a mouse model. Carcinogenesis 2012, 33, 1375–1383. [Google Scholar] [CrossRef]
- Williams, C.; DiLeo, A.; Niv, Y.; Gustafsson, J. Estrogen receptor beta as target for colorectal cancer prevention. Cancer Lett. 2016, 372, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Voigt, R.M.; Forsyth, C.B.; Keshavarzian, A. Circadian rhythms: A regulator of gastrointestinal health and dysfunction. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lin, Y.; Yuan, X.; Li, F.; Guo, L.; Wu, B. REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat. Commun. 2018, 9, 4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weintraub, Y.; Cohen, S.; Chapnik, N.; Ben-Tov, A.; Yerushalmy-Feler, A.; Dotan, I.; Tauman, R.; Froy, O. Clock Gene Disruption Is an Initial Manifestation of Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2020, 18, 115–122.e1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Garrett, S.; Carroll, R.E.; Xia, Y.; Sun, J. Vitamin D receptor upregulates tight junction protein claudin-5 against colitis-associated tumorigenesis. Mucosal Immunol. 2022, 15, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Landskron, G.; Dubois-Camacho, K.; Orellana-Serradell, O.; De la Fuente, M.; Parada-Venegas, D.; Bitrán, M.; Diaz-Jimenez, D.; Tang, S.; Cidlowski, J.A.; Li, X.; et al. Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis. Cells 2022, 11, 1905. [Google Scholar] [CrossRef]
- Li, X.; Yang, A.; Wen, P.; Yuan, Y.; Xiao, Z.; Shi, H.; Wang, R. Nuclear receptor subfamily 3 group c member 2 (NR3C2) is downregulated due to hypermethylation and plays a tumor-suppressive role in colon cancer. Mol. Cell Biochem. 2022; online ahead of print. [Google Scholar] [CrossRef]
- Guo, X.; Yue, L.; Li, M.; Dai, A.; Sun, J.; Fang, L.; Zhao, H.; Sun, Q. Nuclear receptor estrogen-related receptor gamma suppresses colorectal cancer aggressiveness by regulating Wnt/β-catenin signalling. Carcinogenesis, 2022; online ahead of print. [Google Scholar] [CrossRef]
- Lo Sasso, G.; Bovenga, F.; Murzilli, S.; Salvatore, L.; Di Tullio, G.; Martelli, N.; D’Orazio, A.; Rainaldi, S.; Vacca, M.; Mangia, A.; et al. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology 2013, 144, 1497–1507.e13. [Google Scholar] [CrossRef]
- Jakobsson, T.; Vedin, L.L.; Hassan, T.; Venteclef, N.; Greco, D.; D’Amato, M.; Treuter, E.; Gustafsson, J.; Steffensen, K.R. The oxysterol receptor LXRβ protects against DSS- and TNBS-induced colitis in mice. Mucosal Immunol. 2014, 7, 1416–1428. [Google Scholar] [CrossRef]
- Kurnaz-Gomleksiz, O.; Torun, B.C.; Isbir, T.; Bulut, T.; Sokucu, N.; Yilmaz-Aydogan, H.; Canbay, E. The Role of PPAR-gamma C161T Polymorphism in Colorectal Cancer Susceptibility. In Vivo 2022, 36, 1911–1915. [Google Scholar] [CrossRef]
- Niu, B.; Liu, J.; Lv, B.; Lin, J.; Li, X.; Wu, C.; Jiang, X.; Zeng, Z.; Zhang, X.K.; Zhou, H. Interplay between transforming growth factor-β and Nur77 in dual regulations of inhibitor of differentiation 1 for colonic tumorigenesis. Nat. Commun. 2021, 12, 2809. [Google Scholar] [CrossRef]
- Crnčec, I.; Modak, M.; Gordziel, C.; Svinka, J.; Scharf, I.; Moritsch, S.; Pathria, P.; Schlederer, M.; Kenner, L.; Timelthaler, G.; et al. STAT1 is a sex-specific tumor suppressor in colitis-associated colorectal cancer. Mol. Oncol. 2018, 12, 514–528. [Google Scholar] [CrossRef]
- Avalle, L.; Pensa, S.; Regis, G.; Novelli, F.; Poli, V. STAT1 and STAT3 in tumorigenesis: A matter of balance. Jakstat 2012, 1, 65–72. [Google Scholar] [CrossRef]
- Meers, G.K.; Bohnenberger, H.; Reichardt, H.M.; Lühder, F.; Reichardt, S.D. Impaired resolution of DSS-induced colitis in mice lacking the glucocorticoid receptor in myeloid cells. PLoS ONE 2018, 13, e0190846. [Google Scholar] [CrossRef]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef]
- Merten, M.; Greiner, J.F.W.; Niemann, T.; Grosse Venhaus, M.; Kronenberg, D.; Stange, R.; Wähnert, D.; Kaltschmidt, C.; Vordemvenne, T.; Kaltschmidt, B. Human Sex Matters: Y-Linked Lysine Demethylase 5D Drives Accelerated Male Craniofacial Osteogenic Differentiation. Cells 2022, 11, 823. [Google Scholar] [CrossRef]
- Jambhekar, A.; Anastas, J.N.; Shi, Y. Histone Lysine Demethylase Inhibitors. Cold Spring Harb. Perspect. Med. 2017, 7, a026484. [Google Scholar] [CrossRef]
- Lee, S.A.; Yang, K.J.Z.; Brun, P.J.; Silvaroli, J.A.; Yuen, J.J.; Shmarakov, I.; Jiang, H.; Feranil, J.B.; Li, X.; Lackey, A.I.; et al. Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight. Sci. Adv. 2020, 6, eaay8937. [Google Scholar] [CrossRef]
- Zeng, J.; Ge, Z.; Wang, L.; Li, Q.; Wang, N.; Björkholm, M.; Jia, J.; Xu, D. The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology 2010, 138, 981–992. [Google Scholar] [CrossRef]
- Ahnstedt, H.; Patrizz, A.; Chauhan, A.; Roy-O’Reilly, M.; Furr, J.W.; Spychala, M.S.; D’Aigle, J.; Blixt, F.W.; Zhu, L.; Bravo Alegria, J.; et al. Sex differences in T cell immune responses, gut permeability and outcome after ischemic stroke in aged mice. Brain Behav. Immun. 2020, 87, 556–567. [Google Scholar] [CrossRef]
- Lee, B.W.; Yap, H.K.; Chew, F.T.; Quah, T.C.; Prabhakaran, K.; Chan, G.S.; Wong, S.C.; Seah, C.C. Age- and sex-related changes in lymphocyte subpopulations of healthy Asian subjects: From birth to adulthood. Cytometry 1996, 26, 8–15. [Google Scholar] [CrossRef]
- Lisse, I.M.; Aaby, P.; Whittle, H.; Jensen, H.; Engelmann, M.; Christensen, L.B. T-lymphocyte subsets in West African children: Impact of age, sex, and season. J. Pediatr. 1997, 130, 77–85. [Google Scholar] [CrossRef]
- Uppal, S.S.; Verma, S.; Dhot, P.S. Normal values of CD4 and CD8 lymphocyte subsets in healthy indian adults and the effects of sex, age, ethnicity, and smoking. Cytometry B Clin. Cytom. 2003, 52, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Rathod, K.S.; Kapil, V.; Velmurugan, S.; Khambata, R.S.; Siddique, U.; Khan, S.; Van Eijl, S.; Gee, L.C.; Bansal, J.; Pitrola, K.; et al. Accelerated resolution of inflammation underlies sex differences in inflammatory responses in humans. J. Clin. Investig. 2017, 127, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hases, L.; Birgersson, M.; Indukuri, R.; Archer, A.; Williams, C. Colitis Induces Sex-Specific Intestinal Transcriptomic Responses in Mice. Int. J. Mol. Sci. 2022, 23, 10408. https://doi.org/10.3390/ijms231810408
Hases L, Birgersson M, Indukuri R, Archer A, Williams C. Colitis Induces Sex-Specific Intestinal Transcriptomic Responses in Mice. International Journal of Molecular Sciences. 2022; 23(18):10408. https://doi.org/10.3390/ijms231810408
Chicago/Turabian StyleHases, Linnea, Madeleine Birgersson, Rajitha Indukuri, Amena Archer, and Cecilia Williams. 2022. "Colitis Induces Sex-Specific Intestinal Transcriptomic Responses in Mice" International Journal of Molecular Sciences 23, no. 18: 10408. https://doi.org/10.3390/ijms231810408





