p27kip1 Modulates the Morphology and Phagocytic Activity of Microglia
Abstract
1. Introduction
2. Results
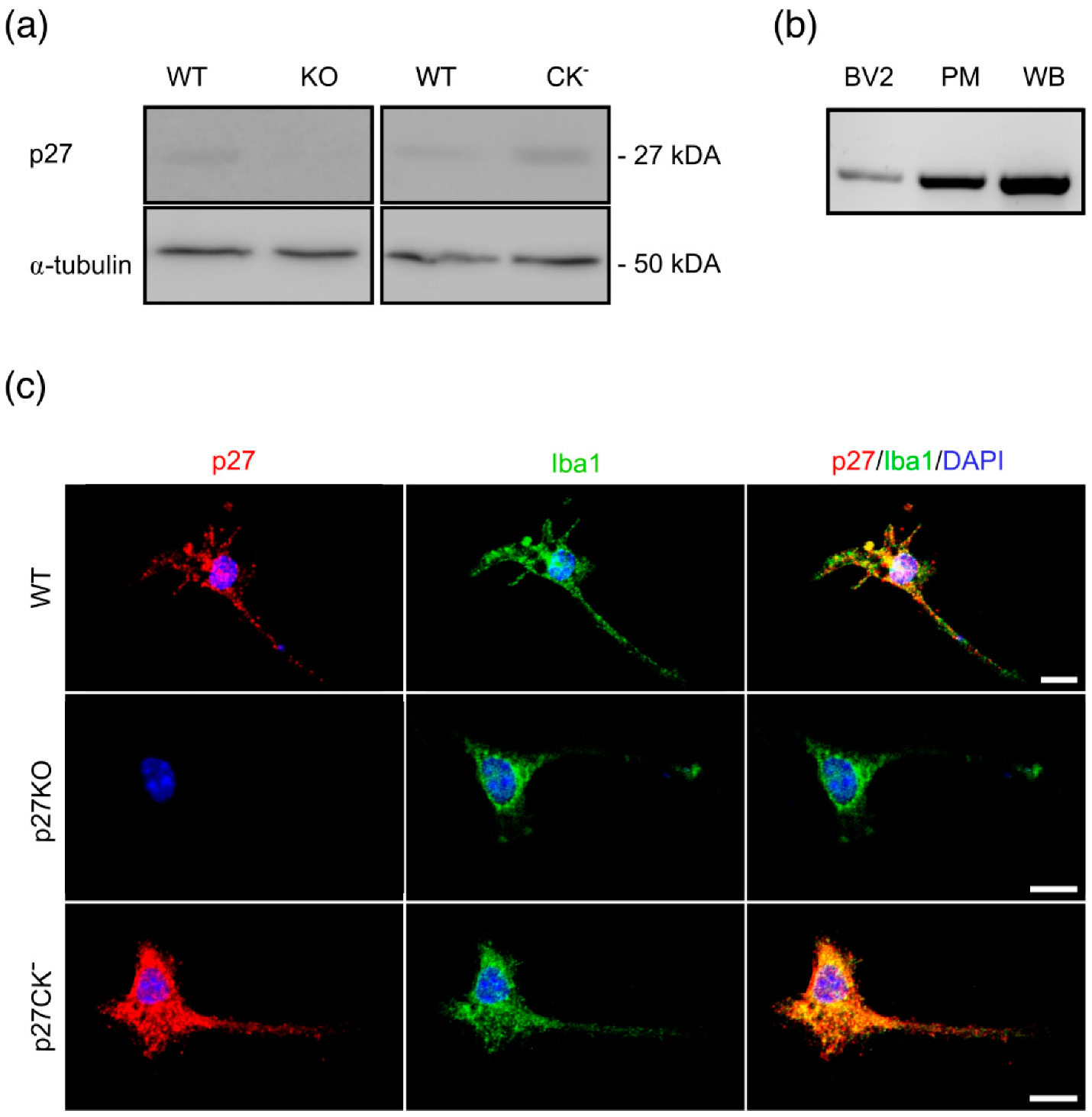
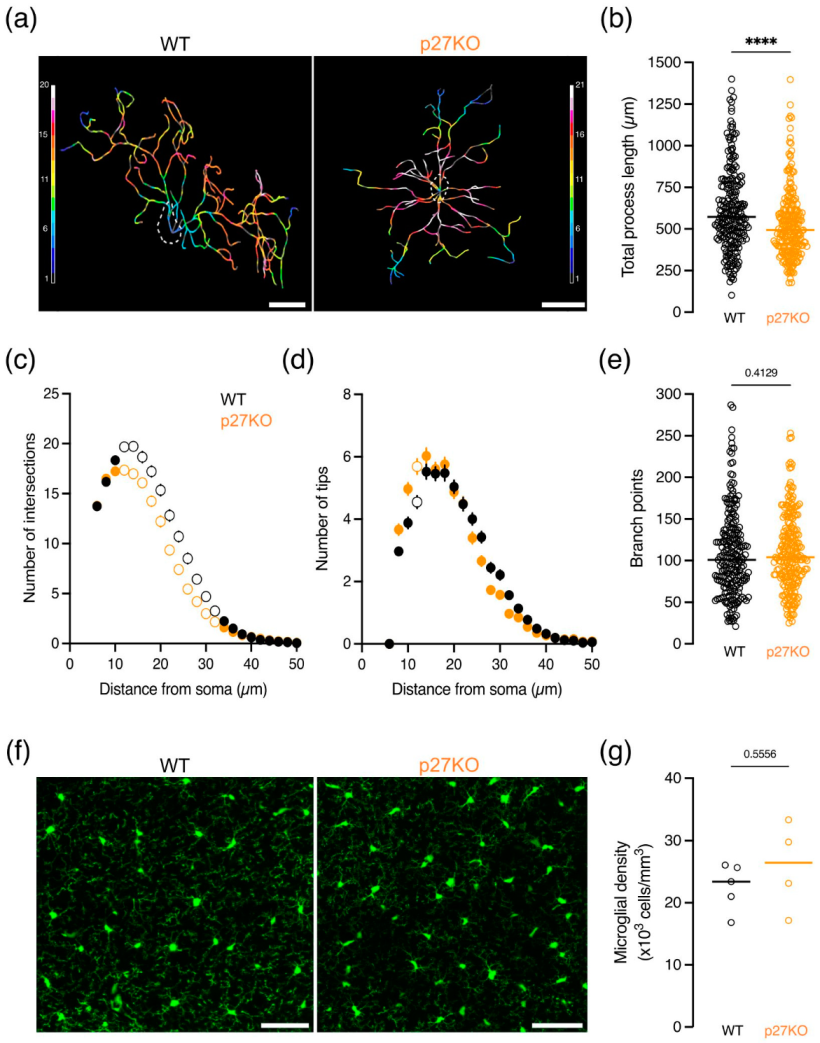
2.1. p27kip1 Is Expressed in Microglial Cytoplasm and Regulates Microglial Morphology
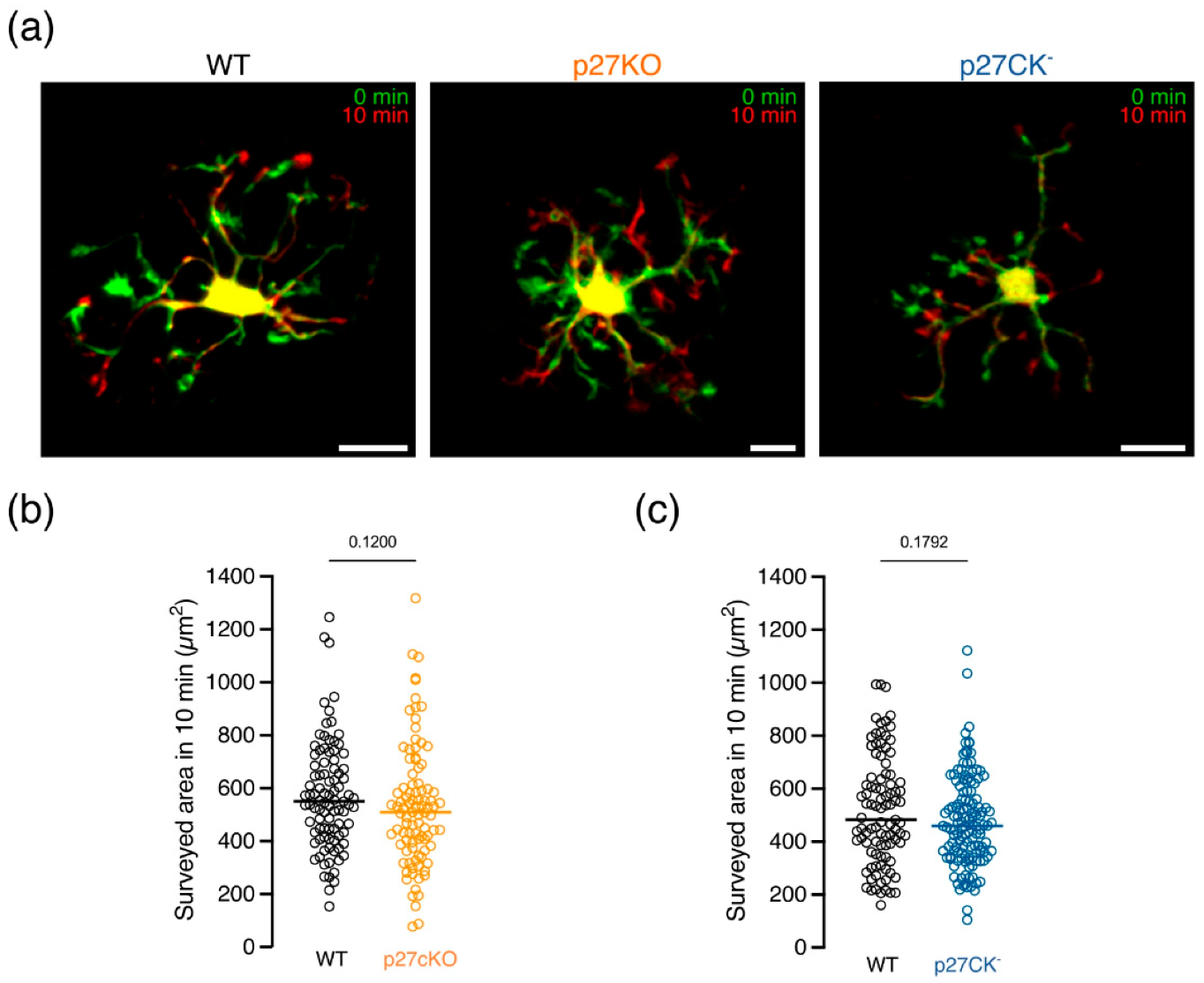
2.2. Microglial Brain Surveillance Is Not Altered upon Loss of p27kip1 or Knock-In of a Cell Cycle Dead Variant of p27kip1
2.3. Conditional Loss of p27kip1 Alters Specific Parameters of Microglial Migration Ex Vivo
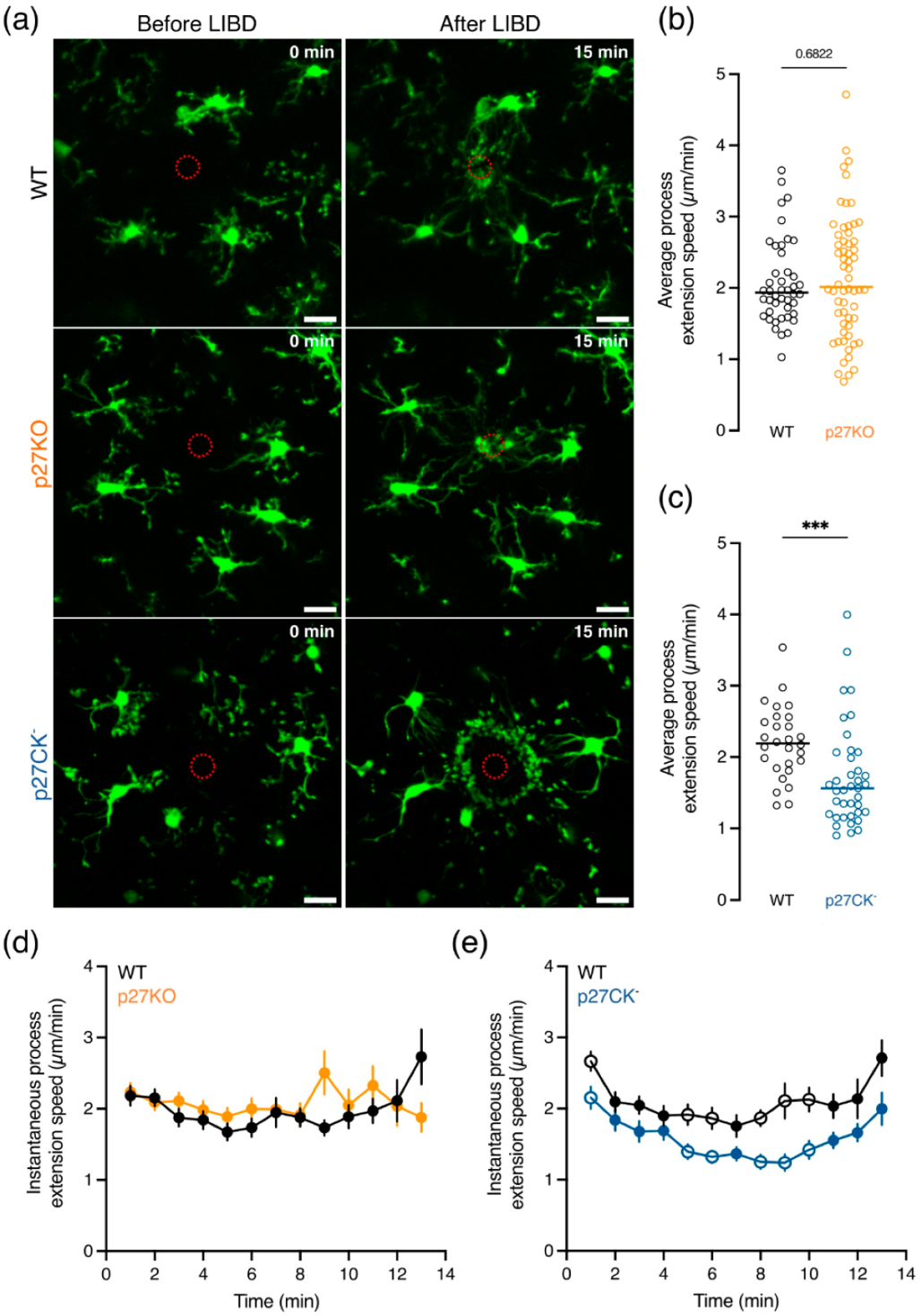
2.4. Process Extension Speed towards a Cortical Lesion Is Decreased upon Expression of the Cell Cycle Dead Variant of p27kip1
2.5. The Phagocytic Capacity of Microglia Is Altered In Vitro upon Loss of p27kip1 or Knock-In of Its Cell Cycle Dead Variant
3. Discussion
3.1. p27kip1 May Act as a Regulator of Morphological Complexity without Modulating Microglial Brain Surveillance or Inflammatory Response
3.2. Emerging Roles for p27kip1 in Microglial Phagocytosis
4. Conclusions
5. Materials and Methods
5.1. Contact for Reagents and Resource Sharing
5.2. Experimental Model and Subject Details
Mouse Genetics
5.3. Method Details
5.3.1. Primary Microglia Isolation and Culture
5.3.2. Immunofluorescence
5.3.3. Western Blotting
5.3.4. Scratch Migration Assay
5.3.5. Quantification of p27kip1 Expression in Microglia after Pro- and Anti-Inflammatory Stimulation
5.3.6. Nitric Oxide Assay
5.3.7. Phagocytosis Assay
5.3.8. Sholl and Microglial Density Analysis in Fixed Brain Slices
5.3.9. Microglial Dynamics in Acute Adolescent Brain Slices Using Two-Photon Imaging
5.3.10. Microglial Migration in Acute Embryonic Brain Slices Using Two-Photon Imaging
5.4. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef] [PubMed]
- Mosser, C.A.; Baptista, S.; Arnoux, I.; Audinat, E. Microglia in CNS development: Shaping the brain for the future. Prog. Neurobiol. 2016, 149–150, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Smolders, S.M.; Kessels, S.; Vangansewinkel, T.; Rigo, J.M.; Legendre, P.; Brone, B. Microglia: Brain cells on the move. Prog. Neurobiol. 2019, 178, 101612. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef]
- Salter, M.W.; Stevens, B. Microglia emerge as central players in brain disease. Nat. Med. 2017, 23, 1018–1027. [Google Scholar] [CrossRef]
- Swinnen, N.; Smolders, S.; Avila, A.; Notelaers, K.; Paesen, R.; Ameloot, M.; Brône, B.; Legendre, P.; Rigo, J.M. Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia 2013, 61, 150–163. [Google Scholar] [CrossRef]
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005, 8, 752–758. [Google Scholar] [CrossRef]
- Lawson, L.J.; Perry, V.H.; Dri, P.; Gordon, S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 1990, 39, 151–170. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef]
- Badimon, A.; Strasburger, H.J.; Ayata, P.; Chen, X.; Nair, A.; Ikegami, A.; Hwang, P.; Chan, A.T.; Graves, S.M.; Uweru, J.O.; et al. Negative feedback control of neuronal activity by microglia. Nature 2020, 586, 417–423. [Google Scholar] [CrossRef]
- Umpierre, A.D.; Wu, L.J. How microglia sense and regulate neuronal activity. Glia 2021, 69, 1637–1653. [Google Scholar] [CrossRef] [PubMed]
- Casano, A.M.; Peri, F. Microglia: Multitasking specialists of the brain. Dev. Cell 2015, 32, 469–477. [Google Scholar] [CrossRef]
- Gilmore, J.H.; Kraft, A.D. Microglia in the developing brain: A potential target with lifetime effects. North 2012, 29, 1883–1889. [Google Scholar] [CrossRef]
- Marín-Teva, J.L.; Dusart, I.; Colin, C.; Gervais, A.; Van Rooijen, N.; Mallat, M. Microglia promote the death of developing purkinje cells. Neuron 2004, 41, 535–547. [Google Scholar] [CrossRef]
- Bernier, L.P.; Bohlen, C.J.; York, E.M.; Choi, H.B.; Kamyabi, A.; Dissing-Olesen, L.; Hefendehl, J.K.; Collins, H.Y.; Stevens, B.; Barres, B.A.; et al. Nanoscale Surveillance of the Brain by Microglia via cAMP-Regulated Filopodia. Cell Rep. 2019, 27, 2895–2908e4. [Google Scholar] [CrossRef] [PubMed]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef]
- Franco-Bocanegra, D.K.; McAuley, C.; Nicoll, J.A.R.; Boche, D. Molecular Mechanisms of Microglial Motility: Changes in Ageing and Alzheimer’s Disease. Cells 2019, 8, 639. [Google Scholar] [CrossRef]
- Ohsawa, K.; Kohsaka, S. Dynamic motility of microglia: Purinergic modulation of microglial movement in the normal and pathological brain. Glia 2011, 59, 1793–1799. [Google Scholar] [CrossRef]
- Godin, J.D.; Nguyen, L. Novel functions of core cell cycle regulators in neuronal migration. Adv. Exp. Med. Biol. 2014, 800, 59–74. [Google Scholar] [CrossRef]
- Nacusi, L.P.; Sheaff, R.J. Akt1 sequentially phosphorylates p27kip1 within a conserved but non-canonical region. Cell Div. 2006, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.S.; Ma, L.; Pledger, W.J. p27Kip1 inhibits the cell cycle through non-canonical G1/S phase-specific gatekeeper mechanism. Cell Cycle 2015, 14, 3954–3964. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, G.; Belletti, B.; Nicoloso, M.S.; Schiappacassi, M.; Vecchione, A.; Spessotto, P.; Morrione, A.; Canzonieri, V.; Colombatti, A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell 2005, 7, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Assoian, R.K.; Roberts, J.M. Regulation of the cytoskeleton: An oncogenic function for CDK inhibitors? Nat. Rev. Cancer 2004, 4, 948–955. [Google Scholar] [CrossRef]
- Fero, M.L.; Rivkin, M.; Tasch, M.; Porter, P.; Carow, C.E.; Firpo, E.; Polyak, K.; Tsai, L.H.; Broudy, V.; Perlmutter, R.M.; et al. A syndrome of multiorgan hyperplasia with features of gigantism tumorigenesis and female sterility in p27kip1 deficient mice. Cell 1996, 85, 733–744. [Google Scholar] [CrossRef]
- Polyak, K.; Kato, J.Y.; Solomon, M.J.; Sherr, C.J.; Massague, J.; Roberts, J.M.; Koff, A. p27kip1 a cyclin CDK inhibitor links transforming growth factorB and contact inhibition to cell cycle arrest. Genes Dev. 1996, 8, 9–22. [Google Scholar] [CrossRef]
- Kiyokawa, H.; Kineman, R.D.; Manova-Todorova, K.O.; Soares, V.C.; Hoffman, E.S.; Ono, M.; Khanam, D.; Hayday, A.C.; Frohman, L.A.; Koff, A. Enhanced growth of mice lacking the cyclin dependent kinase inhibitor function of p27kip. Cell 1996, 85, 721–732. [Google Scholar] [CrossRef]
- Bachs, O.; Gallastegui, E.; Orlando, S.; Bigas, A.; Morante-Redolat, J.M.; Serratosa, J.; Fariñas, I.; Aligué, R.; Pujol, M.J. Role of p27kip1 as a transcriptional regulator. Oncotarget 2018, 9, 26259–26278. [Google Scholar] [CrossRef]
- Sharma, S.S.; Pledger, W.J. The non-canonical functions of p27(Kip1) in normal and tumor biology. Cell Cycle 2016, 15, 1189–1201. [Google Scholar] [CrossRef]
- Belletti, B.; Pellizzari, I.; Berton, S.; Fabris, L.; Wolf, K.; Lovat, F.; Schiappacassi, M.; D’Andrea, S.; Nicoloso, M.S.; Lovisa, S.; et al. p27kip1 Controls Cell Morphology and Motility by Regulating Microtubule-Dependent Lipid Raft Recycling. Cell Morphology and Motility by Regulating Microtubule-Dependent Lipid Raft Recycling. Mol. Cell. Biol. 2010, 30, 2229–2240. [Google Scholar] [CrossRef][Green Version]
- Berton, S.; Belletti, B.; Wolf, K.; Canzonieri, V.; Lovat, F.; Vecchione, A.; Colombatti, A.; Friedl, P.; Baldassarre, G. The tumor suppressor functions of p27(kip1) include control of the mesenchymal/amoeboid transition. Mol. Cell. Biol. 2009, 29, 5031–5045. [Google Scholar] [CrossRef] [PubMed]
- Grimmler, M.; Wang, Y.; Mund, T.; Cilenšek, Z.; Keidel, E.M.; Waddell, M.B.; Jäkel, H.; Kullmann, M.; Kriwacki, R.W.; Hengst, L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 2007, 128, 269–280. [Google Scholar] [CrossRef]
- Besson, A.; Gurian-West, M.; Chen, X.; Kelly-Spratt, K.S.; Kemp, C.J.; Roberts, J.M. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006, 20, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Besson, A.; Heng, J.I.T.; Schuurmans, C.; Teboul, L.; Parras, C.; Philpott, A.; Roberts, J.M.; Guillemot, F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006, 20, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Casaccia-Bonnefil, P.; Tikoo, R.; Kiyokawa, H.; Friedrich, V., Jr.; Chao, M.V.; Koff, A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip. Genes Dev. 1997, 11, 2335–2346. [Google Scholar] [CrossRef] [PubMed]
- Durand, B.; Gao, F.; Raff, M. Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. EMBO J. 1997, 16, 306–317. [Google Scholar] [CrossRef]
- Li, N.; Zhao, C.-T.; Wang, Y.; Yuan, X.-B. The Transcription Factor Cux1 Regulates Dendritic Morphology of Cortical Pyramidal Neurons. PLoS ONE 2010, 5, e10596. [Google Scholar] [CrossRef]
- Godin, J.D.; Thomas, N.; Laguesse, S.; Malinouskaya, L.; Close, P.; Malaise, O.; Purnelle, A.; Raineteau, O.; Campbell, K.; Fero, M.; et al. p27Kip1 Is a Microtubule-Associated Protein that Promotes Microtubule Polymerization during Neuron Migration. Dev. Cell 2012, 23, 729–744. [Google Scholar] [CrossRef]
- Morelli, G.; Even, A.; Gladwyn-Ng, I.; Le Bail, R.; Shilian, M.; Godin, J.D.; Peyre, E.; Hassan, B.A.; Besson, A.; Rigo, J.-M.; et al. p27Kip1 Modulates Axonal Transport by Regulating α-Tubulin Acetyltransferase 1 Stability. Cell Rep. 2018, 23, 2429–2442. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Cheng, C.; Chen, Y.; Shi, S.; Qin, J.; Xiao, F.; Zhou, D.; Lu, M.; Lu, Q.; et al. A Relationship between p27kip1and Skp2 after Adult Brain Injury: Implications for Glial Proliferation. J. Neurotrauma 2010, 27, 361–371. [Google Scholar] [CrossRef]
- Madry, C.; Attwell, D. Receptors, Ion Channels, and Signaling Mechanisms Underlying Microglial Dynamics. J. Biol. Chem. 2015, 290, 12443–12450. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Gurian-West, M.; Schmidt, A.; Hall, A.; Roberts, J.M. p27Kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev. 2004, 18, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Vlach, J.; Hennecke, S.; Amati, B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip. EMBO J. 1997, 16, 5334–5344. [Google Scholar] [CrossRef] [PubMed]
- Sholl, D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953, 87, 387–406. [Google Scholar]
- Sholl, D.A.; Uttley, A.M. Pattern Discrimination and the Visual Cortex. Nature 1953, 171, 387–388. [Google Scholar] [CrossRef]
- Umpierre, A.D.; Bystrom, L.L.; Ying, Y.; Liu, Y.U.; Worrell, G.; Wu, L.-J. Microglial calcium signaling is attuned to neuronal activity in awake mice. eLife 2020, 9, e56502. [Google Scholar] [CrossRef]
- Greig, L.C.; Woodworth, M.; Galazo, M.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef]
- Gorelik, R.; Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 2014, 9, 1931–1943. [Google Scholar] [CrossRef]
- Gorelik, R.; Gautreau, A. The Arp2/3 inhibitory protein arpin induces cell turning by pausing cell migration. Cytoskeleton 2015, 72, 362–371. [Google Scholar] [CrossRef]
- Smolders, S.M.-T.; Swinnen, N.; Kessels, S.; Arnauts, K.; Smolders, S.; Le Bras, B.; Rigo, J.-M.; Legendre, P.; Brône, B. Age-specific function of α5β1 integrin in microglial migration during early colonization of the developing mouse cortex. Glia 2017, 65, 1072–1088. [Google Scholar] [CrossRef]
- Haynes, S.E.; Hollopeter, G.; Yang, G.; Kurpius, D.; Dailey, M.E.; Gan, W.-B.; Julius, D. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 2006, 9, 1512–1519. [Google Scholar] [CrossRef]
- Ohsawa, K.; Irino, Y.; Sanagi, T.; Nakamura, Y.; Suzuki, E.; Inoue, K.; Kohsaka, S. P2Y12receptor-mediated integrin-β1 activation regulates microglial process extension induced by ATP. Glia 2010, 58, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Shen, Q.; Xu, P.; Luo, J.; Tang, Y. Phagocytosis of Microglia in the Central Nervous System Diseases. Mol. Neurobiol. 2014, 49, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Schafer, D.P.; Lehrman, E.K.; Kautzman, A.G.; Koyama, R.; Mardinly, A.R.; Yamasaki, R.; Ransohoff, R.M.; Greenberg, M.E.; Barres, B.A.; Stevens, B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 2012, 74, 691–705. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.A.; Boddeke, H.W.G.M.; Kettenmann, H. Microglia in Physiology and Disease. Annu. Rev. Physiol. 2017, 79, 619–643. [Google Scholar] [CrossRef]
- Prinz, M.; Tay, T.L.; Wolf, Y.; Jung, S. Microglia: Unique and common features with other tissue macrophages. Acta Neuropathol. 2014, 128, 319–331. [Google Scholar] [CrossRef]
- Tusell, J.M.; Ejarque-Ortiz, A.; Mancera, P.; Solà, C.; Saura, J.; Serratosa, J. Upregulation of p21Cip1in activated glial cells. Glia 2009, 57, 524–534. [Google Scholar] [CrossRef]
- Kawauchi, T.; Chihama, K.; Nabeshima, Y.-I.; Hoshino, M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat. Cell Biol. 2006, 8, 17–26. [Google Scholar] [CrossRef]
- Izquierdo, P.; Attwell, D.; Madry, C. Ion Channels and Receptors as Determinants of Microglial Function. Trends Neurosci. 2019, 42, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Thion, M.S.; Garel, S. On place and time: Microglia in embryonic and perinatal brain development. Curr. Opin. Neurobiol. 2017, 47, 121–130. [Google Scholar] [CrossRef]
- Thion, M.S.; Ginhoux, F.; Garel, S. Microglia and early brain development: An intimate journey. Science 2018, 362, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Chánez-Paredes, S.; Montoya-García, A.; Schnoor, M. Cellular and pathophysiological consequences of Arp2/3 complex inhibition: Role of inhibitory proteins and pharmacological compounds. Cell Mol. Life Sci. 2019, 76, 3349–3361. [Google Scholar] [CrossRef] [PubMed]
- Díez-Juan, A.; Andrés, V. Coordinate Control of Proliferation and Migration by the p27 Kip1/Cyclin-Dependent Kinase/Retinoblastoma Pathway in Vascular Smooth Muscle Cells and Fibroblasts. Circ. Res. 2003, 92, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Marx, S.O.; Chen, H.-J.; Poon, M.; Marks, A.R.; Rabbani, L.E. Role for p27 Kip1 in Vascular Smooth Muscle Cell Migration. Circulation 2001, 103, 2967–2972. [Google Scholar] [CrossRef] [PubMed]
- McAllister, S.S.; Becker-Hapak, M.; Pintucci, G.; Pagano, M.; Dowdy, S.F. Novel p27 kip1 C-Terminal Scatter Domain Mediates Rac-Dependent Cell Migration Independent of Cell Cycle Arrest Functions. Mol. Cell. Biol. 2003, 23, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.G.; Peyre, E.; Adhikari, M.H.; Tielens, S.; Tanco, S.; Van Damme, P.; Magno, L.; Krusy, N.; Agirman, G.; Magiera, M.M.; et al. Cell-Intrinsic Control of Interneuron Migration Drives Cortical Morphogenesis. Cell 2018, 172, 1063–1078.e19. [Google Scholar] [CrossRef]
- Hattori, Y. The behavior and functions of embryonic microglia. Anat. Sci. Int. 2022, 97, 1–14. [Google Scholar] [CrossRef]
- Hermann, D.M.; Gunzer, M. Modulating Microglial Cells for Promoting Brain Recovery and Repair. Front. Cell. Neurosci. 2020, 14, 627987. [Google Scholar] [CrossRef]
- Itoh, Y.; Masuyama, N.; Nakayama, K.; Nakayama, K.I.; Gotoh, Y. The Cyclin-dependent Kinase Inhibitors p57 and p27 Regulate Neuronal Migration in the Developing Mouse Neocortex. J. Biol. Chem. 2007, 282, 390–396. [Google Scholar] [CrossRef]
- Tury, A.; Mairet-Coello, G.; DiCicco-Bloom, E. The Cyclin-Dependent Kinase Inhibitor p57Kip2 Regulates Cell Cycle Exit, Differentiation, and Migration of Embryonic Cerebral Cortical Precursors. Cereb. Cortex 2011, 21, 1840–1856. [Google Scholar] [CrossRef]
- Sahai, E.; Olson, M.; Marshall, C.J. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001, 20, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Gallastegui, E.; Domuro, C.; Serratosa, J.; Larrieux, A.; Sin, L.; Martínez, J.; Besson, A.; Morante-Redolat, J.M.; Orlando, S.; Aligué, R.; et al. p27Kip1 regulates alpha-synuclein expression. Oncotarget 2018, 9, 16368–16379. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sierra, A.; Abiega, O.; Shahraz, A.; Neumann, H. Janus-faced microglia: Beneficial and detrimental consequences of microglial phagocytosis. Front. Cell. Neurosci. 2013, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Caron, E.; Hall, A. Identification of Two Distinct Mechanisms of Phagocytosis Controlled by Different Rho GTPases. Science 1998, 282, 1717–1721. [Google Scholar] [CrossRef]
- Chimini, G.; Chavrier, P. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat. Cell Biol. 2000, 2, E191–E196. [Google Scholar] [CrossRef]
- Nowosad, A.; Creff, J.; Jeannot, P.; Culerrier, R.; Codogno, P.; Manenti, S.; Nguyen, L.; Besson, A. p27 controls autophagic vesicle trafficking in glucose-deprived cells via the regulation of ATAT1-mediated microtubule acetylation. Cell Death Dis. 2021, 12, 481. [Google Scholar] [CrossRef]
- Plaza-Zabala, A.; Sierra-Torre, V.; Sierra, A. Autophagy and Microglia: Novel Partners in Neurodegeneration and Aging. Int. J. Mol. Sci. 2017, 18, 598. [Google Scholar] [CrossRef]
- Quraish, R.U.; Sudou, N.; Nomura-Komoike, K.; Sato, F.; Fujieda, H. p27KIP1 loss promotes proliferation and phagocytosis but prevents epithelial-mesenchymal transition in RPE cells after photoreceptor damage. Mol. Vis. 2016, 22, 1103–1121. [Google Scholar]
- Scheiblich, H.; Bicker, G. Regulation of Microglial Phagocytosis by RhoA/ROCK-Inhibiting Drugs. Cell. Mol. Neurobiol. 2017, 37, 461–473. [Google Scholar] [CrossRef]
- Socodato, R.; Portugal, C.C.; Canedo, T.; Rodrigues, A.; Almeida, T.O.; Henriques, J.F.; Vaz, S.H.; Magalhães, J.; Silva, C.M.; Baptista, F.I.; et al. Microglia Dysfunction Caused by the Loss of Rhoa Disrupts Neuronal Physiology and Leads to Neurodegeneration. Cell Rep. 2020, 31, 107796. [Google Scholar] [CrossRef]
- Chu, I.; Sun, J.; Arnaout, A.; Kahn, H.; Hanna, W.; Narod, S.; Sun, P.; Tan, C.-K.; Hengst, L.; Slingerland, J. p27 Phosphorylation by Src Regulates Inhibition of Cyclin E-Cdk. Cell 2007, 128, 281–294. [Google Scholar] [CrossRef]
- Pippa, R.; Espinosa, L.; Gundem, G.; García-Escudero, R.; Dominguez, A.; Orlando, S.; Gallastegui, E.; Saiz, C.; Besson, A.; Pujol, M.J.; et al. p27Kip1 represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene 2012, 31, 4207–4220. [Google Scholar] [CrossRef]
- Jung, S.; Aliberti, J.; Graemmel, P.; Sunshine, M.J.; Kreutzberg, G.W.; Sher, A.; Littman, D.R. Analysis of Fractalkine Receptor CX3CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Mol. Cell. Biol. 2000, 20, 4106–4114. [Google Scholar] [CrossRef]
- Chien, W.-M.; Rabin, S.; Macias, E.; de Marval, P.L.M.; Garrison, K.; Orthel, J.; Rodriguez-Puebla, M.; Fero, M.L. Genetic mosaics reveal both cell-autonomous and cell-nonautonomous function of murine p27 Kip. Proc. Natl. Acad. Sci. USA 2006, 103, 4122–4127. [Google Scholar] [CrossRef]
- Yona, S.; Kim, K.-W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef]
- Scholzen, T.; Gerdes, J. The KI-67 protein: From the known and the unknown. J. Cell. Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Kyrargyri, V.; Madry, C.; Rifat, A.; Arancibia-Carcamo, I.L.; Jones, S.P.; Chan, V.T.T.; Xu, Y.; Robaye, B.; Attwell, D. P2Y 13 receptors regulate microglial morphology, surveillance, and resting levels of interleukin 1β release. Glia 2020, 68, 328–344. [Google Scholar] [CrossRef]
- Beeken, J.; Mertens, M.; Stas, N.; Kessels, S.; Aerts, L.; Janssen, B.; Mussen, F.; Pinto, S.; Vennekens, R.; Rigo, J.; et al. Acute inhibition of transient receptor potential vanilloid-type 4 cation channel halts cytoskeletal dynamism in microglia. Glia 2022, 20, 1–12. [Google Scholar] [CrossRef]
- Xiao, H.; Peng, H. APP2: Automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics 2013, 29, 1448–1454. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Eyo, U.; Peng, J.; Swiatkowski, P.; Mukherjee, A.; Bispo, A.; Wu, L.-J. Neuronal Hyperactivity Recruits Microglial Processes via Neuronal NMDA Receptors and Microglial P2Y12 Receptors after Status Epilepticus. J. Neurosci. 2014, 34, 10528–10540. [Google Scholar] [CrossRef] [PubMed]
- Schiefer, J.; Kampe, K.; Dodt, H.; Zieglgänsberger, W.; Kreutzberg, G. Microglial motility in the rat facial nucleus following peripheral axotomy. J. Neurocytol. 1999, 28, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Thevenaz, P.; Ruttimann, U.; Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998, 7, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for Cell and Particle Tracking. Methods Enzymol. 2012, 504, 183–200. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beeken, J.; Kessels, S.; Rigo, J.-M.; Alpizar, Y.A.; Nguyen, L.; Brône, B. p27kip1 Modulates the Morphology and Phagocytic Activity of Microglia. Int. J. Mol. Sci. 2022, 23, 10432. https://doi.org/10.3390/ijms231810432
Beeken J, Kessels S, Rigo J-M, Alpizar YA, Nguyen L, Brône B. p27kip1 Modulates the Morphology and Phagocytic Activity of Microglia. International Journal of Molecular Sciences. 2022; 23(18):10432. https://doi.org/10.3390/ijms231810432
Chicago/Turabian StyleBeeken, Jolien, Sofie Kessels, Jean-Michel Rigo, Yeranddy A. Alpizar, Laurent Nguyen, and Bert Brône. 2022. "p27kip1 Modulates the Morphology and Phagocytic Activity of Microglia" International Journal of Molecular Sciences 23, no. 18: 10432. https://doi.org/10.3390/ijms231810432
APA StyleBeeken, J., Kessels, S., Rigo, J.-M., Alpizar, Y. A., Nguyen, L., & Brône, B. (2022). p27kip1 Modulates the Morphology and Phagocytic Activity of Microglia. International Journal of Molecular Sciences, 23(18), 10432. https://doi.org/10.3390/ijms231810432




