Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley
Abstract
1. Introduction
2. Results
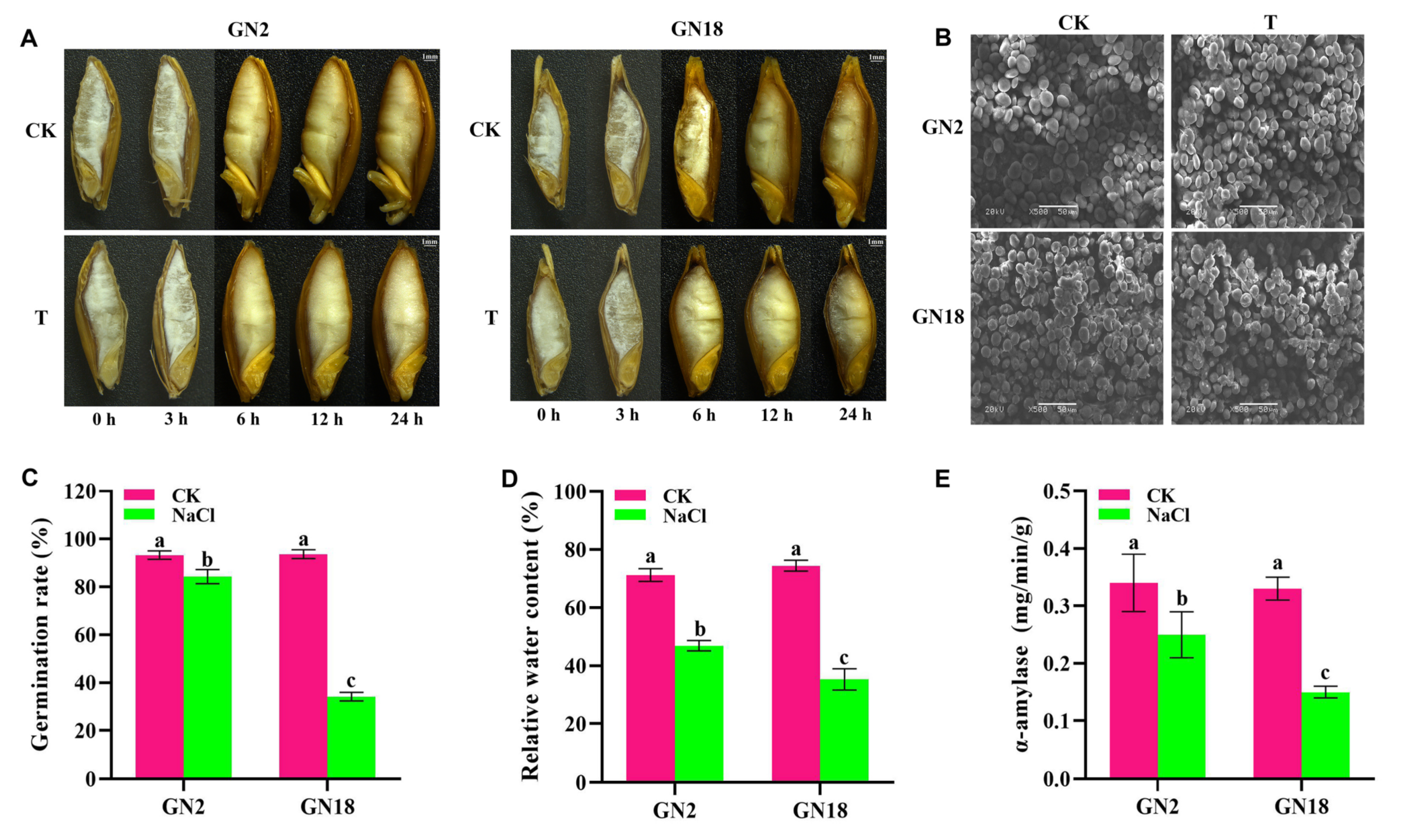
2.1. Structural and Physiological Changes in Response to Salt Stress during Seed Germination
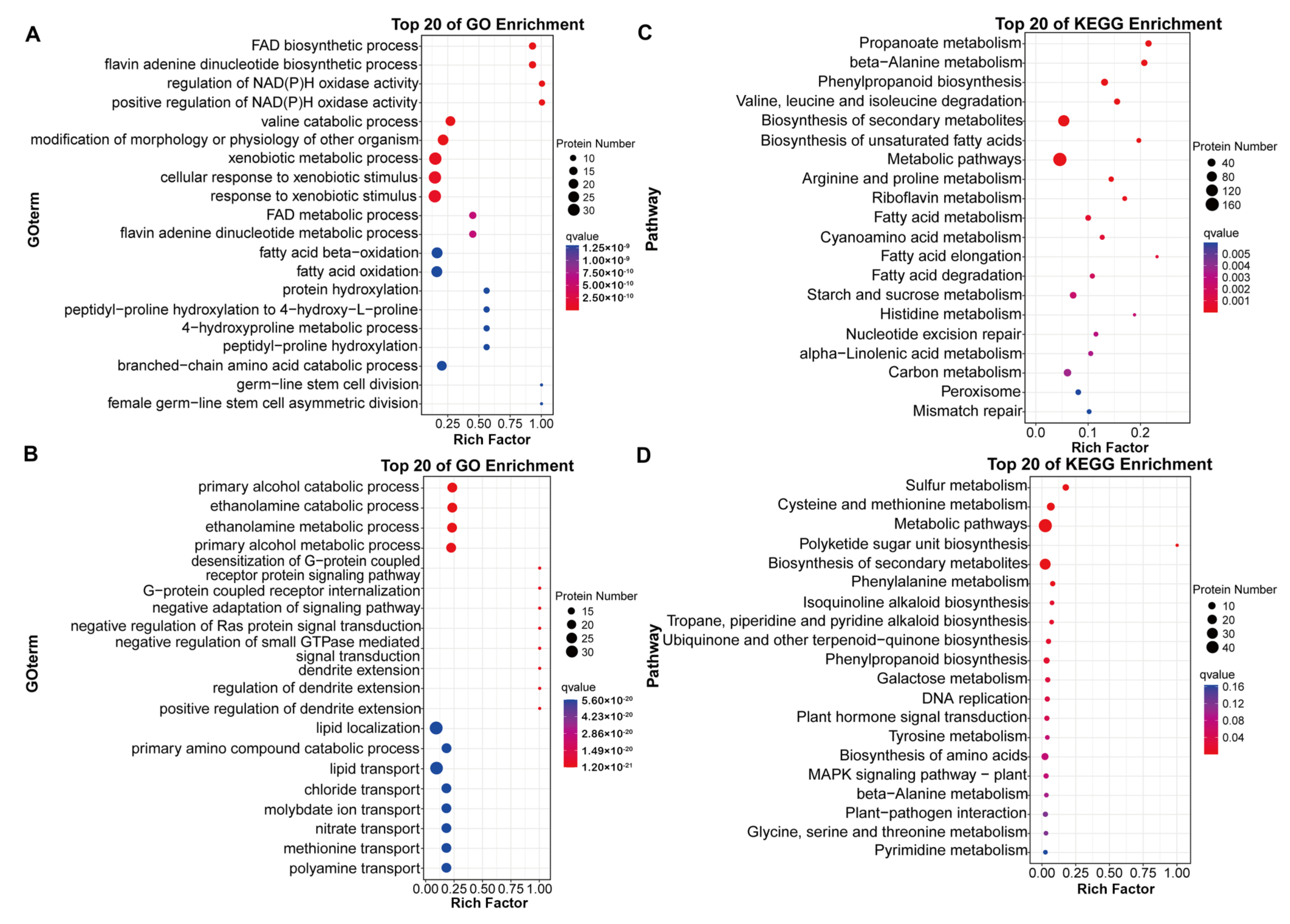
2.2. Analysis of DEPs in Seeds in Response to Salt Stress among Various Cultivars
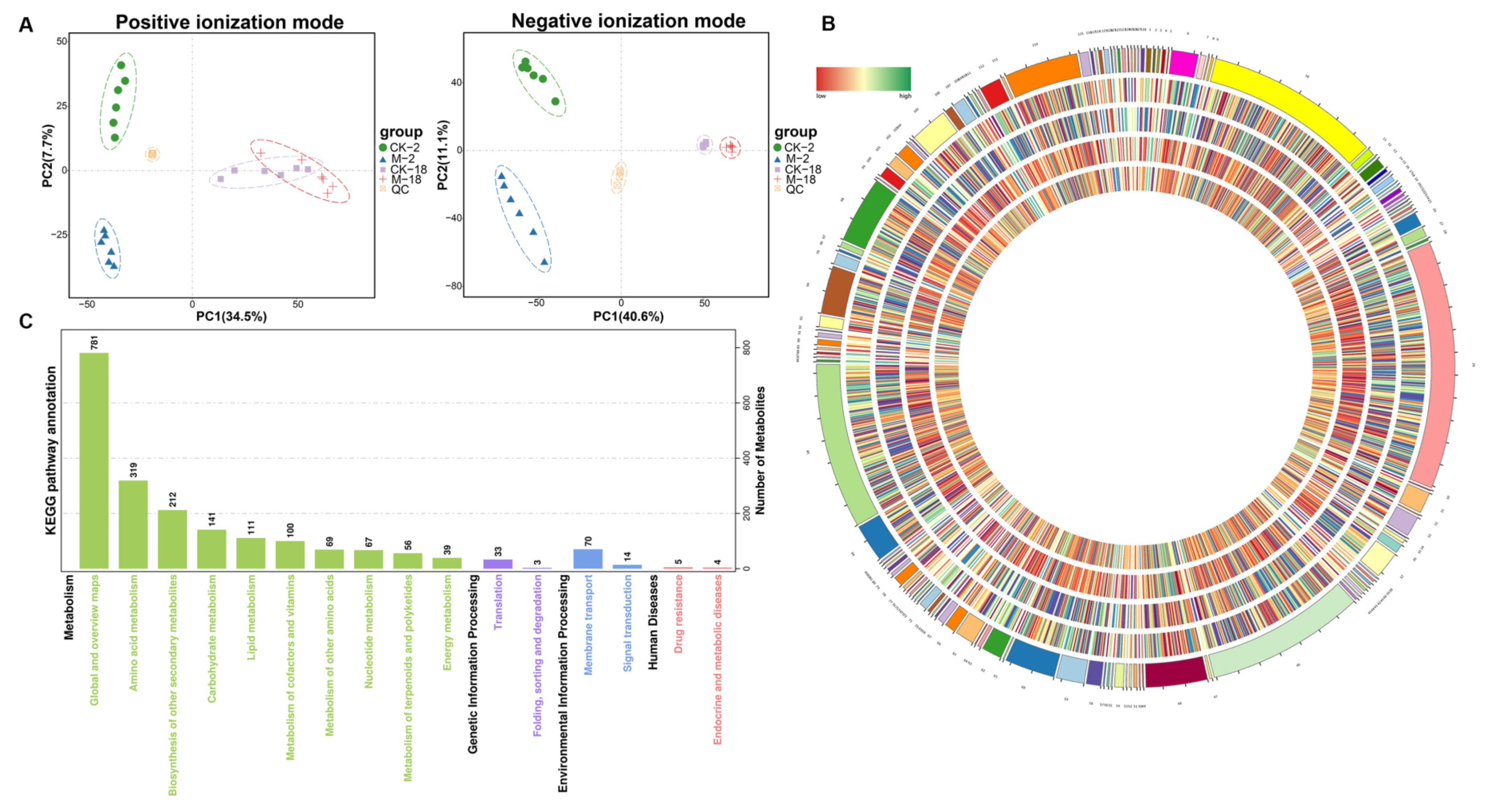
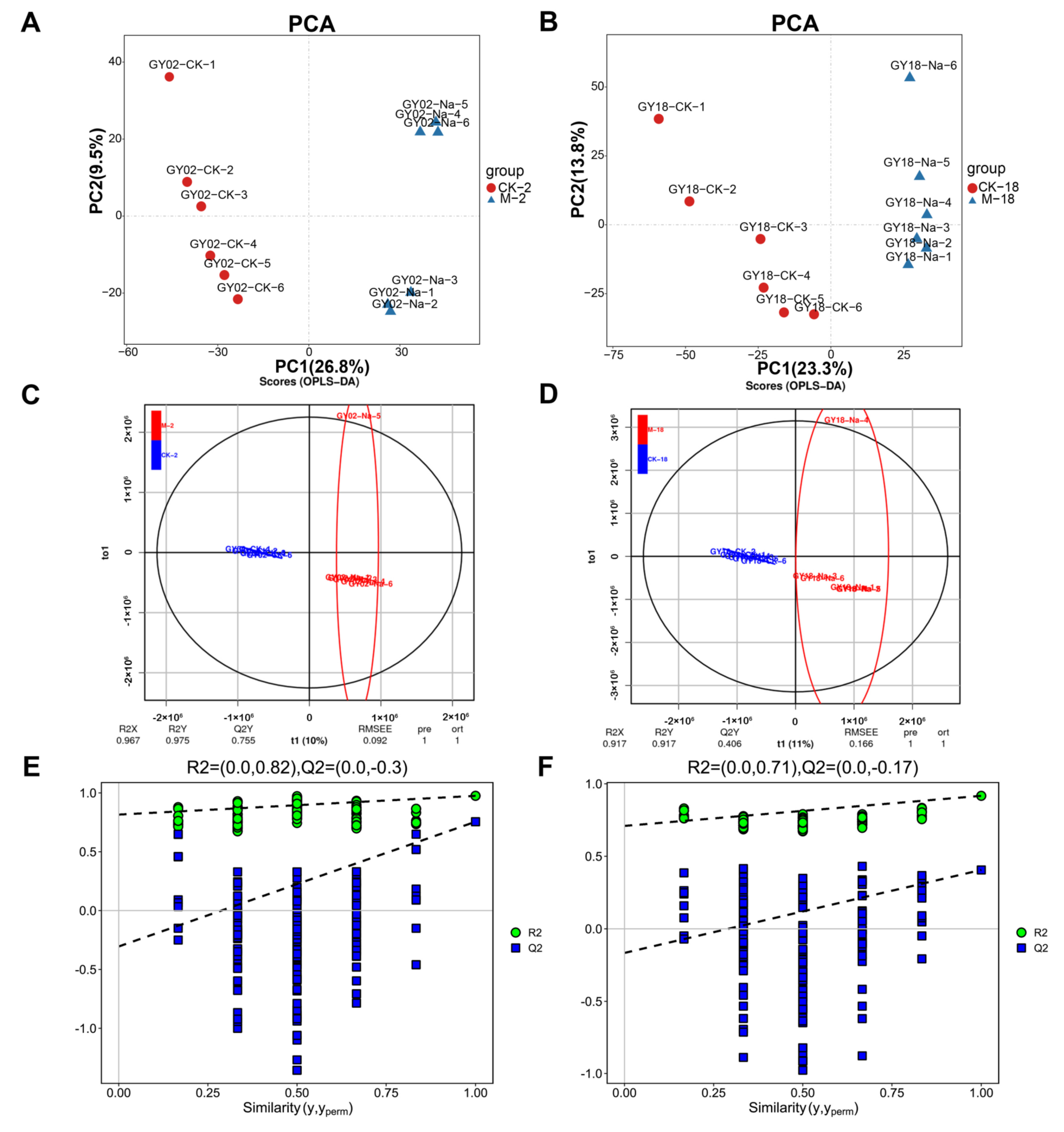
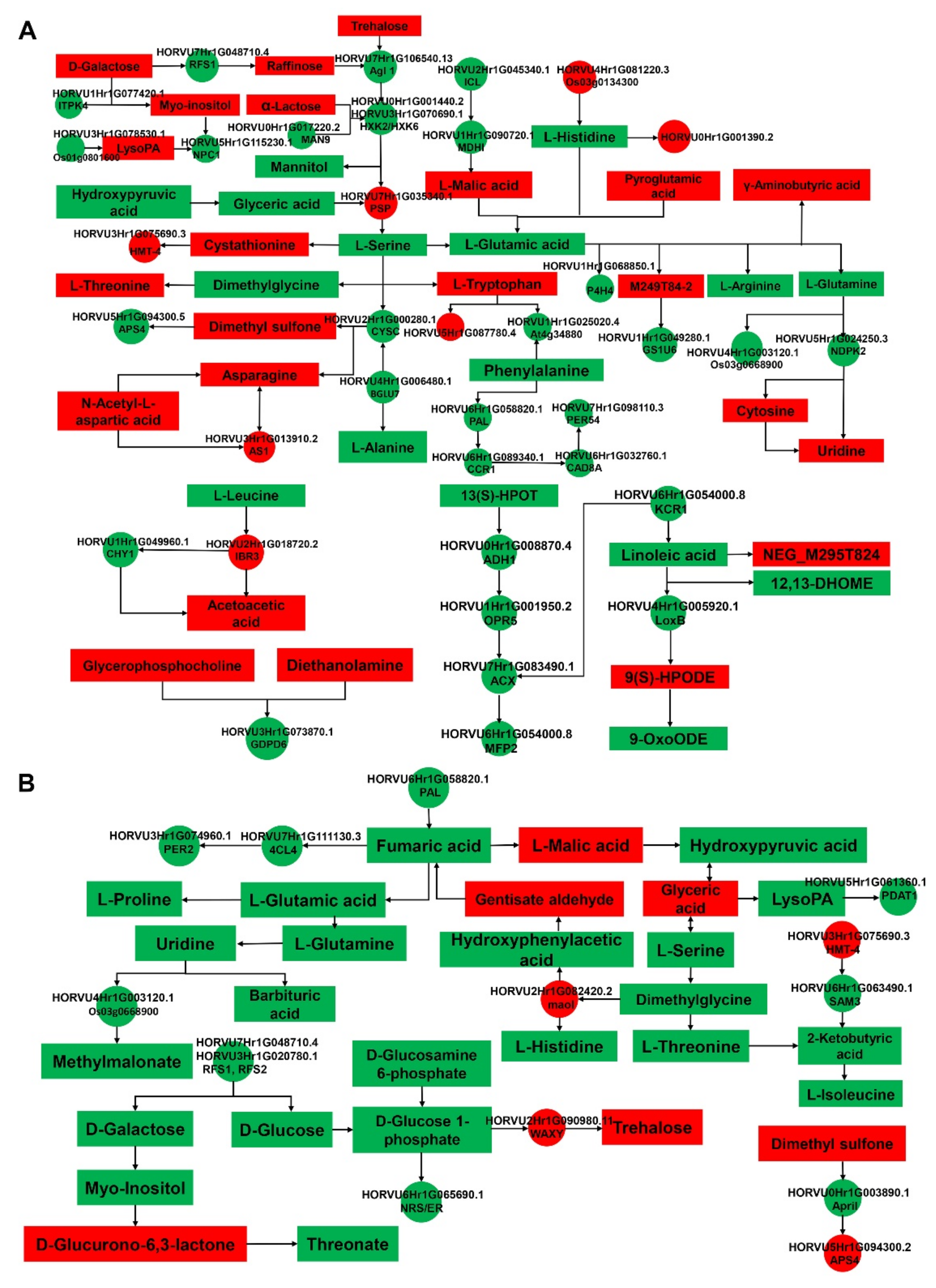
2.3. Metabolic Analyses of Barley Seeds in Response to Salt Stress during the Germination Process
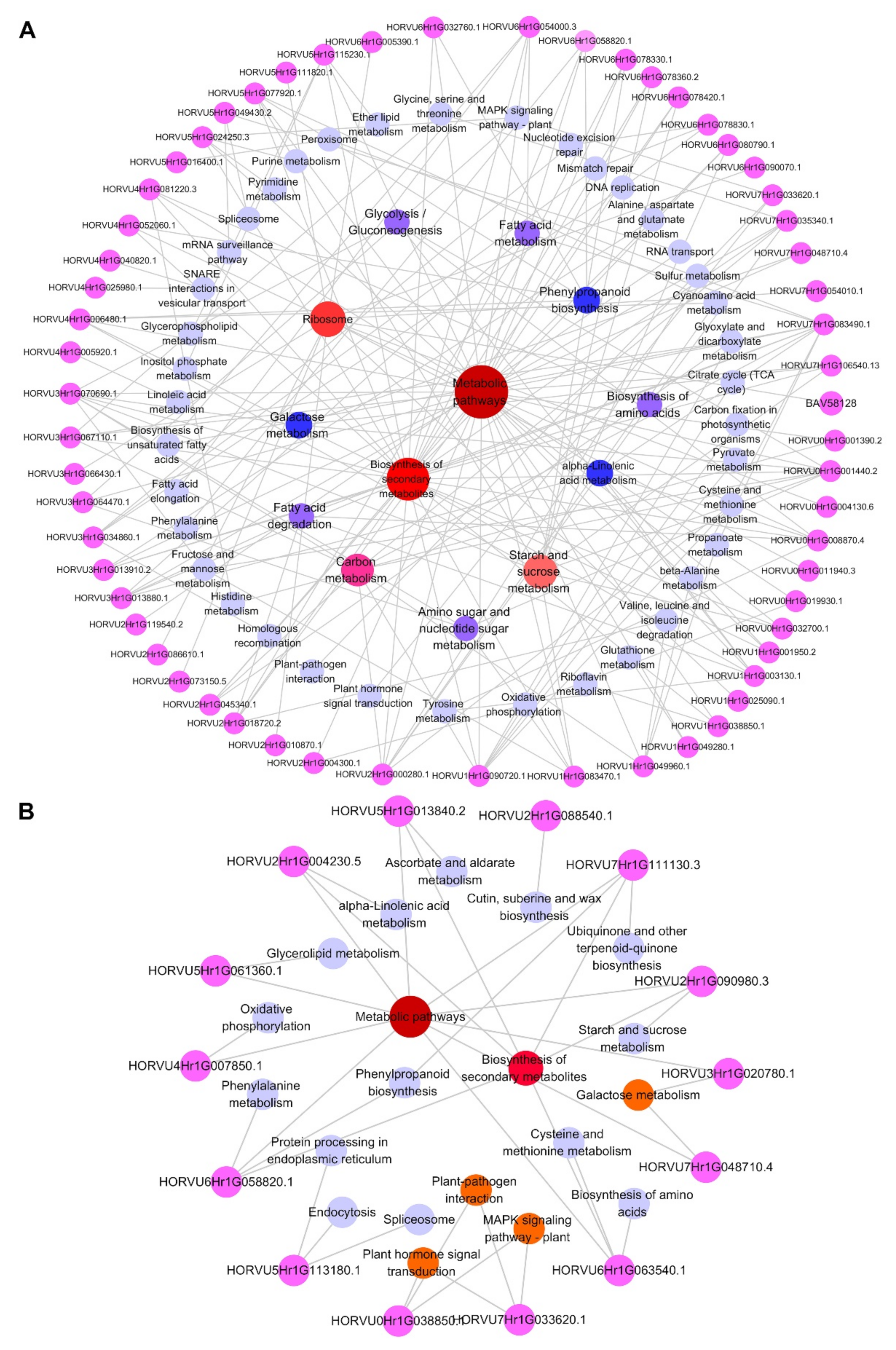
2.4. Integrative Proteomic and Metabolomic Analyses
3. Discussion
3.1. Signal Transduction in Response to Salt Stress
3.2. Amino Acid Metabolism for Salt Tolerance
3.3. Carbohydrate Metabolism for Salt Tolerance
3.4. Effects of Salt Stress on Phytohormone Signal Transduction
3.5. Cell Walls, Biomembranes and Protein Stability under Salt Stress
3.6. Antioxidants under Salt Stress
4. Materials and Methods
4.1. Barley Materials and Seed Germination under Salt Stress
4.2. Measurement of Seed Morphology and Physiological Parameters
4.3. Isolation of Total Proteins and Proteome Analysis
4.4. Detection and Identification of Metabolites and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yan, G. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Butcher, K.; Wick, A.F.; Desutter, T.; Chatterjee, A.; Harmon, J. Soil salinity, a threat to global food security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Rana, M.; Mark, T. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef]
- Xu, E.; Chen, M.; He, H.; Zhan, C.; Cheng, Y.; Zhang, H.; Wang, Z. Proteomic Analysis Reveals Proteins Involved in Seed Imbibition under Salt Stress in Rice. Front. Plant Sci. 2016, 7, 2006. [Google Scholar] [CrossRef]
- Woolhouse, H.W.; Bewley, J.D.; Black, M. Physiology and biochemistry of seeds in relation to germination. Vol. 1: Development, Germination and Growth. J. Ecol. 1980, 68, 315. [Google Scholar] [CrossRef]
- Shu, K.; Qi, Y.; Chen, F.; Meng, Y.; Luo, X.; Shuai, H.; Zhou, W.; Ding, J.; Du, J.; Liu, J.; et al. Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front. Plant Sci. 2017, 8, 1372. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, H.J.; Sun, Q.Q.; Cao, Y.Y.; Li, X.; Zhao, B.; Wu, P.; Guo, Y.D. Proteomic analysis reveals a role of melatonin in promoting cucumber seed germination under high salinity by regulating energy production. Sci. Rep. 2017, 7, 503. [Google Scholar] [CrossRef]
- Limor-Waisberg, K.; Ben-Dor, S. Diversification of Quiescin sulfhydryl oxidase in a preserved framework for redox relay. BMC Evol. Biol. 2013, 13, 70. [Google Scholar] [CrossRef]
- Duan, X.; Wang, Z.; Zhang, Y.; Li, H.; Yang, M.; Yin, H.; Cui, J.; Chai, H.; Gao, Y.; Hu, G.; et al. Overexpression of a Thioredoxin-Protein-Encoding Gene, MsTRX, from Medicago sativa Enhances Salt Tolerance to Transgenic Tobacco. Agronomy 2022, 12, 1467. [Google Scholar] [CrossRef]
- Luo, X.; Dai, Y.; Zheng, C.; Yang, Y.; Chen, W.; Wang, Q.; Chandrasekaran, U.; Du, J.; Liu, W.; Shu, K. The ABI4-RbohD/VTC2 regulatory module promotes Reactive Oxygen Species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021, 229, 950–962. [Google Scholar] [CrossRef]
- Yang, F.; Chen, H.; Liu, C.; Li, L.; Liu, L.; Han, X.; Wan, Z.; Sha, A. Transcriptome profile analysis of two Vicia faba cultivars with contrasting salinity tolerance during seed germination. Sci. Rep. 2020, 10, 7250. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, X.; Wang, Y.; Zhou, S.; Fu, F.; Li, W. A betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus enhances tolerance of Arabidopsis to high salt and drought stresses. Plant Growth Regul. 2017, 83, 265–276. [Google Scholar] [CrossRef]
- Jia, X.M.; Zhu, Y.F.; Hu, Y.; Zhang, R.; Cheng, L.; Zhu, Z.L.; Zhao, T.; Zhang, X.; Wang, Y.X. Integrated physiologic, proteomic, and metabolomic analyses of Malus halliana adaptation to saline-alkali stress. Hortic. Res. 2019, 6, 91. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, H.; Du, H.; Bao, Z.; Shi, Q. Sugar metabolic and n-glycosylated profiles unveil the regulatory mechanism of tomato quality under salt stress. Environ. Exp. Bot. 2020, 177, 104145. [Google Scholar] [CrossRef]
- Wang, W.S.; Zhao, X.Q.; Li, M.; Huang, L.Y.; Xu, J.L.; Zhang, F.; Cui, Y.R.; Fu, B.Y.; Li, Z.K. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. J. Exp. Bot. 2016, 67, 405–419. [Google Scholar] [CrossRef]
- Sami, F.; Yusuf, M.; Faizan, M.; Faraz, A.; Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 2016, 109, 54–61. [Google Scholar] [CrossRef]
- Liu, L.; Wu, X.; Sun, W.; Yu, X.; Qiang, Z. Galactinol synthase confers salt-stress tolerance by regulating the synthesis of galactinol and raffinose family oligosaccharides in poplar. Ind. Crops Prod. 2021, 165, 113432. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, N.; Li, Y.; Zhu, S.; Zhang, S.; Sun, Y.; Zhang, H.X.; Wang, L.; Su, H. Overexpression of PeMIPS1 confers tolerance to salt and copper stresses by scavenging reactive oxygen species in transgenic poplar. Tree Physiol. 2018, 38, 1566–1577. [Google Scholar] [CrossRef]
- Hu, L.; Zhou, K.; Liu, Y.; Yang, S.; Zhang, J.; Gong, X.; Ma, F. Overexpression of MdMIPS1 enhances salt tolerance by improving osmosis, ion balance, and antioxidant activity in transgenic apple. Plant Sci. 2020, 301, 110654. [Google Scholar] [CrossRef]
- Keyvan, A.; Setsuko, K. Crop and medicinal plants proteomics in response to salt stress. Front. Plant Sci. 2013, 4, 8. [Google Scholar] [CrossRef]
- Lai, Y.; Zhang, D.; Wang, J.; Wang, J.; Ren, P.; Yao, L.; Si, E.; Kong, Y.; Wang, H. Integrative transcriptomic and proteomic analyses of molecular mechanism responding to salt stress during seed germination in hulless barley. Int. J. Mol. Sci. 2020, 21, 359. [Google Scholar] [CrossRef]
- Boussadia, O.; Mariem, F.B.; Mechri, B.; Boussetta, W.; Braham, M.; Hadj, S. Response to drought of two olive tree cultivars (cv Koroneki and Meski). Sci. Hortic. 2008, 116, 388–393. [Google Scholar] [CrossRef]
- Doerr, A. DIA mass spectrometry. Nat. Methods 2014, 12, 35. [Google Scholar] [CrossRef]
- Buts, K.; Michielssens, S.; Hertog, M.L.; Hayakawa, E.; Cordewener, J.; America, A.H.; Nicolai, B.M.; Carpentier, S.C. Improving the identification rate of data independent label-free quantitative proteomics experiments on non-model crops: A case study on apple fruit. J. Proteom. 2014, 105, 31–45. [Google Scholar] [CrossRef]
- Wang, Y.; Sang, Z.; Xu, S.; Xu, Q.; Zeng, X.; Jabu, D.; Yuan, H. Comparative proteomics analysis of Tibetan hull-less barley under osmotic stress via data-independent acquisition mass spectrometry. GigaScience 2020, 9, giaa019. [Google Scholar] [CrossRef]
- Zou, J.; Yu, H.; Yu, Q.; Jin, X.; Cao, L.; Wang, M.; Wang, M.; Ren, C.; Zhang, Y. Physiological and UPLC-MS/MS widely targeted metabolites mechanisms of alleviation of drought stress-induced soybean growth inhibition by melatonin. Ind. Crops Prod. 2021, 163, 113323. [Google Scholar] [CrossRef]
- Pan, J.; Li, Z.; Dai, S.; Ding, H.; Wang, Q.; Li, X.; Ding, G.; Wang, P.; Guan, Y.; Liu, W. Integrative analyses of transcriptomics and metabolomics upon seed germination of foxtail millet in response to salinity. Sci. Rep. 2020, 10, 13660. [Google Scholar] [CrossRef]
- Isayenkov, S.V. Genetic sources for the development of salt tolerance in crops. Plant Growth Regul. 2019, 89, 1–17. [Google Scholar] [CrossRef]
- Wang, M.; Ding, Y.; Wang, Q.; Wang, P.; Han, Y.; Gu, Z.; Yang, R. NaCl treatment on physio-biochemical metabolism and phenolics accumulation in barley seedlings. Food Chem. 2020, 331, 127282. [Google Scholar] [CrossRef] [PubMed]
- Latef, A.A.H.A.; Alhmad, M.F.A.; Kordrostami, M.; Abo-Baker, A.B.; Zakir, A. Inoculation with Azospirillum lipoferum or Azotobacter chroococcum Reinforces Maize Growth by Improving Physiological Activities Under Saline Conditions. J. Plant Growth Regul. 2020, 39, 1293–1306. [Google Scholar] [CrossRef]
- Lavergne, F.D.; Broeckling, C.D.; Brown, K.J.; Cockrell, D.M.; Haley, S.D.; Peairs, F.B.; Pearce, S.; Wolfe, L.M.; Jahn, C.E.; Heuberger, A.L. Differential Stem Proteomics and Metabolomics Profiles for Four Wheat Cultivars in Response to the Insect Pest Wheat Stem Sawfly. J. Proteome Res. 2020, 19, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Vo, K.T.X.; Rahman, M.M.; Rahman, M.M.; Trinh, K.T.T.; Kim, S.T.; Jeon, J.S. Proteomics and Metabolomics Studies on the Biotic Stress Responses of Rice: An Update. Rice 2021, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [PubMed]
- Bush, D.S. Calcium Regulation in Plant Cells and its Role in Signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 95–122. [Google Scholar] [CrossRef]
- Manishankar, P.; Wang, N.; Köster, P.; Alatar, A.A.; Kudla, J. Calcium Signaling during Salt Stress and in the Regulation of Ion Homeostasis. J. Exp. Bot. 2018, 17, 4215–4226. [Google Scholar] [CrossRef]
- Lu, J.; Du, J.; Tian, L.; Li, M.; Zhang, X.; Zhang, S.; Wan, X.; Chen, Q. Divergent Response Strategies of CsABF Facing Abiotic Stress in Tea Plant: Perspectives from Drought-Tolerance Studies. Front. Plant Sci. 2021, 12, 763843. [Google Scholar] [CrossRef]
- Mucha, E.; Fricke, I.; Schaefer, A.; Wittinghofer, A.; Berken, A. Rho proteins of plants–functional cycle and regulation of cytoskeletal dynamics. Eur. J. Cell Biol. 2011, 90, 934–943. [Google Scholar] [CrossRef] [PubMed]
- Bloch, D.; Yalovsky, S. Cell polarity signaling. Curr. Opin. Plant Biol. 2013, 16, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Takahashi, Y.; Scavo, A.; Brandt, B.; Nguyen, D.; Rieu, P.; Schroeder, J.I. Abscisic acid-induced degradation of Arabidopsis guanine nucleotide exchange factor requires calcium-dependent protein kinases. Proc. Natl. Acad. Sci. USA 2018, 15, E4522–E4531. [Google Scholar] [CrossRef]
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 2009, 458, 422–429. [Google Scholar] [CrossRef]
- Chen, B.; Lin, L.; Lu, Y.; Peng, J.; Zheng, H.; Yang, Q.; Rao, S.; Wu, G.; Li, J.; Chen, Z.; et al. Ubiquitin-Like protein 5 interacts with the silencing suppressor p3 of rice stripe virus and mediates its degradation through the 26S proteasome pathway. PLoS Pathog. 2020, 16, e1008780. [Google Scholar] [CrossRef]
- Patel, M.; Milla-Lewis, S.; Zhang, W.; Templeton, K.; Reynolds, W.C.; Richardson, K.; Biswas, M.; Zuleta, M.C.; Dewey, R.E.; Qu, R.; et al. Overexpression of ubiquitin-like LpHUB1 gene confers drought tolerance in perennial ryegrass. Plant Biotechnol. J. 2015, 13, 689–699. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, C.; Shi, Z.; Kou, X. The Amino Acid Metabolic and Carbohydrate Metabolic Pathway Play Important Roles during Salt-Stress Response in Tomato. Front. Plant Sci. 2017, 8, 1231. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Tohidfar, M.; Mortazavian, S.M.M.; Sabbatini, P. Betaine Aldehyde Dehydrogenase (BADH) vs. Flavodoxin (Fld): Two Important Genes for Enhancing Plants Stress Tolerance and Productivity. Front. Plant Sci. 2021, 12, 650215. [Google Scholar] [CrossRef]
- Kerepesi, I.; Galiba, G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Sci. 2000, 40, 482–487. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Zhao, Y.; Yang, Y.; Wang, Y.; Siddique, K.H.M. Integrated transcriptomics and metabolomics analysis to characterize alkali stress responses in canola (Brassica napus L.). Plant Physiol. Biochem. 2021, 166, 605–620. [Google Scholar] [CrossRef]
- Kang, J.S.; Frank, J.; Kang, C.H.; Kajiura, H.; Vikram, M.; Ueda, A.; Kim, S.; Bahk, J.D.; Triplett, B.; Fujiyama, K.; et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 2008, 105, 5933–5938. [Google Scholar] [CrossRef]
- Chung, C.Y.; Majewska, N.I.; Wang, Q.; Paul, J.T.; Betenbaugh, M.J. SnapShot: N-Glycosylation Processing Pathways across Kingdoms. Cell 2017, 171, 258–258.e1. [Google Scholar] [CrossRef]
- Palma, F.; Tejera, N.A.; Lluch, C. Nodule carbohydrate metabolism and polyols involvement in the response of medicago sativa to salt stress. Environ. Exp. Bot. 2013, 85, 43–49. [Google Scholar] [CrossRef]
- De Zélicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- He, Y.; Yang, B.; He, Y.; Zhan, C.; Cheng, Y.; Zhang, J.; Zhang, H.; Cheng, J.; Wang, Z. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019, 97, 1089–1104. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of aba in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, Y.G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J.K. Understanding and improving salt tolerance in plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, M.; Jia, J.; Zhao, X.; Huang, X.; Ji, E.; Ni, L.; Jiang, M. An Atypical Late Embryogenesis Abundant Protein OsLEA5 Plays a Positive Role in ABA-Induced Antioxidant Defense in Oryza sativa L. Plant Cell Physiol. 2018, 59, 916–929. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, J.; Sun, L.; Yang, X.; Li, D. Group 3 LEA Protein, ZmLEA3, Is Involved in Protection from Low Temperature Stress. Front. Plant Sci. 2016, 7, 1011. [Google Scholar] [CrossRef] [PubMed]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.R.; Firouzabadi, F.N.; Shobbar, Z.S. Transcriptome response of roots to salt stress in a salinity-tolerant bread wheat cultivar. PLoS ONE 2019, 14, e0213305. [Google Scholar] [CrossRef] [PubMed]
- Espelund, M.; Saebøe-Larssen, S.; Hughes, D.W.; Galau, G.A.; Larsen, F.; Jakobsen, K.S. Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic acid and osmotic stress. Plant J. 1992, 2, 241–252. [Google Scholar] [CrossRef]
- Xiang, D.J.; Man, L.L.; Zhang, C.L.; Liu, P.; Li, Z.G.; Zheng, G.C. A new Em-like protein from Lactuca sativa, LsEm1, enhances drought and salt stress tolerance in Escherichia coli and rice. Protoplasma 2018, 255, 1089–1106. [Google Scholar] [CrossRef]
- Dermendjiev, G.; Schnurer, M.; Weiszmann, J.; Wilfinger, S.; Ott, E.; Gebert, C.; Weckwerth, W.; Ibl, V. Tissue-Specific Proteome and Subcellular Microscopic Analyses Reveal the Effect of High Salt Concentration on Actin Cytoskeleton and Vacuolization in Aleurone Cells during Early Germination of Barley. Int. J. Mol. Sci. 2021, 22, 9642. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Abedini, R.; GhaneGolmohammadi, F.; PishkamRad, R.; Pourabed, E.; Jafarnezhad, A.; Shobbar, Z.S.; Shahbazi, M. Plant dehydrins: Shedding light on structure and expression patterns of dehydrin gene family in barley. J. Plant Res. 2017, 130, 747–763. [Google Scholar] [CrossRef]
- Zhu, Y.; Hu, X.; Duan, Y.; Li, S.; Wang, Y.; Rehman, A.U.; He, J.; Zhang, J.; Hua, D.; Yang, L.; et al. The Arabidopsis Nodulin Homeobox Factor AtNDX Interacts with AtRING1A/B and Negatively Regulates Abscisic Acid Signaling. Plant Cell 2020, 32, 703–721. [Google Scholar] [CrossRef]
- Alizadeh, M.; Hoy, R.; Lu, B.; Song, L. Team effort: Combinatorial control of seed maturation by transcription factors. Curr. Opin. Plant Biol. 2021, 63, 102091. [Google Scholar] [CrossRef]
- Melnick, A.; Ahmad, K.F.; Arai, S.; Polinger, A.; Ball, H.; Borden, K.L.; Carlile, G.W.; Prive, G.G.; Licht, J.D. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 2000, 20, 6550–6567. [Google Scholar] [CrossRef]
- Wan, X.; Peng, L.; Xiong, J.; Li, X.; Wang, J.; Li, X.; Yang, Y. AtSIBP1, a Novel BTB Domain-Containing Protein, Positively Regulates Salt Signaling in Arabidopsis thaliana. Plants 2019, 8, 573. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Zhang, M.; Lv, D.; Ge, P.; Bian, Y.; Chen, G.; Zhu, G.; Li, X.; Yan, Y. Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteom. 2014, 109, 290–308. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Li, D.; Sun, Y.; Li, Y.; Luo, Q.; Liu, Z.; Wang, J.; Li, X.; Zhang, H.; et al. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discov. 2018, 4, 30. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Long, S.; Zhao, C. Maintenance of Cell Wall Integrity under High Salinity. Int. J. Mol. Sci. 2021, 22, 3260. [Google Scholar] [CrossRef]
- Zhao, C.; Jiang, W.; Zayed, O.; Liu, X.; Tang, K.; Nie, W.; Li, Y.; Xie, S.; Li, Y.; Long, T.; et al. The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 2020, 8, nwaa149. [Google Scholar] [CrossRef]
- Baumberger, N.; Doesseger, B.; Guyot, R.; Diet, A.; Parsons, R.L.; Clark, M.A.; Simmons, M.P.; Bedinger, P.; Goff, S.A.; Ringli, C.; et al. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003, 131, 1313–1326. [Google Scholar] [CrossRef]
- Zhao, C.; Zayed, O.; Yu, Z.; Jiang, W.; Zhu, P.; Hsu, C.C.; Zhang, L.; Tao, W.A.; Lozano-Durán, R.; Zhu, J.K. Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, 13123–13128. [Google Scholar] [CrossRef]
- Quan, X.; Liu, J.; Zhang, N.; Xie, C.; Li, H.; Xia, X.; He, W.; Qin, Y. Genome-Wide Association Study Uncover the Genetic Architecture of Salt Tolerance-Related Traits in Common Wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 663941. [Google Scholar] [CrossRef]
- Nakamura, Y.; Ngo, A.H. Non-specific phospholipase C (NPC): An emerging class of phospholipase C in plant growth and development. J. Plant Res. 2020, 133, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Song, T.; Wallrad, L.; Kudla, J.; Wang, X.; Zhang, W. Tissue-specific accumulation of pH-sensing phosphatidic acid determines plant stress tolerance. Nat. Plants 2019, 5, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.M. The plasma membrane transport systems and adaptation to salinity. J. Plant Physiol. 2014, 171, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, D.; Li, M.; Shi, L. Metabolic Profiles Reveal Changes in Wild and Cultivated Soybean Seedling Leaves under Salt Stress. PLoS ONE 2016, 11, e0159622. [Google Scholar] [CrossRef]
- Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 2017, 37, 42–47. [Google Scholar] [CrossRef]
- Sui, N.; Wang, Y.; Liu, S.; Yang, Z.; Wang, F.; Wan, S. Transcriptomic and Physiological Evidence for the Relationship between Unsaturated Fatty Acid and Salt Stress in Peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Saha, A.; Das, S.; Moin, M.; Dutta, M.; Bakshi, A.; Madhav, M.S.; Kirti, P.B. Genome-Wide Identification and Comprehensive Expression Profiling of Ribosomal Protein Small Subunit (RPS) Genes and their Comparative Analysis with the Large Subunit (RPL) Genes in Rice. Front. Plant Sci. 2017, 8, 1553. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.; Li, C.; Ren, P.; Yao, L.; Li, B.; Meng, Y.; Ma, X.; Si, E.; Yang, K.; et al. Dynamic Responses of Barley Root Succinyl-Proteome to Short-Term Phosphate Starvation and Recovery. Front. Plant Sci. 2021, 12, 649147. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Karunadasa, S.S.; Kurepa, J.; Shull, T.E.; Smalle, J.A. Cytokinin–induced protein synthesis suppresses growth and osmotic stress tolerance. New Phytol. 2020, 227, 50–64. [Google Scholar] [CrossRef]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef]
- Kim, K.Y.; Park, S.W.; Chung, Y.S.; Chung, C.H.; Kim, J.I.; Lee, J.H. Molecular cloning of low-temperature-inducible ribosomal proteins from soybean. J. Exp. Bot. 2004, 55, 1153–1155. [Google Scholar] [CrossRef]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, W.; Chan, Z.; Wu, Y. Endogenous Cytokinin Overproduction Modulates ROS Homeostasis and Decreases Salt Stress Resistance in Arabidopsis Thaliana. Front. Plant Sci. 2015, 6, 1004. [Google Scholar] [CrossRef]
- Gelhaye, E.; Rouhier, N.; Navrot, N.; Jacquot, J.P. The plant thioredoxin system. Cell Mol. Life Sci. 2005, 62, 24–35. [Google Scholar] [CrossRef]
- Vanacker, H.; Guichard, M.; Bohrer, A.S.; Issakidis-Bourguet, E. Redox Regulation of Monodehydroascorbate Reductase by Thioredoxin y in Plastids Revealed in the Context of Water Stress. Antioxidants 2018, 7, 183. [Google Scholar] [CrossRef]
- Lemaire, S.D.; Michelet, L.; Zaffagnini, M.; Massot, V.; Issakidis-Bourguet, E. Thioredoxins in chloroplasts. Curr. Genet. 2007, 51, 343–365. [Google Scholar] [CrossRef]
- Geigenberger, P.; Thormahlen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef]
- Aswad, D.W. Stoichiometric methylation of porcine adrenocorticotropin by protein carboxyl methyltransferase requires deamidation of asparagine 25. Evidence for methylation at the alpha-carboxyl group of atypical L-isoaspartyl residues. J. Biol. Chem. 1984, 259, 10714–10721. [Google Scholar] [CrossRef]
- Murray, E.D., Jr.; Clarke, S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. Detection of a new site of methylation at isomerized L-aspartyl residues. J. Biol. Chem. 1984, 259, 10722–10732. [Google Scholar] [CrossRef]
- Biterge, B.; Richter, F.; Mittler, G.; Schneider, R. Methylation of histone H4 at aspartate 24 by protein L-isoaspartate O-methyltransferase (PCMT1) links histone modifications with protein homeostasis. Sci. Rep. 2014, 4, 6674. [Google Scholar] [CrossRef]
- Petla, B.P.; Kamble, N.U.; Kumar, M.; Verma, P.; Ghosh, S.; Singh, A.; Rao, V.; Salvi, P.; Kaur, H.; Saxena, S.C.; et al. Rice PROTEIN l-ISOASPARTYL METHYLTRANSFERASE isoforms differentially accumulate during seed maturation to restrict deleterious isoAsp and reactive oxygen species accumulation and are implicated in seed vigor and longevity. New Phytol. 2016, 211, 627–645. [Google Scholar] [CrossRef]
- Sangwongchai, W.; Krusong, K.; Thitisaksakul, M. Salt tolerance at vegetative stage is partially associated with changes in grain quality and starch physicochemical properties of rice exposed to salinity stress at reproductive stage. J. Sci. Food Agric. 2022, 102, 370–382. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Fujita, M. Salicylic acid alleviates copper toxicity in rice (Oryza sativa L.) seedlings by up-regulating antioxidative and glyoxalase systems. Ecotoxicology 2013, 22, 959–973. [Google Scholar] [CrossRef]
- Zhang, C.W.; Wei, Y.P.; Xiao, D.; Gao, L.W.; Lyu, S.W.; Hou, X.L.; Bouuema, G. Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. J. Proteom. 2016, 144, 1–10. [Google Scholar] [CrossRef]
- Kim, Y.J.; Chambers, A.G.; Cecchi, F.; Hembrough, T. Targeted data-independent acquisition for mass spectrometric detection of RAS mutations in formalin-fixed, paraffin-embedded tumor biopsies. J. Proteom. 2018, 189, 91–96. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Yang, S.; Yuan, Y. Lignin Involvement in Programmed Changes in Peach-Fruit Texture Indicated by Metabolite and Transcriptome Analyses. J. Agric. Food Chem. 2018, 66, 12627–12640. [Google Scholar] [CrossRef]
- Ying, S.; Su, M.; Wu, Y.; Zhou, L.; Fu, R.; Li, Y.; Guo, H.; Luo, J.; Wang, S.; Zhang, Y. Trichome regulator SlMIXTA-like directly manipulates primary metabolism in tomato fruit. Plant Biotechnol. J. 2020, 18, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Luo, X.; Zhan, X.; Hao, J.; Zhang, L.; Song, Y.B.; Shen, C.; Dong, M. Comparative metabolomics reveals the metabolic variations between two endangered Taxus species (T. fuana and T. yunnanensis) in the Himalayas. BMC Plant Biol. 2018, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, K.; Yao, L.; Ma, Z.; Li, C.; Si, E.; Li, B.; Meng, Y.; Ma, X.; Shang, X.; et al. Metabolomics Analyses Provide Insights into Nutritional Value and Abiotic Stress Tolerance in Halophyte Halogeton glomeratus. Front. Plant Sci. 2021, 12, 703255. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Wang, J.; Yao, L.; Li, B.; Ma, X.; Si, E.; Yang, K.; Li, C.; Shang, X.; Meng, Y.; et al. Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley. Int. J. Mol. Sci. 2022, 23, 10515. https://doi.org/10.3390/ijms231810515
Chen Y, Wang J, Yao L, Li B, Ma X, Si E, Yang K, Li C, Shang X, Meng Y, et al. Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley. International Journal of Molecular Sciences. 2022; 23(18):10515. https://doi.org/10.3390/ijms231810515
Chicago/Turabian StyleChen, Yiyou, Juncheng Wang, Lirong Yao, Baochun Li, Xiaole Ma, Erjing Si, Ke Yang, Chengdao Li, Xunwu Shang, Yaxiong Meng, and et al. 2022. "Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley" International Journal of Molecular Sciences 23, no. 18: 10515. https://doi.org/10.3390/ijms231810515
APA StyleChen, Y., Wang, J., Yao, L., Li, B., Ma, X., Si, E., Yang, K., Li, C., Shang, X., Meng, Y., & Wang, H. (2022). Combined Proteomic and Metabolomic Analysis of the Molecular Mechanism Underlying the Response to Salt Stress during Seed Germination in Barley. International Journal of Molecular Sciences, 23(18), 10515. https://doi.org/10.3390/ijms231810515






