Metabolic and Proteomic Profiles Associated with Immune Responses Induced by Different Inactivated SARS-CoV-2 Vaccine Candidates
Abstract
:1. Introduction
2. Results
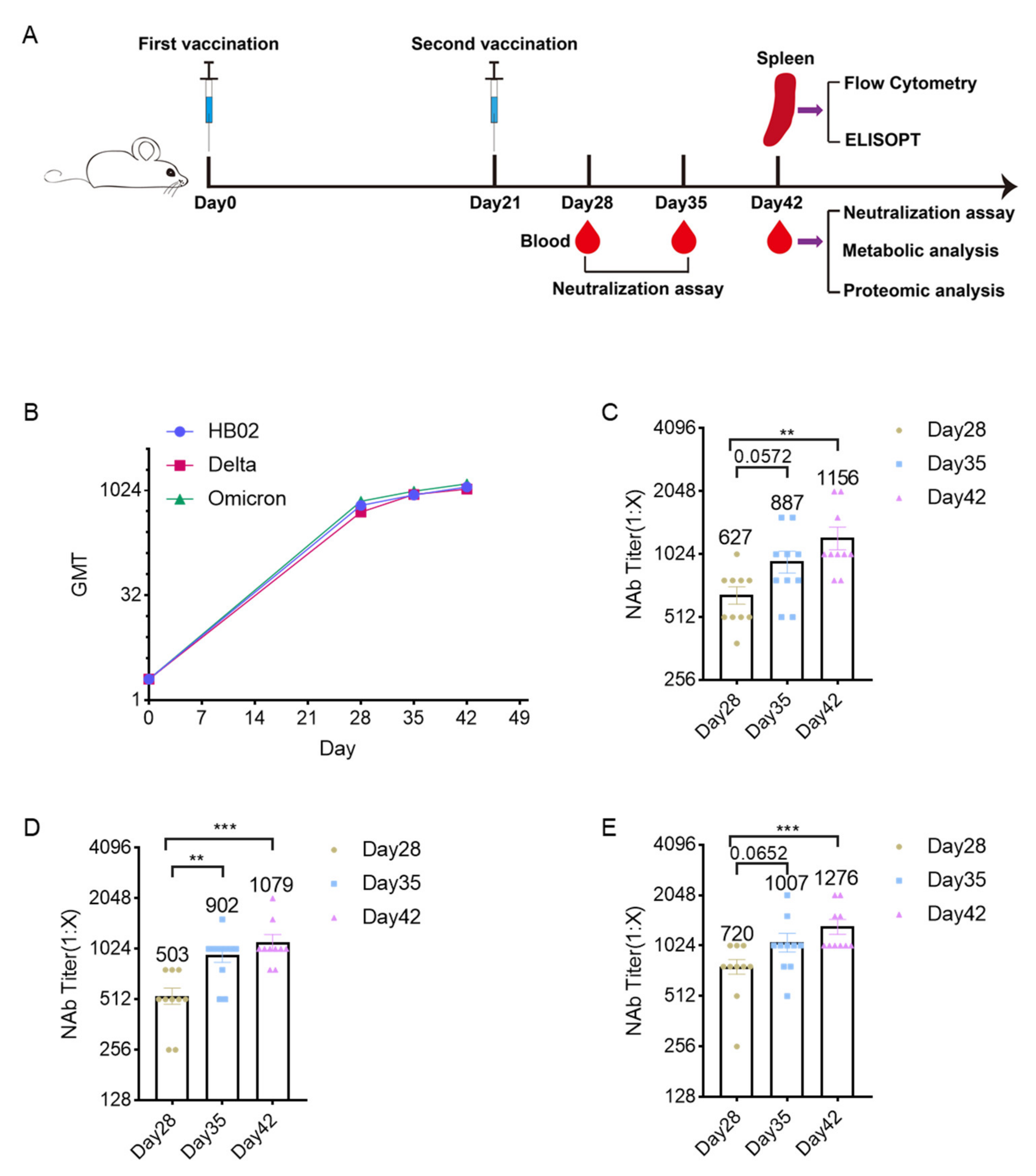
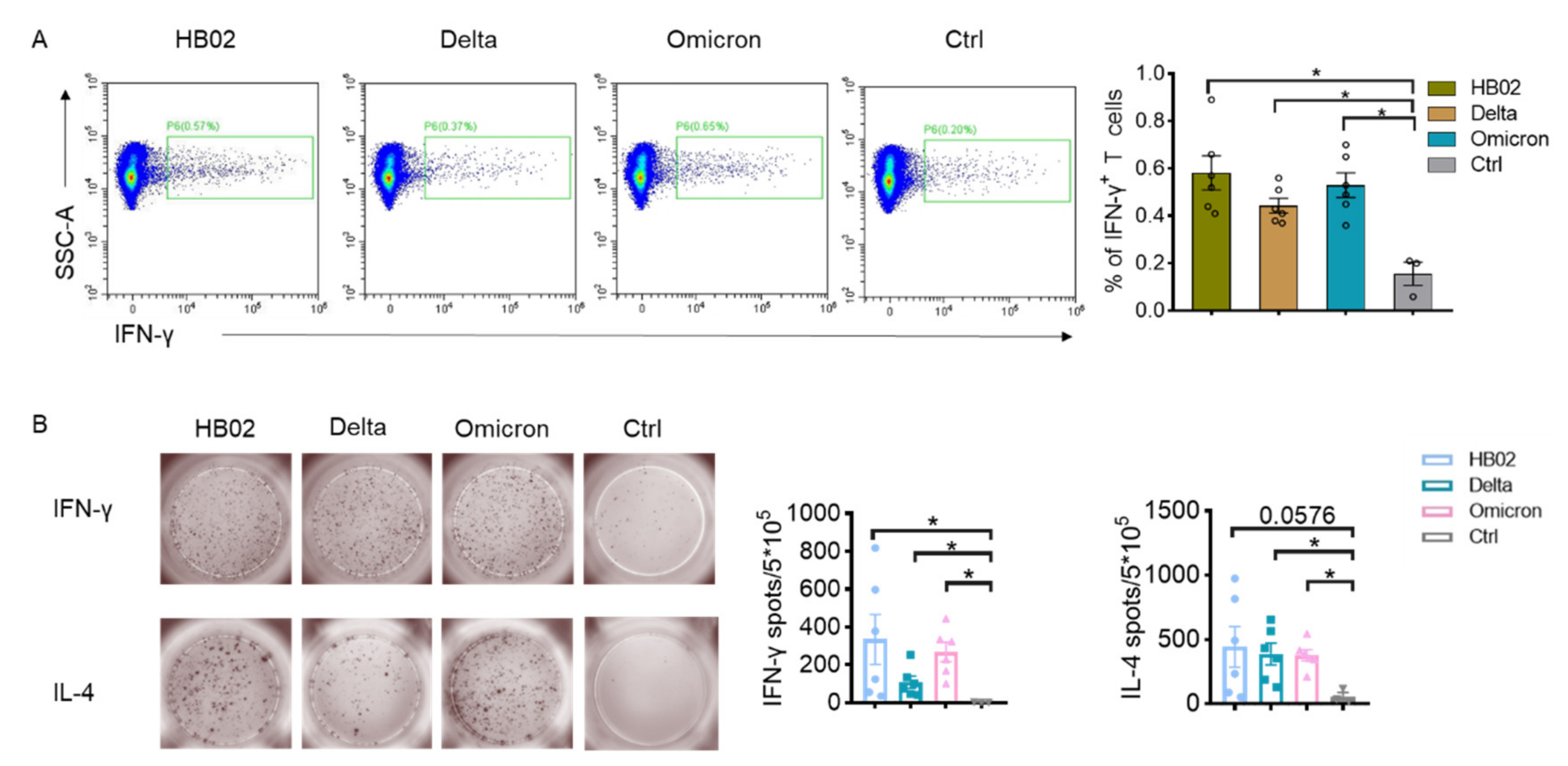
2.1. Immunogenicity Analysis
2.2. Metabolic Profiles
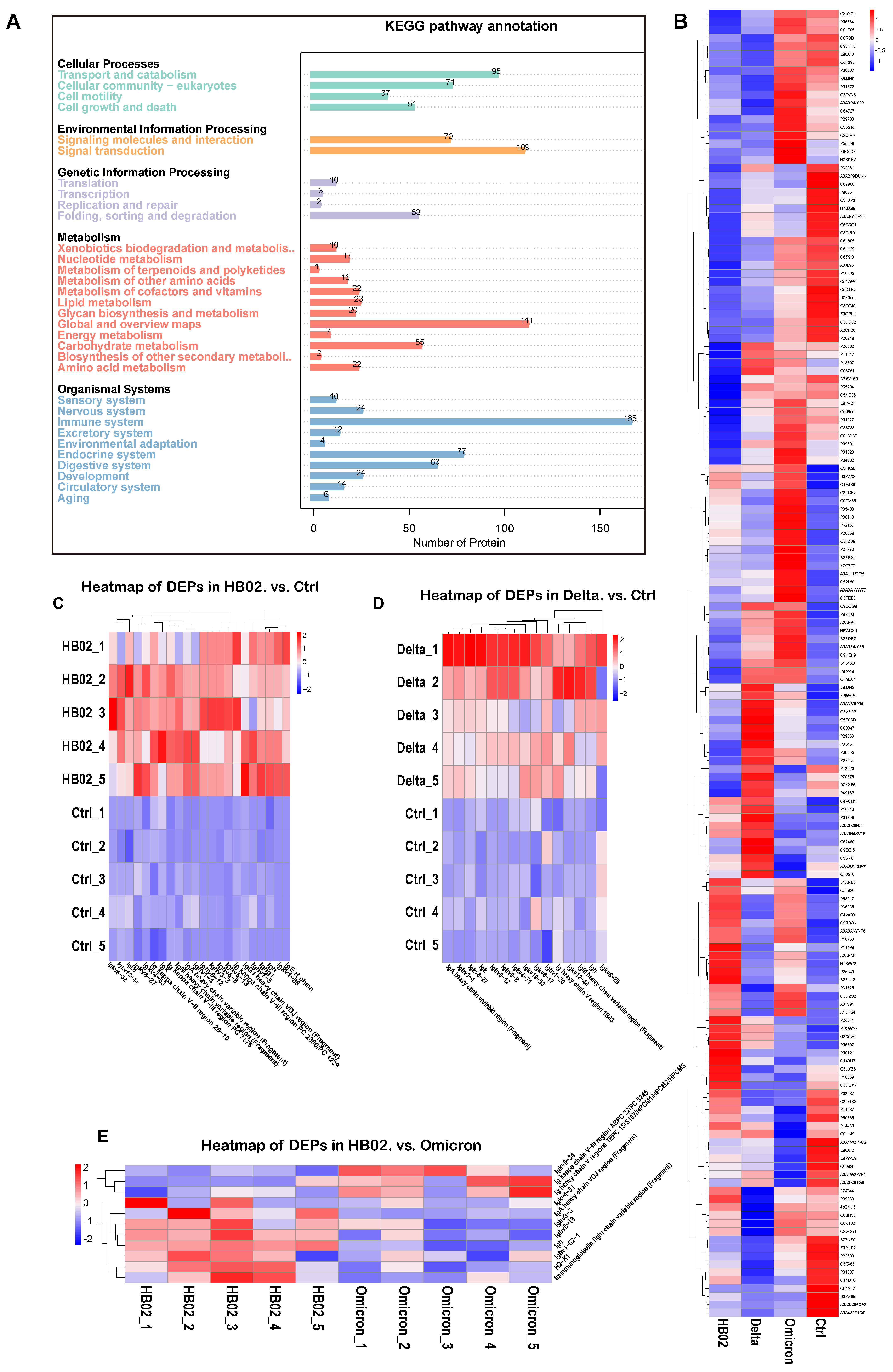
2.3. Proteomic Profiles
3. Discussion
4. Materials and Methods
4.1. Vaccine
4.2. Animal Models
4.3. Neutralization Assay
4.4. Flow Cytometry
4.5. ELISPOT Assay
4.6. Metabolic Analysis
4.7. Proteomic Analysis
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Feng, W.; Zong, W.; Wang, F.; Ju, S. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review. Mol. Cancer 2020, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Fiolet, T.; Kherabi, Y.; MacDonald, C.J.; Ghosn, J.; Peiffer-Smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2022, 28, 202–221. [Google Scholar] [CrossRef] [PubMed]
- Moghadas, S.M.; Vilches, T.N.; Zhang, K.; Wells, C.R.; Shoukat, A.; Singer, B.H.; Meyers, L.A.; Neuzil, K.M.; Langley, J.M.; Fitzpatrick, M.C. The impact of vaccination on COVID-19 outbreaks in the United States. Medrxiv Prepr. Serv. Health Sci. 2021, 73, 2257–2264. [Google Scholar]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults: A Randomized Clinical Trial. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef]
- Forchette, L.; Sebastian, W.; Liu, T. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr. Med. Sci. 2021, 41, 1037–1051. [Google Scholar] [CrossRef]
- Raeven, R.H.M.; van Riet, E.; Meiring, H.D.; Metz, B.; Kersten, G.F.A. Systems vaccinology and big data in the vaccine development chain. Immunology 2019, 156, 33–46. [Google Scholar] [CrossRef]
- Hagan, T.; Nakaya, H.I.; Subramaniam, S.; Pulendran, B. Systems vaccinology: Enabling rational vaccine design with systems biological approaches. Vaccine 2015, 33, 5294–5301. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Luu, L.D.W.; Chen, S.; Jin, F.; Wang, S.; Huang, X.; Wang, L.; Zhou, X.; Chen, X. Proteomic and Metabolomic Signatures Associated With the Immune Response in Healthy Individuals Immunized with an Inactivated SARS-CoV-2 Vaccine. Front. Immunol. 2022, 13, 848961. [Google Scholar] [CrossRef]
- Kim, K.B.; Lee, B.M. Metabolomics, a New Promising Technology for Toxicological Research. Toxicol. Res. 2009, 25, 59–69. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Carvalho, A.; La Rocca, C.; Palma, C.; Rodrigues, F.; Silvestre, R.; Kleinnijenhuis, J.; Lachmandas, E.; Gonçalves, L.G.; Belinha, A.; et al. Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell. Rep. 2016, 17, 2562–2571. [Google Scholar] [CrossRef] [Green Version]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Garout, M.A.; Al-Qaaneh, A.M.; Sule, A.A.; Tirupathi, R.; Mutair, A.A.; Alhumaid, S.; Hasan, A.; Dhawan, M.; et al. Diverse Immunological Factors Influencing Pathogenesis in Patients with COVID-19: A Review on Viral Dissemination, Immunotherapeutic Options to Counter Cytokine Storm and Inflammatory Responses. Pathogens 2021, 10, 565. [Google Scholar] [CrossRef]
- Smith, W.L.; Urade, Y.; Jakobsson, P.J. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 2011, 111, 5821–5865. [Google Scholar] [CrossRef]
- Pan, X.Y.; Wang, L.; You, H.M.; Cheng, M.; Yang, Y.; Huang, C.; Li, J. Alternative activation of macrophages by prostacyclin synthase ameliorates alcohol induced liver injury. Lab. Investig. 2021, 101, 1210–1224. [Google Scholar] [CrossRef]
- Bao, Y.S.; Zhang, P.; Xie, R.J.; Wang, M.; Wang, Z.Y.; Zhou, Z.; Zhai, W.J.; Feng, S.Z.; Han, M.Z. The regulation of CD4+ T cell immune responses toward Th2 cell development by prostaglandin E2. Int. Immunopharmacol. 2011, 11, 1599–1605. [Google Scholar] [CrossRef]
- Kalinski, P. Regulation of immune responses by prostaglandin E2. J. Immunol. 2012, 188, 21–28. [Google Scholar] [CrossRef]
- Aoki, R.; Aoki-Yoshida, A.; Suzuki, C.; Takayama, Y. Indole-3-Pyruvic Acid, an Aryl Hydrocarbon Receptor Activator, Suppresses Experimental Colitis in Mice. J. Immunol. 2018, 201, 3683–3693. [Google Scholar] [CrossRef]
- Garai, J.; Lóránd, T. Macrophage migration inhibitory factor (MIF) tautomerase inhibitors as potential novel anti-inflammatory agents: Current developments. Curr. Med. Chem. 2009, 16, 1091–1114. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Wang, T.; Gnanaprakasam JN, R.; Chen, X.; Kang, S.; Xu, X.; Sun, H.; Liu, L.; Rodgers, H.; Miller, E.; Cassel, T.A.; et al. Inosine is an alternative carbon source for CD8(+)-T-cell function under glucose restriction. Nat. Metab. 2020, 2, 635–647. [Google Scholar] [CrossRef]
- Depelchin, A.; Letesson, J.J. Adrenaline influence on the immune response. I. Accelerating or suppressor effects according to the time of application. Immunol. Lett. 1981, 3, 199–205. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Endothelial PECAM-1 and its function in vascular physiology and atherogenic pathology. Exp. Mol. Pathol. 2016, 100, 409–415. [Google Scholar] [CrossRef]
- Newman, P.J. The biology of PECAM-1. J. Clin. Investig. 1997, 99, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Hata, H.; Yoshimoto, T.; Hayashi, N.; Hada, T.; Nakanishi, K. IL-18 together with anti-CD3 antibody induces human Th1 cells to produce Th1- and Th2-cytokines and IL-8. Int. Immunol. 2004, 16, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tan, W.; Lou, Z.; Huang, B.; Zhou, W.; Zhao, Y.; Zhang, J.; Liang, H.; Li, N.; Zhu, X.; et al. Immunogenicity Evaluating of the Multivalent COVID-19 Inactivated Vaccine against the SARS-CoV-2 Variants. Vaccines 2022, 10, 956. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wang, Y.; Guan, R.; Lu, M.; Yuan, L.; Xu, W.; Hu, S. Enhanced Immune Responses with Serum Proteomic Analysis of Hu Sheep to Foot-and-Mouth Disease Vaccine Emulsified in a Vegetable Oil Adjuvant. Vaccines 2020, 8, 180. [Google Scholar] [CrossRef]
- Sasaki, E.; Kusunoki, H.; Momose, H.; Furuhata, K.; Hosoda, K.; Wakamatsu, K.; Mizukami, T.; Hamaguchi, I. Changes of urine metabolite profiles are induced by inactivated influenza vaccine inoculations in mice. Sci. Rep. 2019, 9, 16249. [Google Scholar] [CrossRef]
- McClenathan, B.M.; Stewart, D.A.; Spooner, C.E.; Pathmasiri, W.W.; Burgess, J.P.; McRitchie, S.L.; Choi, Y.S.; Sumner, S.C. Metabolites as biomarkers of adverse reactions following vaccination: A pilot study using nuclear magnetic resonance metabolomics. Vaccine 2017, 35, 1238–1245. [Google Scholar] [CrossRef]
- Adamczyk-Poplawska, M.; Markowicz, S.; Jagusztyn-Krynicka, E.K. Proteomics for development of vaccine. J. Proteom. 2011, 74, 2596–2616. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Wan, S.; Zhang, L.; Zhou, F. SARS-CoV-2 Omicron variant: Recent progress and future perspectives. Signal Transduct. Target. Ther. 2022, 7, 141. [Google Scholar] [CrossRef]
- Kupferschmidt, K. Where did ‘weird’ Omicron come from? Science 2021, 374, 1179. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]

| Experiment | Group | Total Number | Male | Female |
|---|---|---|---|---|
| Neutralization analysis | HB02 | 10 | 5 | 5 |
| Delta | 10 | 5 | 5 | |
| Omicron | 10 | 5 | 5 | |
| Immunoassay (FACS, ELISPOT) | HB02 | 6 | 3 | 3 |
| Delta | 6 | 3 | 3 | |
| Omicron | 6 | 3 | 3 | |
| Omic analysis | HB02 | 5 | 3 | 2 |
| Delta | 5 | 3 | 2 | |
| Omicron | 5 | 3 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; He, Y.; Ji, W.; Yang, R.; Zhao, Y.; Li, Y.; Liu, Y.; Ding, L.; Ma, M.; Wang, H.; et al. Metabolic and Proteomic Profiles Associated with Immune Responses Induced by Different Inactivated SARS-CoV-2 Vaccine Candidates. Int. J. Mol. Sci. 2022, 23, 10644. https://doi.org/10.3390/ijms231810644
Yu S, He Y, Ji W, Yang R, Zhao Y, Li Y, Liu Y, Ding L, Ma M, Wang H, et al. Metabolic and Proteomic Profiles Associated with Immune Responses Induced by Different Inactivated SARS-CoV-2 Vaccine Candidates. International Journal of Molecular Sciences. 2022; 23(18):10644. https://doi.org/10.3390/ijms231810644
Chicago/Turabian StyleYu, Shouzhi, Yao He, Wenheng Ji, Rong Yang, Yuxiu Zhao, Yan Li, Yingwei Liu, Ling Ding, Meng Ma, Hui Wang, and et al. 2022. "Metabolic and Proteomic Profiles Associated with Immune Responses Induced by Different Inactivated SARS-CoV-2 Vaccine Candidates" International Journal of Molecular Sciences 23, no. 18: 10644. https://doi.org/10.3390/ijms231810644






