Skeletal Class III Malocclusion Is Associated with ADAMTS2 Variants and Reduced Expression in a Familial Case
Abstract
:1. Introduction
2. Results
2.1. ADAMTS2 Is Associated with Maxillary Development
2.2. Expression of the Mutation of ADAMTS2 in HEK293 and Proband-Derived Dental Pulp Stem Cells (DPSCs)
2.3. Expression and Phenotype of ADAMTS2 during Development
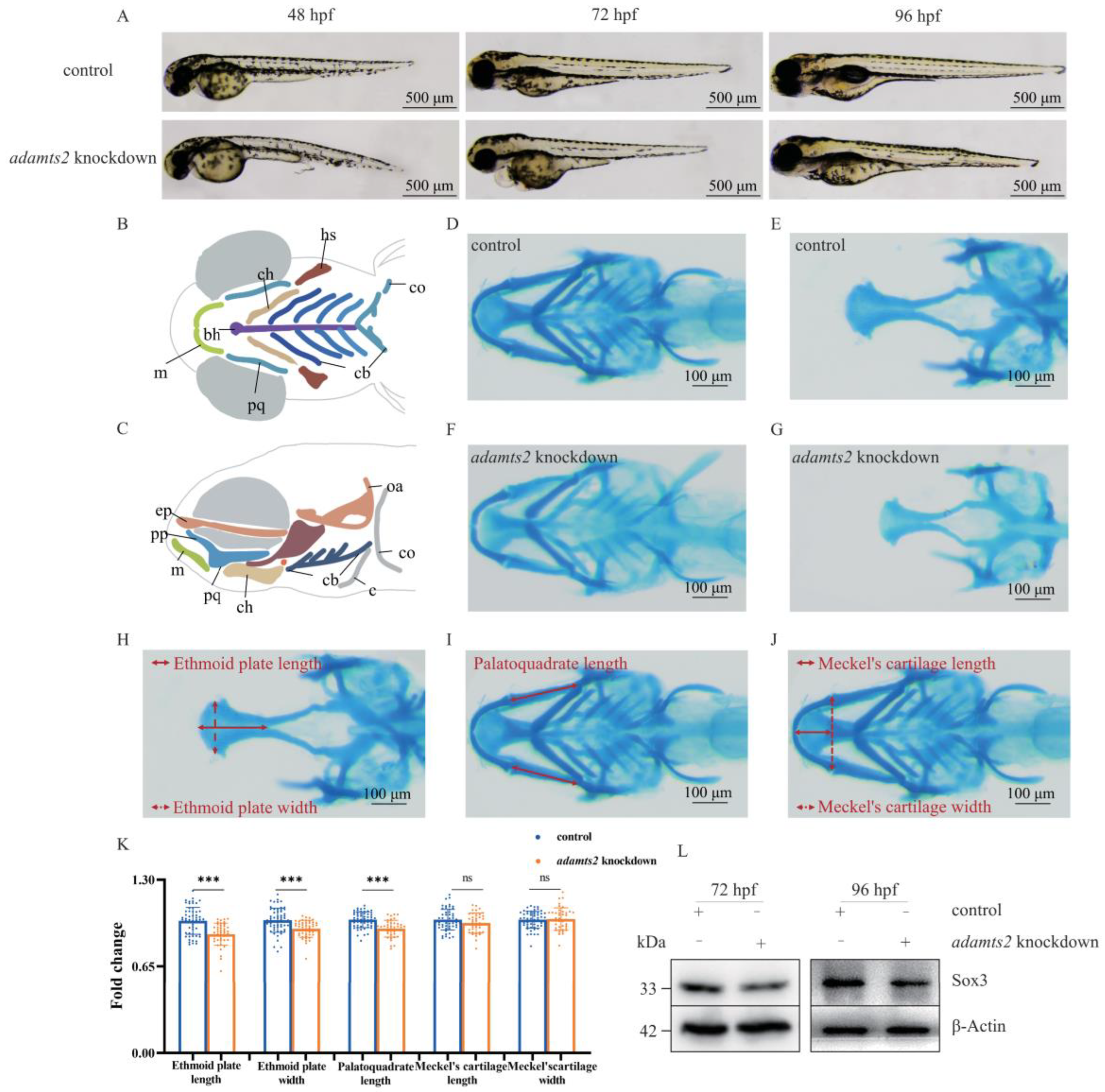
2.4. Adamts2 Deficiency Results in Zebrafish Craniofacial Defects
2.5. ADAMTS2 Deficiency Disrupts EGFR Signaling
3. Discussion
4. Materials and Methods
4.1. Family and Cohort Recruitment
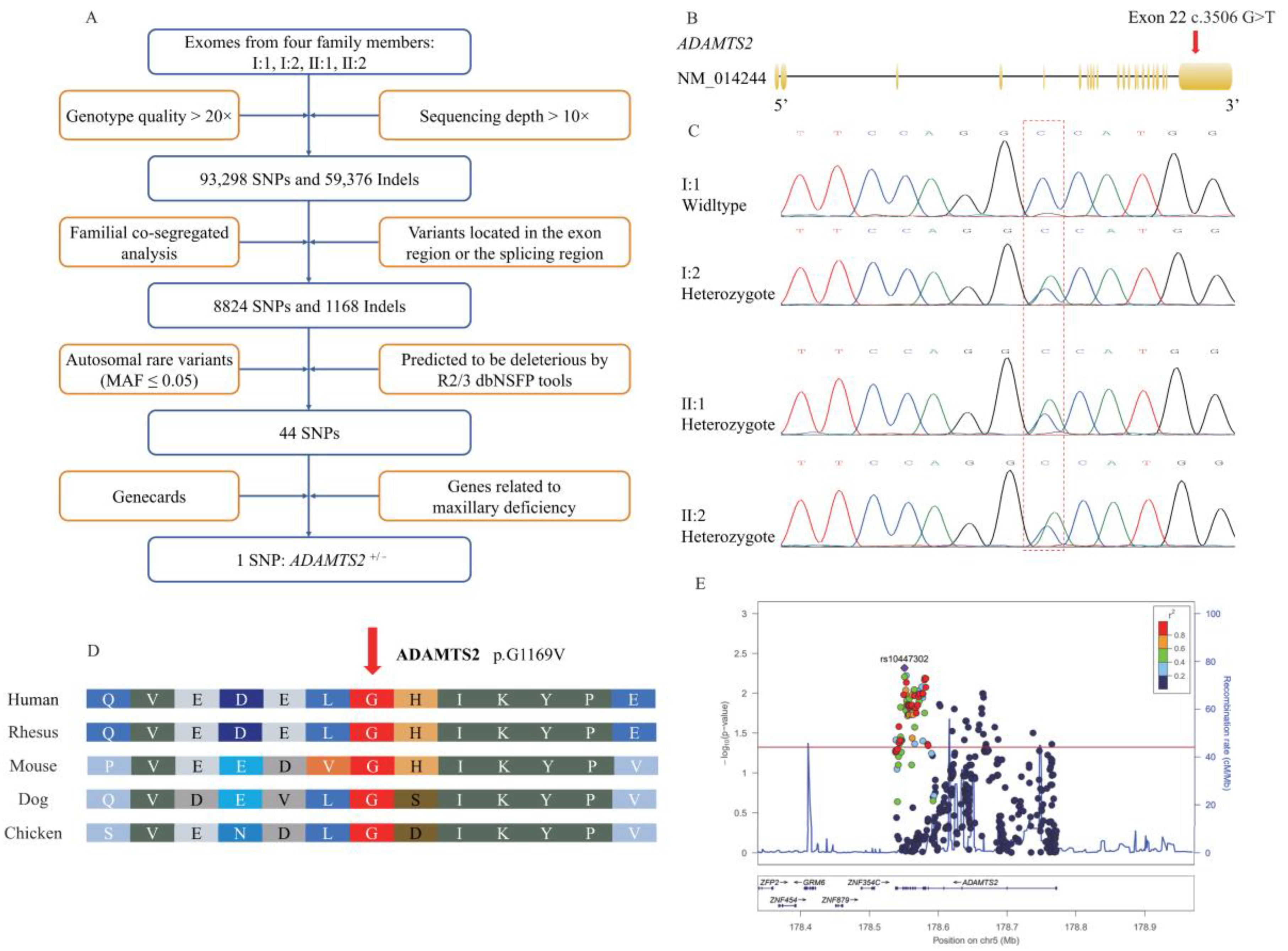
4.2. WES, Causative Variant Screening, Sanger Analysis, Species Conservation Analysis, Protein Structure Prediction, and Tolerance Prediction
4.3. Genotyping, Data Processing, Quality Control (QC) and Variant Selection
4.4. Cell Culture, Plasmid and Small Interference RNA (siRNA) Constructs, and Transfection
4.5. Zebrafish Models
4.6. Alcian Blue Staining
4.7. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
4.8. Western Blot
4.9. Co-Expression, Correlation, and Pathway Enrichment Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Consent to Participate
Consent for Publication
References
- Dehesa-Santos, A.; Iber-Diaz, P.; Iglesias-Linares, A. Genetic factors contributing to skeletal class III malocclusion: A systematic review and meta-analysis. Clin. Oral Investig. 2021, 25, 1587–1612. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.K.; Ye, X.; Taub, P.J. Review of the Genetic Basis of Jaw Malformations. J. Pediatr. Genet. 2016, 5, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Eslami, S.; Faber, J.; Fateh, A.; Sheikholaemmeh, F.; Grassia, V.; Jamilian, A. Treatment decision in adult patients with class III malocclusion: Surgery versus orthodontics. Prog. Orthod. 2018, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Xie, C.; Yang, H.; Wu, C.; Ren, A. Prevalence of malocclusion in Chinese schoolchildren from 1991 to 2018: A systematic review and meta-analysis. Int. J. Paediatr. Dent 2020, 30, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Sato, H.; Suga, H.; Takemoto, Y.; Inada, E.; Saitoh, I.; Kakuno, E.; Kanomi, R.; Yamasaki, Y. Relationships among nasal resistance, adenoids, tonsils, and tongue posture and maxillofacial form in Class II and Class III children. Am. J. Orthod. Dentofac. Orthop. Off. Publ. Am. Assoc. Orthod. Its Const. Soc. Am. Board Orthod. 2017, 151, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Meng, M.; Law, C.S.; Rao, Y.; Zhou, X. Common dental diseases in children and malocclusion. Int. J. Oral Sci. 2018, 10, 7. [Google Scholar] [CrossRef]
- Chai, Y.; Maxson, R.E. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006, 235, 2353–2375. [Google Scholar] [CrossRef]
- Yuan, Y.; Chai, Y. Regulatory mechanisms of jaw bone and tooth development. Curr. Top. Dev. Biol. 2019, 133, 91–118. [Google Scholar] [CrossRef]
- Doraczynska-Kowalik, A.; Nelke, K.H.; Pawlak, W.; Sasiadek, M.M.; Gerber, H. Genetic Factors Involved in Mandibular Prognathism. J. Craniofac. Surg. 2017, 28, e422–e431. [Google Scholar] [CrossRef]
- Oh, J.; Wang, C.J.; Poole, M.; Kim, E.; Davis, R.C.; Nishimura, I.; Pae, E.K. A genome segment on mouse chromosome 12 determines maxillary growth. J. Dent. Res. 2007, 86, 1203–1206. [Google Scholar] [CrossRef]
- Nikopensius, T.; Saag, M.; Jagomagi, T.; Annilo, T.; Kals, M.; Kivistik, P.A.; Milani, L.; Metspalu, A. A missense mutation in DUSP6 is associated with Class III malocclusion. J. Dent. Res. 2013, 92, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, Q.; Gu, M.; Li, X.; Yu, J.; Zhang, Y.B. Identification of a Mutation in FGF23 Involved in Mandibular Prognathism. Sci. Rep. 2015, 5, 11250. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Samocha, K.E.; Robinson, E.B.; Sanders, S.J.; Stevens, C.; Sabo, A.; McGrath, L.M.; Kosmicki, J.A.; Rehnström, K.; Mallick, S.; Kirby, A.; et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014, 46, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Gulick, J.; Colbert, M.C.; Robbins, J. Protein tyrosine phosphatase activity in the neural crest is essential for normal heart and skull development. Proc. Natl. Acad. Sci. USA 2009, 106, 11270–11275. [Google Scholar] [CrossRef]
- Alansary, M.; Drummond, B.; Coates, D. Immunocytochemical characterization of primary teeth pulp stem cells from three stages of resorption in serum-free medium. Dent. Traumatol. 2021, 37, 90–102. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Firth, H.V.; Richards, S.M.; Bevan, A.P.; Clayton, S.; Corpas, M.; Rajan, D.; Van Vooren, S.; Moreau, Y.; Pettett, R.M.; Carter, N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009, 84, 524–533. [Google Scholar] [CrossRef]
- Swartz, M.E.; Sheehan-Rooney, K.; Dixon, M.J.; Eberhart, J.K. Examination of a palatogenic gene program in zebrafish. Dev. Dyn. 2011, 240, 2204–2220. [Google Scholar] [CrossRef]
- Rizzoti, K.; Lovell-Badge, R. SOX3 activity during pharyngeal segmentation is required for craniofacial morphogenesis. Development 2007, 134, 3437–3448. [Google Scholar] [CrossRef] [Green Version]
- Linder, M.; Hecking, M.; Glitzner, E.; Zwerina, K.; Holcmann, M.; Bakiri, L.; Ruocco, M.G.; Tuckermann, J.; Schett, G.; Wagner, E.F.; et al. EGFR controls bone development by negatively regulating mTOR-signaling during osteoblast differentiation. Cell Death Differ. 2018, 25, 1094–1106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yao, X.; Yan, G.; Xu, Y.; Yan, J.; Zou, W.; Wang, G. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun. 2016, 7, 11149. [Google Scholar] [CrossRef] [PubMed]
- Kusuyama, J.; Amir, M.S.; Albertson, B.G.; Bandow, K.; Ohnishi, T.; Nakamura, T.; Noguchi, K.; Shima, K.; Semba, I.; Matsuguchi, T. JNK inactivation suppresses osteogenic differentiation, but robustly induces osteopontin expression in osteoblasts through the induction of inhibitor of DNA binding 4 (Id4). FASEB J. 2019, 33, 7331–7347. [Google Scholar] [CrossRef] [PubMed]
- Tokuzawa, Y.; Yagi, K.; Yamashita, Y.; Nakachi, Y.; Nikaido, I.; Bono, H.; Ninomiya, Y.; Kanesaki-Yatsuka, Y.; Akita, M.; Motegi, H.; et al. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS Genet. 2010, 6, e1001019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ji, D.; Li, L.; Yang, S.; Zhang, H.; Duan, X. ClC-7 Regulates the Pattern and Early Development of Craniofacial Bone and Tooth. Theranostics 2019, 9, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tamasi, J.; Lu, X.; Zhu, J.; Chen, H.; Tian, X.; Lee, T.-C.; Threadgill, D.W.; Kream, B.E.; Kang, Y.; et al. Epidermal growth factor receptor plays an anabolic role in bone metabolism in vivo. J. Bone Miner. Res. 2011, 26, 1022–1034. [Google Scholar] [CrossRef]
- Zhu, J.; Shimizu, E.; Zhang, X.; Partridge, N.C.; Qin, L. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J. Cell Biochem. 2011, 112, 1749–1760. [Google Scholar] [CrossRef]
- Lees-Shepard, J.B.; Flint, K.; Fisher, M.; Omi, M.; Richard, K.; Antony, M.; Chen, P.J.; Yadav, S.; Threadgill, D.; Maihle, N.J.; et al. Cross-talk between EGFR and BMP signals regulates chondrocyte maturation during endochondral ossification. Dev. Dyn. 2022, 251, 75–94. [Google Scholar] [CrossRef]
- Miettinen, P.J.; Chin, J.R.; Shum, L.; Slavkin, H.C.; Shuler, C.F.; Derynck, R.; Werb, Z. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat. Genet. 1999, 22, 69–73. [Google Scholar] [CrossRef]
- Küchler, E.C.; Reis, C.L.B.; Carelli, J.; Scariot, R.; Nelson-Filho, P.; Coletta, R.D.; Paza, A.O.; Matsumoto, M.A.N.; Proff, P.; Kirschneck, C. Potential interactions among single nucleotide polymorphisms in bone- and cartilage-related genes in skeletal malocclusions. Orthod. Craniofac. Res. 2021, 24, 277–287. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Ahmed, N.; Bertels, K.; Al-Ars, Z. GPU accelerated sequence alignment with traceback for GATK HaplotypeCaller. BMC Genom. 2019, 20, 184. [Google Scholar] [CrossRef]
- Ma, L.; Lou, S.; Miao, Z.; Yao, S.; Yu, X.; Kan, S.; Zhu, G.; Yang, F.; Zhang, C.; Zhang, W.; et al. Identification of novel susceptibility loci for non-syndromic cleft lip with or without cleft palate. J. Cell. Mol. Med. 2020, 24, 13669–13678. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Rooryck, C.; Diaz-Font, A.; Osborn, D.P.S.; Chabchoub, E.; Hernandez-Hernandez, V.; Shamseldin, H.; Kenny, J.; Waters, A.; Jenkins, D.; Kaissi, A.A.; et al. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat. Genet. 2011, 43, 197–203. [Google Scholar] [CrossRef]
- Sun, R.; Wang, Y.; Jin, M.; Chen, L.; Cao, Y.; Chen, F. Identification and Functional Studies of MYO1H for Mandibular Prognathism. J. Dent. Res. 2018, 97, 1501–1509. [Google Scholar] [CrossRef]
- Lou, S.; Ma, L.; Kan, S.; Yu, X.; Wang, Y.; Yang, F.; Zhu, G.; Fan, L.; Li, D.; Wang, H.; et al. Association Study of Genetic Variants in Autophagy Pathway and Risk of Non-syndromic Cleft Lip With or Without Cleft Palate. Front. Cell Dev. Biol. 2020, 8, 576. [Google Scholar] [CrossRef]
- Wang, Y.; Li, D.; Xu, Y.; Ma, L.; Lu, Y.; Wang, Z.; Wang, L.; Zhang, W.; Pan, Y. Functional Effects of SNPs in MYH9 and Risks of Nonsyndromic Orofacial Clefts. J. Dent. Res. 2018, 97, 388–394. [Google Scholar] [CrossRef]
- Hamidouche, Z.; Fromigué, O.; Ringe, J.; Häupl, T.; Vaudin, P.; Pagès, J.-C.; Srouji, S.; Livne, E.; Marie, P.J. Priming integrin alpha5 promotes human mesenchymal stromal cell osteoblast differentiation and osteogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 18587–18591. [Google Scholar] [CrossRef]
- Timmons, J.A.; Szkop, K.J.; Gallagher, I.J. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol. 2015, 16, 186. [Google Scholar] [CrossRef] [Green Version]
- Brinkley, J.F.; Fisher, S.; Harris, M.P.; Holmes, G.; Hooper, J.E.; Jabs, E.W.; Jones, K.L.; Kesselman, C.; Klein, O.D.; Maas, R.L.; et al. The FaceBase Consortium: A comprehensive resource for craniofacial researchers. Development 2016, 143, 2677–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
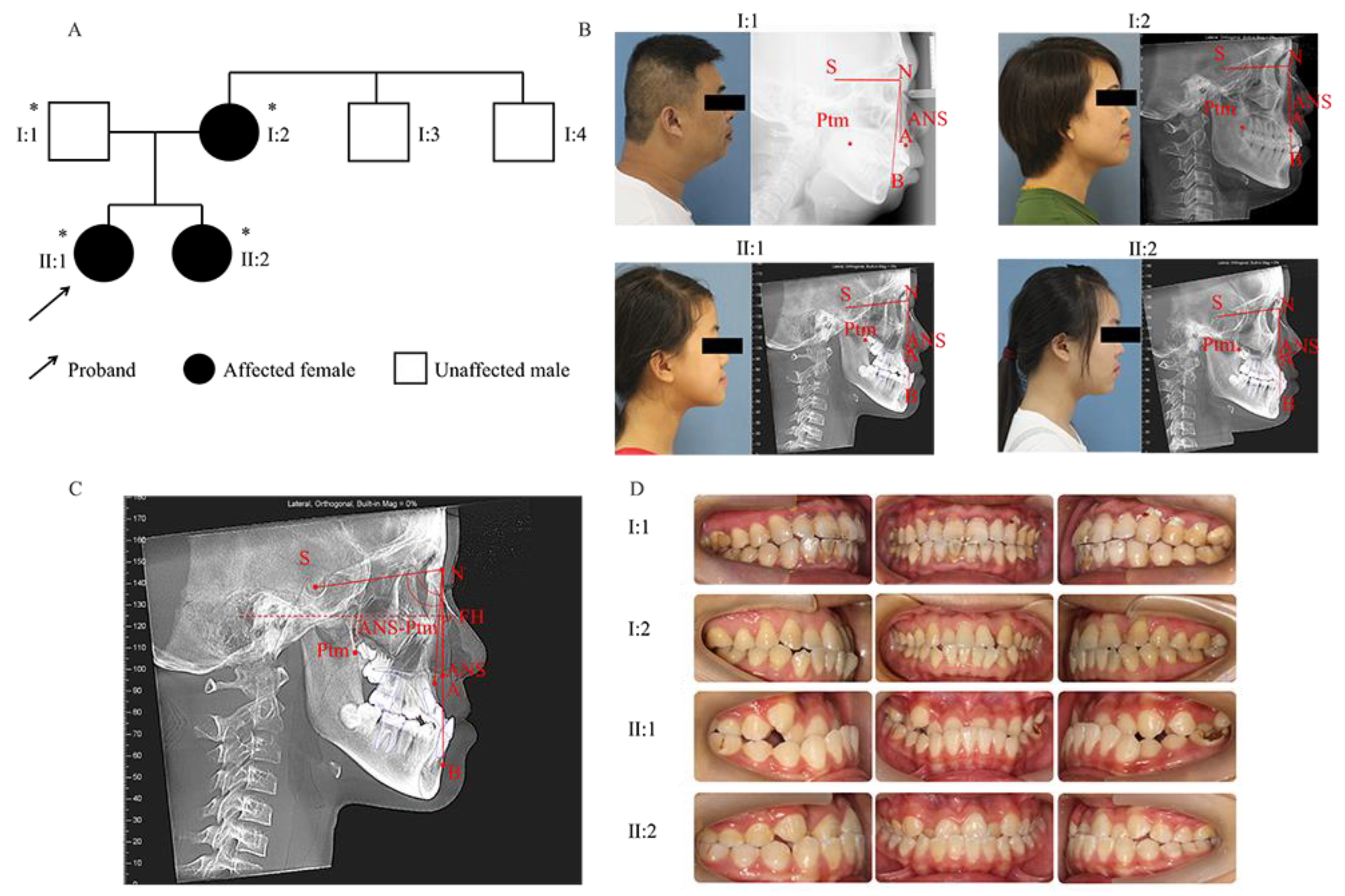

| Cephalometric Variables | I:1 | I:2 | II:1 | II:2 | Norms * |
|---|---|---|---|---|---|
| ANB(°) | 1.2 | −1.3 | −3.6 | −3.1 | 2.7 ± 2.0 |
| SNA(°) | 81.8 | 78.3 | 78.3 | 78.4 | 82.8 ± 4.0 |
| SNB(°) | 80.6 | 79.5 | 81.9 | 81.5 | 80.1 ± 3.9 |
| ANS-Ptm (mm) | 52.0 | 42.9 | 42.3 | 43.1 | Male permanent dentition: 52.1 ± 2.8 Female permanent dentition: 49.9 ± 2.1 Early female permanent dentition: 47.7 ± 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, S.; Zhou, X.; Vona, B.; Fan, L.; Zhang, C.; Li, D.; Yuan, H.; Du, Y.; Ma, L.; Pan, Y. Skeletal Class III Malocclusion Is Associated with ADAMTS2 Variants and Reduced Expression in a Familial Case. Int. J. Mol. Sci. 2022, 23, 10673. https://doi.org/10.3390/ijms231810673
Yao S, Zhou X, Vona B, Fan L, Zhang C, Li D, Yuan H, Du Y, Ma L, Pan Y. Skeletal Class III Malocclusion Is Associated with ADAMTS2 Variants and Reduced Expression in a Familial Case. International Journal of Molecular Sciences. 2022; 23(18):10673. https://doi.org/10.3390/ijms231810673
Chicago/Turabian StyleYao, Siyue, Xi Zhou, Barbara Vona, Liwen Fan, Chengcheng Zhang, Dandan Li, Hua Yuan, Yifei Du, Lan Ma, and Yongchu Pan. 2022. "Skeletal Class III Malocclusion Is Associated with ADAMTS2 Variants and Reduced Expression in a Familial Case" International Journal of Molecular Sciences 23, no. 18: 10673. https://doi.org/10.3390/ijms231810673






