MicroRNA398: A Master Regulator of Plant Development and Stress Responses
Abstract
:1. Introduction
2. Evolutionary Analysis of MiR398
3. MiR398 Has Both Conserved and Species-Specific Target Genes
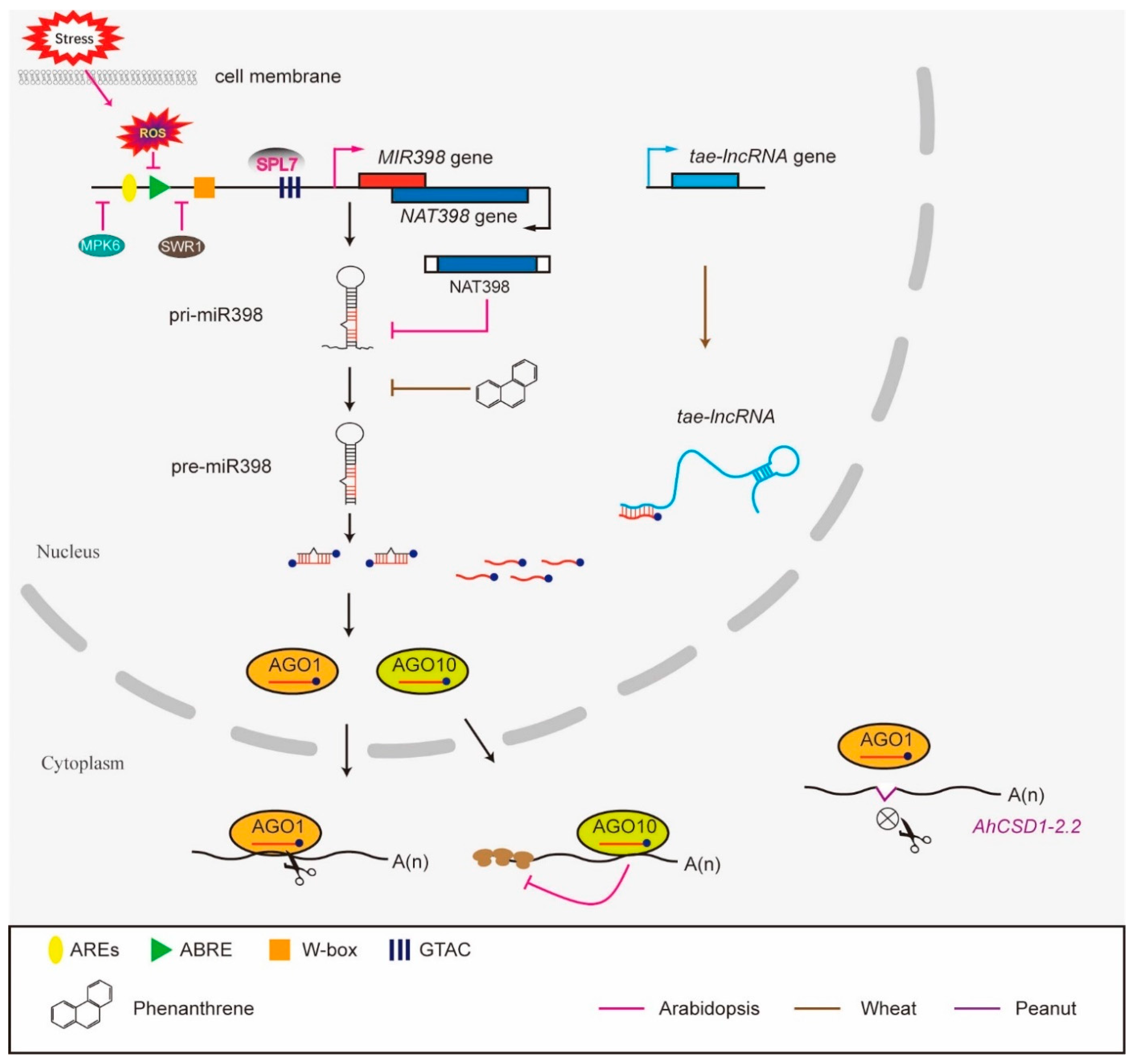
4. Multi-Layered Functional Regulation of MiR398
5. The Mode of Action of MiR398 on Target Genes
6. Role of MiR398 and Its Targets in Abiotic Stress Responses
6.1. Oxidative Stress
6.2. Heavy-Metal, Environmental-Pollutant, High-Light, Ozone and UV Stress
6.3. Salinity and Drought Stress
6.4. Water Deficit and Flood Stress
6.5. Thermal Stress
6.6. MiR398 in Nutrient Homeostasis
| Abiotic Stress | Species | MiR398 | Target Genes | References |
|---|---|---|---|---|
| Salt | Ath | ↓ | CSD1↑, CSD2– | [23] |
| Pte | ↑&↓ | CSD1↓&↑ | [23] | |
| Ghi | ↓ | nd | [47] | |
| Tae | ↓ | nd | [21] | |
| Sly | ↓ | CSD1↑ | [48,49] | |
| Iba | root↓; leaves↑ | nd | [50] | |
| Drought | Osa | ↓ | CSD1↑, CSD2↑ | [43] |
| Ttu | ↑ | nd | [52] | |
| Sly | ↓ | nd | [48] | |
| Ghi | ↓ | nd | [51] | |
| Pvi | – | nd | [53] | |
| Ahy | ↑ | CSD1↓, CSD2↓ | [30] | |
| Water deficit | Mtr | ↑ | COX5b↓ | [54] |
| Psa | ↓ | CSD1↑, COX5b– | [55] | |
| Pvu | ↓ | CSD1↑ | [56] | |
| Flooding | Ath | ↓ | nd | [57] |
| Cold | Ath | ↓ | nd | [6] |
| Cdi | ↓ | CSD1↑, CSD2↑ | [58] | |
| Tae | ↓ | CSD1↑ | [21,28] | |
| Bra | nd | [59] | ||
| Heat | Ath | ↑ | CSD1↓, CSD2↓, CCS1↓, COX5b– | [62] |
| Zma | ↑ | nd | [62] | |
| Pto | ↓ | nd | [64] | |
| Bra | ↓ | CSD1↑ | [63] | |
| Han | root↓; leaves↑ | nd | [65] | |
| Pvi | ↓ | nd | [53] | |
| High light | Ath | ↓ | CSD1↑, CSD2↑ | [11] |
| Osa | ↓ | CSD1↑, CSD2↑ | [43] | |
| Heavy metal | Ath | ↓ | CSD1↑, CSD2↑ | [11] |
| Vvi | ↓ | CSD1↑, CSD2↑ | [40] | |
| Cca | ↓ | CSD1↑, CSD2↑ | [41] | |
| Methyl viologen | Ath | ↓ | CSD1↑, CSD2↑ | [11] |
| Ozone | Ath | ↓ | CSD1↑, CSD2– | [44] |
| UV | Ath | ↑ | nd | [45] |
| Pte | ↑ | nd | [46] | |
| SO2 | Ath | ↓ | CSD1↑, CSD2 | [42] |
| Phenanthrene | Tae | ↓ | CSD1↑, CSD2↑ | [26] |
| Nickel | Rco | ↓ | Cu-Zn/SOD↑ | [39] |
7. Role of MiR398 and Its Targets in Biotic Stress Responses
| Biotic Stresses | Species | MiR398 | Target Genes | References |
|---|---|---|---|---|
| Bacteria | ||||
| Pst DC3000 | Ath | ↓ | CSD1↑, CSD2– | [44] |
| Flg22 | Ath | ↓ | CSD1↑, CSD2↑, COX5b↑ | [74] |
| Ca. L. | Csi | ↓ | nd | [78] |
| Fungus | ||||
| M. oryzae | Osa | ↑ | nd | [79,80] |
| bgh | Hvu | ↓ | HvSOD1↑ | [81] |
| S. scleortiorum | Pvu | ↓ | CSD1↑, NOD19↑ | [17] |
| F. culmorum | Tae | ↑ | nd | [82] |
| Virus | ||||
| TMV | Ath | ↑ | nd | [84] |
| ORMV | Ath | ↑ | nd | [83] |
| TolCNV | Sly | ↑ | CSD1↓, CSD2↓ | [85] |
| PVX/Y | Nbe | ↑ | CSD1↑, CSD2↑ | [86] |
| PMeV | Cpa | ↑ | nd | [87] |
| PSTVd | Sly | ↑ | nd | [88] |
| BaMV | Bdi | ↑ | CSD2↑ | [89] |
| BNYVV | Nbe | ↑ | nd | [90] |
8. Role of MiR398 in Plant Growth and Development
9. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moshelion, M.; Altman, A. Current challenges and future perspectives of plant and agricultural biotechnology. Trends Biotechnol. 2015, 33, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Matsuoka, M. Genetic approaches to crop improvement: Responding to environmental and population changes. Nat. Rev. Genet. 2008, 9, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef]
- Chávez Montes, R.A.; De Fátima Rosas-Cáárdenas, F.; De Paoli, E.; Accerbi, M.; Rymarquis, L.A.; Mahalingam, G.; Marsch-Martínez, N.; Meyers, B.C.; Green, P.J.; de Folter, S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 2014, 5, 3722. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.-K. Novel and Stress-Regulated MicroRNAs and Other Small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W. Conservation and divergence in plant microRNAs. Plant Mol. Biol. 2011, 80, 3–16. [Google Scholar] [CrossRef]
- Bonnet, E.; Wuyts, J.; Rouzé, P.; Van de Peer, Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 11511–11516. [Google Scholar] [CrossRef]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD Gene Family in Tomato: Identification, Phylogenetic Relationships, and Expression Patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Kapoor, A.; Zhu, J.-K. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Higashi, Y.; Takechi, K.; Takano, H.; Takio, S. Involvement of microRNA in copper deficiency-induced repression of chloroplastic CuZn-superoxide dismutase genes in the moss Physcomitrella patens. Plant. Cell Physiol. 2013, 54, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Beauclair, L.; Yu, A.; Bouche, N. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant. J. 2010, 62, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-F.; Zheng, Y.; Addo-Quaye, C.; Zhang, L.; Saini, A.; Jagadeeswaran, G.; Axtell, M.J.; Zhang, W.; Sunkar, R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010, 62, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Brousse, C.; Liu, Q.; Beauclair, L.; Deremetz, A.; Axtell, M.J.; Bouché, N. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014, 42, 5270–5279. [Google Scholar] [CrossRef]
- Huen, A.; Bally, J.; Smith, P. Identification and characterisation of microRNAs and their target genes in phosphate-starved Nicotiana benthamiana by small RNA deep sequencing and 5’RACE analysis. BMC Genom. 2018, 19, 940. [Google Scholar] [CrossRef]
- Naya, L.; Paul, S.; Valdés-López, O.; Mendoza-Soto, A.B.; Nova-Franco, B.; Sosa-Valencia, G.; Reyes, J.L.; Hernandez, G. Regulation of Copper Homeostasis and Biotic Interactions by MicroRNA 398b in Common Bean. PLoS ONE 2014, 9, e84416. [Google Scholar] [CrossRef]
- Song, Q.-X.; Liu, Y.-F.; Hu, X.-Y.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 2011, 11, 5. [Google Scholar] [CrossRef]
- Chen, C.; Galon, Y.; Ishka, M.R.; Malihi, S.; Shimanovsky, V.; Twito, S.; Rath, A.; Vatamaniuk, O.K.; Miller, G. ASCORBATE PEROXIDASE6 delays the onset of age-dependent leaf senescence. Plant Physiol. 2021, 185, 441–456. [Google Scholar] [CrossRef]
- Cai, H.; Liu, L.; Zhang, M.; Chai, M.; Huang, Y.; Chen, F.; Yan, M.; Su, Z.; Henderson, I.; Palanivelu, R.; et al. Spatiotemporal control of miR398 biogenesis, via chromatin remodeling and kinase signaling, ensures proper ovule development. Plant Cell 2021, 33, 1530–1553. [Google Scholar] [CrossRef]
- Wang, B.; Sun, Y.-F.; Song, N.; Wei, J.-P.; Wang, X.-J.; Feng, H.; Yin, Z.-Y.; Kang, Z.-S. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol. Biochem. 2014, 80, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Wang, W.-X.; Ren, L.; Chen, Q.-J.; Mendu, V.; Willcut, B.; Dinkins, R.; Tang, X.; Tang, G. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of Copper Homeostasis by Micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein–Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Li, J.; Shen, Y.; Zhu, J.; Liu, S.; Zeng, N.; Zhan, X. miR398 is involved in the relief of phenanthrene-induced oxidative toxicity in wheat roots. Environ. Pollut. 2020, 258, 113701. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 535. [Google Scholar] [CrossRef]
- Lu, Q.; Guo, F.; Xu, Q.; Cang, J. LncRNA improves cold resistance of winter wheat by interacting with miR398. Funct. Plant Biol. 2020, 47, 544–555. [Google Scholar] [CrossRef]
- Zhan, J. Get out and stay out: Spatiotemporally regulated miR398 biogenesis enables proper ovule development. Plant Cell 2021, 33, 1403–1404. [Google Scholar] [CrossRef]
- Park, S.Y.; Grabau, E. Bypassing miRNA-mediated gene regulation under drought stress: Alternative splicing affects CSD1 gene expression. Plant. Mol. Biol. 2017, 95, 243–252. [Google Scholar] [CrossRef]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread Translational Inhibition by Plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; et al. MicroRNAs Inhibit the Translation of Target mRNAs on the Endoplasmic Reticulum in Arabidopsis. Cell 2013, 153, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dong, W.; Ren, W.; Zhao, Q.; Wu, F.; He, Y. Cytoplasmic HYL1 modulates miRNA-mediated translational repression. Plant. Cell 2021, 33, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef]
- Iwakawa, H.; Tomari, Y. Molecular Insights into microRNA-Mediated Translational Repression in Plants. Mol. Cell 2013, 52, 591–601. [Google Scholar] [CrossRef]
- Chu, C.-C.; Lee, W.-C.; Guo, W.-Y.; Pan, S.-M.; Chen, L.-J.; Li, H.-M.; Jinn, T.-L. A Copper Chaperone for Superoxide Dismutase That Confers Three Types of Copper/Zinc Superoxide Dismutase Activity in Arabidopsis. Plant Physiol. 2005, 139, 425–436. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant. Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Çelik, Ö.; Akdaş, E.Y. Tissue-specific transcriptional regulation of seven heavy metal stress-responsive miRNAs and their putative targets in nickel indicator castor bean (R. communis L.) plants. Ecotoxicol. Environ. Saf. 2019, 170, 682–690. [Google Scholar] [CrossRef]
- Leng, X.; Wang, P.; Zhu, X.; Li, X.; Zheng, T.; Shangguan, L.; Fang, J. Ectopic expression of CSD1 and CSD2 targeting genes of miR398 in grapevine is associated with oxidative stress tolerance. Funct. Integr. Genom. 2017, 17, 697–710. [Google Scholar] [CrossRef]
- Sun, Z.; Shu, L.; Zhang, W.; Wang, Z. Cca-miR398 increases copper sulfate stress sensitivity via the regulation of CSD mRNA transcription levels in transgenic Arabidopsis thaliana. PeerJ 2020, 8, e9105. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yi, H.; Xue, M.; Yi, M. miR398 and miR395 are involved in response to SO2 stress in Arabidopsis thaliana. Ecotoxicology 2017, 26, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Bian, L.; Xie, H.; Liang, J. miR398 regulation in rice of the responses to abiotic and biotic stresses depends on CSD1 and CSD2 expression. Funct. Plant Biol. 2011, 38, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Saini, A.; Sunkar, R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 2009, 229, 1009–1014. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, G.; Zhang, W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007, 3, 103. [Google Scholar] [CrossRef]
- Jia, X.; Ren, L.; Chen, Q.-J.; Li, R.; Tang, G. UV-B-responsive microRNAs in Populus tremula. J. Plant Physiol. 2009, 166, 2046–2057. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Q.; Zhang, B. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 2013, 530, 26–32. [Google Scholar] [CrossRef]
- Luan, Y.; Wang, W.; Liu, P. Identification and functional analysis of novel and conserved microRNAs in tomato. Mol. Biol. Rep. 2014, 41, 5385–5394. [Google Scholar] [CrossRef]
- He, Y.; Zhou, J.X.; Hu, Y.F.; Fang, C.Y.; Yu, Y.J.; Yang, J.; Zhu, B.A.; Ruan, Y.L.; Zhu, Z.J. Overexpression of sly-miR398b increased salt sensitivity likely via regulating antioxidant system and photosynthesis in tomato. Environ. Exp. Bot. 2021, 181, 104273. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, P.; Kang, H.; Liu, L.; Cao, Q.; Sun, J.; Dong, T.; Zhu, M.; Li, Z.; Xu, T. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genom. 2020, 21, 164. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2014, 66, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2010, 233, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Hivrale, V.; Zheng, Y.; Puli, C.O.R.; Jagadeeswaran, G.; Gowdu, K.; Kakani, V.G.; Barakat, A.; Sunkar, R. Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 2016, 242, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; dos Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2009, 231, 705–716. [Google Scholar] [CrossRef]
- Jovanović, Ž.; Stanisavljević, N.; Mikić, A.; Radović, S.; Maksimović, V. Water deficit down-regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiol. Biochem. 2014, 83, 26–31. [Google Scholar] [CrossRef]
- De La Rosa, C.; Covarrubias, A.A.; Reyes, J.L. A dicistronic precursor encoding miR398 and the legume-specific miR2119 coregulates CSD1 and ADH1 mRNAs in response to water deficit. Plant Cell Environ. 2019, 42, 133–144. [Google Scholar] [CrossRef]
- Loreti, E.; Betti, F.; Ladera-Carmona, M.J.; Fontana, F.; Novi, G.; Valeri, M.C.; Perata, P. ARGONAUTE1 and ARGONAUTE4 Regulate Gene Expression and Hypoxia Tolerance. Plant Physiol. 2020, 182, 287–300. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J.; Song, A.; Chen, S.; Shan, H.; Luo, H.; Gu, C.; Sun, J.; Zhu, L.; Fang, W.; et al. Ambient temperature enhanced freezing tolerance of Chrysanthemum dichrum CdICE1 Arabidopsis via miR398. BMC Biol. 2013, 11, 121. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 52. [Google Scholar] [CrossRef]
- Lee, H.; Yoo, S.J.; Lee, J.H.; Kim, W.; Yoo, S.K.; Fitzgerald, H.; Carrington, J.; Ahn, J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010, 38, 3081–3093. [Google Scholar] [CrossRef]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant. J. 2013, 74, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Guan, Q.; Zhu, J. Downregulation of CSD2 by a heat-inducible miR398 is required for thermotolerance in Arabidopsis. Plant. Signal. Behav. 2013, 8, e24952. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, H.; Lu, Y.; De Ruiter, M.; Cariaso, M.; Prins, M.; Van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2011, 63, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ren, Y.; Zhang, Y.; Xu, J.; Sun, F.; Zhang, Z.; Wang, Y. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 2012, 504, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Khaksefidi, R.E.; Mirlohi, S.; Khalaji, F.; Fakhari, Z.; Shiran, B.; Fallahi, H.; Rafiei, F.; Budak, H.; Ebrahimie, E. Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus. Front. Plant Sci. 2015, 6, 741. [Google Scholar] [CrossRef]
- Jamla, M.; Joshi, S.; Patil, S.; Tripathi, B.N.; Kumar, V. MicroRNAs modulating nutrient homeostasis: A sustainable approach for developing biofortified crops. Protoplasma 2022. online ahead of print. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Perea-Garcia, A.; Andres-Borderia, A.; Huijser, P.; Penarrubia, L. The copper-microRNA pathway is integrated with developmental and environmental stress responses in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 9547. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Burkhead, J.L.; Gogolin, K.A.; Andres-Colas, N.; Bodecker, J.R.; Puig, S.; Penarrubia, L.; Pilon, M. AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Lett. 2005, 579, 2307–2312. [Google Scholar] [CrossRef]
- Hacisalihoglu, G.; Hart, J.J.; Wang, Y.-H.; Cakmak, I.; Kochian, L.V. Zinc Efficiency Is Correlated with Enhanced Expression and Activity of Zinc-Requiring Enzymes in Wheat. Plant Physiol. 2003, 131, 595–602. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Y.; Shi, D.; Liu, X.; Qin, J.; Ge, Q.; Xu, L.; Pan, X.; Li, W.; Zhu, Y.; et al. Spatial-temporal analysis of zinc homeostasis reveals the response mechanisms to acute zinc deficiency in Sorghum bicolor. New Phytol. 2013, 200, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.-R. Identification of Nutrient-Responsive Arabidopsis and Rapeseed MicroRNAs by Comprehensive Real-Time Polymerase Chain Reaction Profiling and Small RNA Sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Xu, K.; Chen, A.; Zhu, Y.; Tang, G.; Xu, G. Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant. 2010, 138, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, H.; Li, Y.; Wan, X.; Ma, Z.; Cao, J.; Li, Z.; He, F.; Wang, Y.; Wan, L.; et al. Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol. Biol. 2018, 96, 473–492. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Chakrabarti, S.K. Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato. PLoS ONE 2020, 15, e0233076. [Google Scholar] [CrossRef]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome-Wide Identification of MicroRNAs in Response to Low Nitrate Availability in Maize Leaves and Roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.-M. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, R.; Albrecht, U.; Padmanabhan, C.; Wang, A.; Coffey, M.D.; Girke, T.; Wang, Z.; Close, T.J.; Roose, M.; et al. Small RNA Profiling Reveals Phosphorus Deficiency as a Contributing Factor in Symptom Expression for Citrus Huanglongbing Disease. Mol. Plant 2013, 6, 301–310. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Y.-G.; Shi, Y.; Wu, L.; Xu, Y.-J.; Huang, F.; Guo, X.-Y.; Zhang, Y.; Fan, J.; Zhao, J.-Q.; et al. Multiple Rice MicroRNAs Are Involved in Immunity against the Blast Fungus Magnaporthe oryzae. Plant Physiol. 2013, 164, 1077–1092. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.L.; Zhu, Y.; Yang, X.M.; Zhang, K.N.; Xiao, Z.Y.; Wang, H.; Zhao, J.H.; Zhang, L.L.; Li, G.B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New. Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Meng, Y.; Wise, R.P. Mla- and Rom1 -mediated control of microRNA398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytol. 2014, 201, 1396–1412. [Google Scholar] [CrossRef] [PubMed]
- Salamon, S.; Zok, J.; Gromadzka, K.; Blaszczyk, L. Expression Patterns of miR398, miR167, and miR159 in the Interaction between Bread Wheat (Triticum aestivum L.) and Pathogenic Fusarium culmorum and Beneficial Trichoderma Fungi. Pathogens 2021, 10, 1461. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Hollunder, J.; Niehl, A.; Kørner, C.J.; Gereige, D.; Windels, D.; Arnold, A.; Kuiper, M.; Vazquez, F.; Pooggin, M.; et al. Specific Impact of Tobamovirus Infection on the Arabidopsis Small RNA Profile. PLoS ONE 2011, 6, e19549. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Manacorda, C.A.; Tohge, T.; Conti, G.; Rodriguez, M.C.; Nunes-Nesi, A.; Villanueva, S.; Fernie, A.R.; Carrari, F.; Asurmendi, S. Metabolic and miRNA Profiling of TMV Infected Plants Reveals Biphasic Temporal Changes. PLoS ONE 2011, 6, e28466. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Haq, Q.M.; Mukherjee, S.K. MicroRNA profiling of tomato leaf curl New Delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, R.; García-Marcos, A.; Barajas, D.; Martiáñez, J.; Tenllado, F. PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res. 2012, 165, 231–235. [Google Scholar] [CrossRef]
- Abreu, P.M.V.; Gaspar, C.G.; Buss, D.S.; Ventura, J.A.; Ferreira, P.C.G.; Fernandes, P.M.B. Carica papaya MicroRNAs Are Responsive to Papaya meleira virus Infection. PLoS ONE 2014, 9, e103401. [Google Scholar] [CrossRef]
- Suzuki, T.; Ikeda, S.; Kasai, A.; Taneda, A.; Fujibayashi, M.; Sugawara, K.; Okuta, M.; Maeda, H.; Sano, T. RNAi-mediated down-regulation of Dicer-Like 2 and 4 changes the response of ‘Moneymaker’ tomato to potato spindle tuber viroid infection from tolerance to lethal systemic necrosis, accompanied by up-regulation of miR398, 398a-3p and production of excessive amount of reactive oxygen species. Viruses 2019, 11, 344. [Google Scholar]
- Lin, K.-Y.; Wu, S.-Y.; Hsu, Y.-H.; Lin, N.-S. MiR398-regulated antioxidants contribute to Bamboo mosaic virus accumulation and symptom manifestation. Plant Physiol. 2021, 188, 593–607. [Google Scholar] [CrossRef]
- Liu, J.; Fan, H.; Wang, Y.; Han, C.; Wang, X.; Yu, J.; Li, D.; Zhang, Y. Genome-Wide microRNA Profiling Using Oligonucleotide Microarray Reveals Regulatory Networks of microRNAs in Nicotiana benthamiana During Beet Necrotic Yellow Vein Virus Infection. Viruses 2020, 12, 310. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.-K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Song, Q.; Zuo, Z.-F.; Liu, L. MicroRNA398: A Master Regulator of Plant Development and Stress Responses. Int. J. Mol. Sci. 2022, 23, 10803. https://doi.org/10.3390/ijms231810803
Li J, Song Q, Zuo Z-F, Liu L. MicroRNA398: A Master Regulator of Plant Development and Stress Responses. International Journal of Molecular Sciences. 2022; 23(18):10803. https://doi.org/10.3390/ijms231810803
Chicago/Turabian StyleLi, Jing, Qiaoqiao Song, Zhi-Fang Zuo, and Lin Liu. 2022. "MicroRNA398: A Master Regulator of Plant Development and Stress Responses" International Journal of Molecular Sciences 23, no. 18: 10803. https://doi.org/10.3390/ijms231810803






