In-Plant Persistence and Systemic Transport of Nicotiana benthamiana Retrozyme RNA
Abstract
1. Introduction
2. Results
2.1. Retrozymes in the N. benthamiana Genome
2.2. Retrozymes in the N. benthamiana Transcriptome
2.3. Retrozyme Transcriptional Silencing
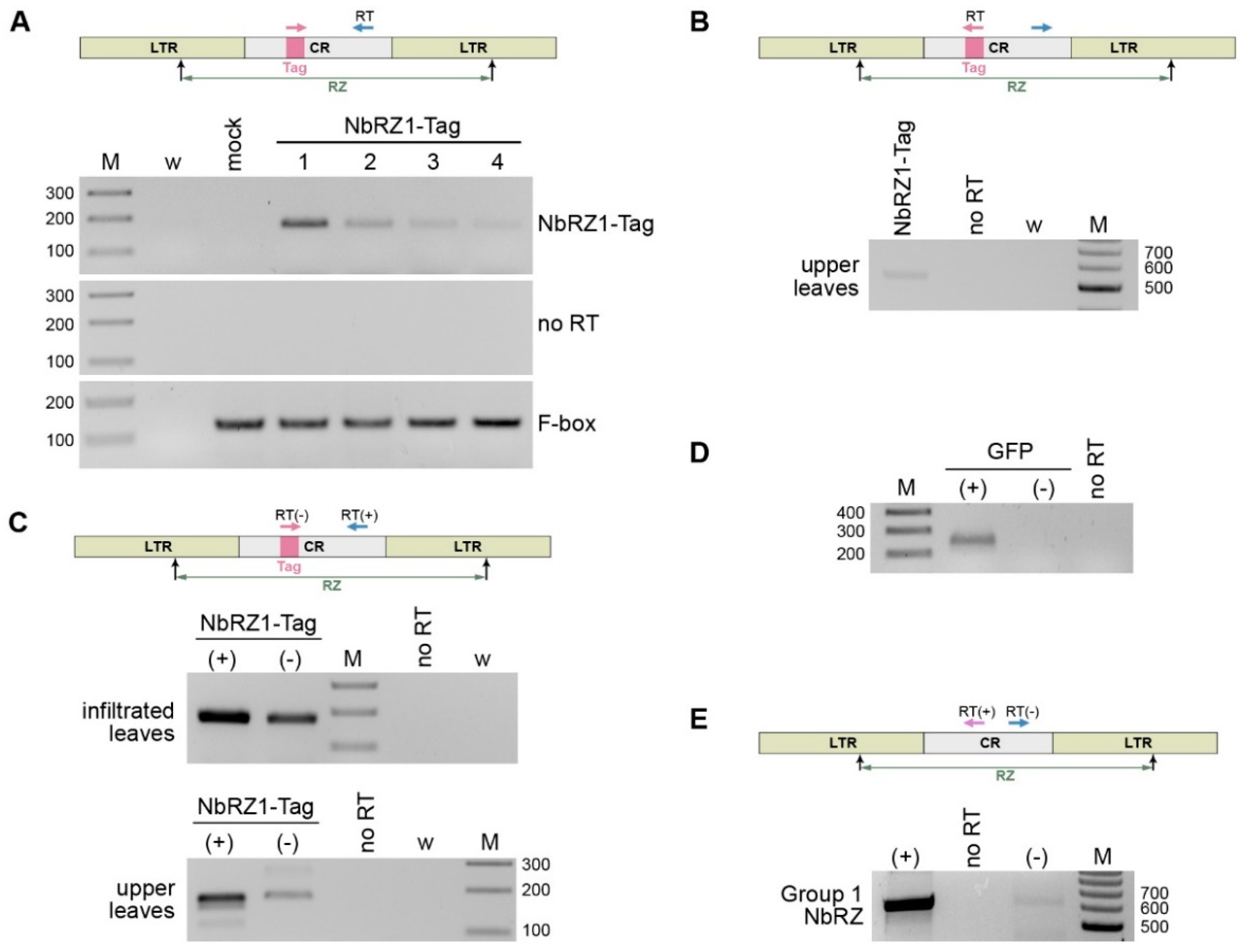
2.4. Retrozyme Replication and Systemic Transport in N. benthamiana
3. Discussion
4. Materials and Methods
4.1. RNA Detection in N. benthamiana Plants
4.2. High-Throughput Sequencing
4.3. 5′-RACE
4.4. Bisulfite Sequencing
4.5. Molecular Cloning and Recombinant Constructs
4.6. Agroinfiltration of Plants
4.7. Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeck, W.R.; Sharpless, N.E. Detecting and Characterizing Circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of Form and Function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Kadener, S. CircRNAs in Cancer. Curr. Opin. Genet. Dev. 2018, 48, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Wu, S.; Huang, S.; Liu, M.; Gao, B. Advances in the Identification of Circular RNAs and Research Into CircRNAs in Human Diseases. Front. Genet. 2021, 12, 665233. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, E.; Sole, C.; Manterola, L.; Iparraguirre, L.; Otaegui, D.; Lawrie, C.H. CircRNAs and Cancer: Biomarkers and Master Regulators. Semin. Cancer Biol. 2019, 58, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Ain, R. Regulation of Transcription by Circular RNAs. In Circular RNAs; Xiao, J., Ed.; Springer: Singapore, 2018; pp. 81–94. [Google Scholar]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Mumtaz, P.T.; Taban, Q.; Dar, M.A.; Mir, S.; ul Haq, Z.; Zargar, S.M.; Shah, R.A.; Ahmad, S.M. Deep Insights in Circular RNAs: From Biogenesis to Therapeutics. Biol. Proced. Online 2020, 22, 10. [Google Scholar] [CrossRef]
- Zhao, W.; Chu, S.; Jiao, Y. Present Scenario of Circular RNAs (CircRNAs) in Plants. Front. Plant Sci. 2019, 10, 379. [Google Scholar] [CrossRef]
- Zhang, P.; Li, S.; Chen, M. Characterization and Function of Circular RNAs in Plants. Front. Mol. Biosci. 2020, 7, 91. [Google Scholar] [CrossRef]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and Functional Characterization of Tomato CircRNAs Derived from Genes Involved in Fruit Pigment Accumulation. Sci. Rep. 2017, 7, 8594. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Li, X.; Yao, L.; Wu, H.; Baluška, F.; Wan, Y. An Antisense Circular RNA Regulates Expression of RuBisCO Small Subunit Genes in Arabidopsis. Front. Plant Sci. 2021, 12, 665014. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A CircRNA from SEPALLATA3 Regulates Splicing of Its Cognate MRNA through R-Loop Formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Characterization and Cloning of Grape Circular RNAs Identified the Cold Resistance-Related Vv-CircATS1. Plant Physiol. 2019, 180, 966–985. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Jin, L.; Ling, X.; Liu, T.; Chen, T.; Ji, Y.; Yu, W.; Zhang, B. Re-Analysis of Long Non-Coding RNAs and Prediction of CircRNAs Reveal Their Novel Roles in Susceptible Tomato Following TYLCV Infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; Urbina, D.; de la Peña, M. Retrozymes Are a Unique Family of Non-Autonomous Retrotransposons with Hammerhead Ribozymes That Propagate in Plants through Circular RNAs. Genome Biol. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, M.; Ceprián, R.; Cervera, A. A Singular and Widespread Group of Mobile Genetic Elements: RNA Circles with Autocatalytic Ribozymes. Cells 2020, 9, 2555. [Google Scholar] [CrossRef]
- De la Peña, M.; Gago-Zachert, S. A Life of Research on Circular RNAs and Ribozymes: Towards the Origin of Viroids, Deltaviruses and Life. Virus Res. 2022, 314, 198757. [Google Scholar] [CrossRef]
- De la Peña, M.; Cervera, A. Circular RNAs with Hammerhead Ribozymes Encoded in Eukaryotic Genomes: The Enemy at Home. RNA Biol. 2017, 14, 985–991. [Google Scholar] [CrossRef]
- Liu, G.; Jiang, H.; Sun, W.; Zhang, J.; Chen, D.; Murchie, A.I.H. The Function of Twister Ribozyme Variants in Non-LTR Retrotransposition in Schistosoma Mansoni. Nucleic Acids Res. 2021, 49, 10573–10588. [Google Scholar] [CrossRef]
- Eickbush, D.G.; Eickbush, T.H. R2 Retrotransposons Encode a Self-Cleaving Ribozyme for Processing from an RRNA Cotranscript. Mol. Cell. Biol. 2010, 30, 3142–3150. [Google Scholar] [CrossRef]
- Sánchez-Luque, F.J.; López, M.C.; Macias, F.; Alonso, C.; Thomas, M.C. Identification of an Hepatitis Delta Virus-like Ribozyme at the MRNA 5′-End of the L1Tc Retrotransposon from Trypanosoma Cruzi. Nucleic Acids Res. 2011, 39, 8065–8077. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; De la Peña, M. Eukaryotic Penelope-Like Retroelements Encode Hammerhead Ribozyme Motifs. Mol. Biol. Evol. 2014, 31, 2941–2947. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R.; Yushenova, I.A.; Rodriguez, F. Giant Reverse Transcriptase-Encoding Transposable Elements at Telomeres. Mol. Biol. Evol. 2017, 34, 2245–2257. [Google Scholar] [CrossRef] [PubMed]
- Le Grice, S.F.J. “In the Beginning”: Initiation of Minus Strand DNA Synthesis in Retroviruses and LTR-Containing Retrotransposons. Biochemistry 2003, 42, 14349–14355. [Google Scholar] [CrossRef]
- Sabot, F.; Schulman, A.H. Parasitism and the Retrotransposon Life Cycle in Plants: A Hitchhiker’s Guide to the Genome. Heredity 2006, 97, 381–388. [Google Scholar] [CrossRef]
- Landry, P.; Perreault, J.-P. Identification of a Peach Latent Mosaic Viroid Hairpin Able to Act as a Dicer-like Substrate. J. Virol. 2005, 79, 6540–6543. [Google Scholar] [CrossRef]
- Ramesh, S.V.; Yogindran, S.; Gnanasekaran, P.; Chakraborty, S.; Winter, S.; Pappu, H.R. Virus and Viroid-Derived Small RNAs as Modulators of Host Gene Expression: Molecular Insights Into Pathogenesis. Front. Microbiol. 2021, 11, 3170. [Google Scholar] [CrossRef]
- Henderson, I.R.; Zhang, X.; Lu, C.; Johnson, L.; Meyers, B.C.; Green, P.J.; Jacobsen, S.E. Dissecting Arabidopsis Thaliana DICER Function in Small RNA Processing, Gene Silencing and DNA Methylation Patterning. Nat. Genet. 2006, 38, 721–725. [Google Scholar] [CrossRef]
- Bologna, N.G.; Voinnet, O. The Diversity, Biogenesis, and Activities of Endogenous Silencing Small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Atsumi, G.; Matsuo, K.; Fukuzawa, N.; Matsumura, T. Virus-Mediated Targeted DNA Methylation Illuminates the Dynamics of Methylation in an Endogenous Plant Gene. Int. J. Mol. Sci. 2021, 22, 4125. [Google Scholar] [CrossRef] [PubMed]
- Rajeevkumar, S.; Anunanthini, P.; Sathishkumar, R. Epigenetic Silencing in Transgenic Plants. Front. Plant Sci. 2015, 6, 693. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic Regulation in Plant Responses to the Environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Tolstyko, E.A.; Lezzhov, A.A.; Morozov, S.Y.; Solovyev, A.G. Phloem Transport of Structured RNAs: A Widening Repertoire of Trafficking Signals and Protein Factors. Plant Sci. 2020, 299, 110602. [Google Scholar] [CrossRef]
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-Dependent RNA Polymerase Gene in Arabidopsis Is Required for Posttranscriptional Gene Silencing Mediated by a Transgene but Not by a Virus. Cell 2000, 101, 543–553. [Google Scholar] [CrossRef]
- Zakrzewski, F.; Schmidt, M.; van Lijsebettens, M.; Schmidt, T. DNA Methylation of Retrotransposons, DNA Transposons and Genes in Sugar Beet (Beta vulgaris L.). Plant J. 2017, 90, 1156–1175. [Google Scholar] [CrossRef]
- Srikant, T.; Drost, H.-G. How Stress Facilitates Phenotypic Innovation through Epigenetic Diversity. Front. Plant Sci. 2021, 11, 2200. [Google Scholar] [CrossRef]
- Kehr, J.; Kragler, F. Long Distance RNA Movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef]
- Ding, B. The Biology of Viroid-Host Interactions. Annu. Rev. Phytopathol. 2009, 47, 105–131. [Google Scholar] [CrossRef]
- Navarro, B.; Flores, R.; Di Serio, F. Advances in Viroid-Host Interactions. Annu. Rev. Virol. 2021, 8, 305–325. [Google Scholar] [CrossRef]
- Daros, J.A.; Flores, R. Identification of a Retroviroid-like Element from Plants. Proc. Natl. Acad. Sci. USA 1995, 92, 6856–6860. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, K.; Dallmann, G.; Balázs, E. The DNA Form of a Retroviroid-like Element Is Involved in Recombination Events with Itself and with the Plant Genome. Virology 2004, 325, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana Benthamiana Using Quantitative Real-Time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Alieva, N.O.; Chenchik, A.; Lukyanov, S. Amplification of CDNA Ends Using PCR Suppression Effect and Step-Out PCR. Methods Mol. Biol. 2003, 221, 41–50. [Google Scholar] [CrossRef]
- Lezzhov, A.A.; Atabekova, A.K.; Tolstyko, E.A.; Lazareva, E.A.; Solovyev, A.G. RNA Phloem Transport Mediated by Pre-MiRNA and Viral TRNA-like Structures. Plant Sci. 2019, 284, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.G.; Minina, E.A.; Makarova, S.S.; Erokhina, T.N.; Makarov, V.V.; Kaplan, I.B.; Kopertekh, L.; Schiemann, J.; Richert-Pöggeler, K.R.; Morozov, S.Y. Subcellular Localization and Self-Interaction of Plant-Specific Nt-4/1 Protein. Biochimie 2013, 95, 1360–1370. [Google Scholar] [CrossRef]
- Yelina, N.E.; Erokhina, T.N.; Lukhovitskaya, N.I.; Minina, E.; Schepetilnikov, M.V.; Lesemann, D.-E.; Schiemann, J.; Solovyev, G.; Morozov, S.Y. Localization of Poa Semilatent Virus Cysteine-Rich Protein in Peroxisomes Is Dispensable for Its Ability to Suppress RNA Silencing. J. Gen. Virol. 2005, 86, 479–489. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lezzhov, A.A.; Tolstyko, E.A.; Atabekova, A.K.; Chergintsev, D.A.; Morozov, S.Y.; Solovyev, A.G. In-Plant Persistence and Systemic Transport of Nicotiana benthamiana Retrozyme RNA. Int. J. Mol. Sci. 2022, 23, 13890. https://doi.org/10.3390/ijms232213890
Lezzhov AA, Tolstyko EA, Atabekova AK, Chergintsev DA, Morozov SY, Solovyev AG. In-Plant Persistence and Systemic Transport of Nicotiana benthamiana Retrozyme RNA. International Journal of Molecular Sciences. 2022; 23(22):13890. https://doi.org/10.3390/ijms232213890
Chicago/Turabian StyleLezzhov, Alexander A., Eugene A. Tolstyko, Anastasia K. Atabekova, Denis A. Chergintsev, Sergey Y. Morozov, and Andrey G. Solovyev. 2022. "In-Plant Persistence and Systemic Transport of Nicotiana benthamiana Retrozyme RNA" International Journal of Molecular Sciences 23, no. 22: 13890. https://doi.org/10.3390/ijms232213890
APA StyleLezzhov, A. A., Tolstyko, E. A., Atabekova, A. K., Chergintsev, D. A., Morozov, S. Y., & Solovyev, A. G. (2022). In-Plant Persistence and Systemic Transport of Nicotiana benthamiana Retrozyme RNA. International Journal of Molecular Sciences, 23(22), 13890. https://doi.org/10.3390/ijms232213890





