Molecular Characterization of a Rare Case of Monozygotic Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Transfer in Preimplantation Genetic Testing (PGT)
Abstract
1. Introduction
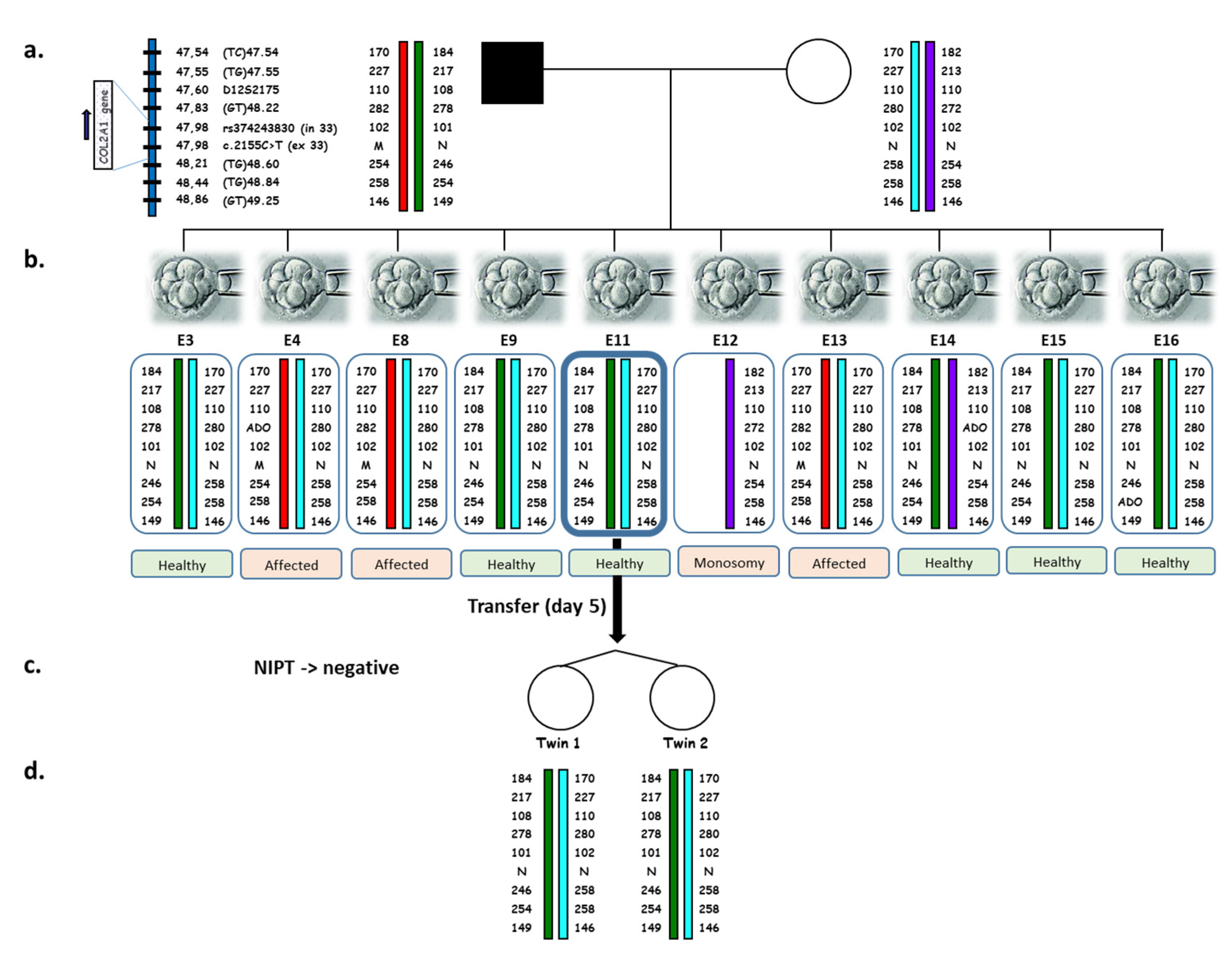
2. Results
- Controlled ovarian stimulation
- Fertilization, embryo culture, and embryo biopsy
- PGT-M
- Embryo transfer
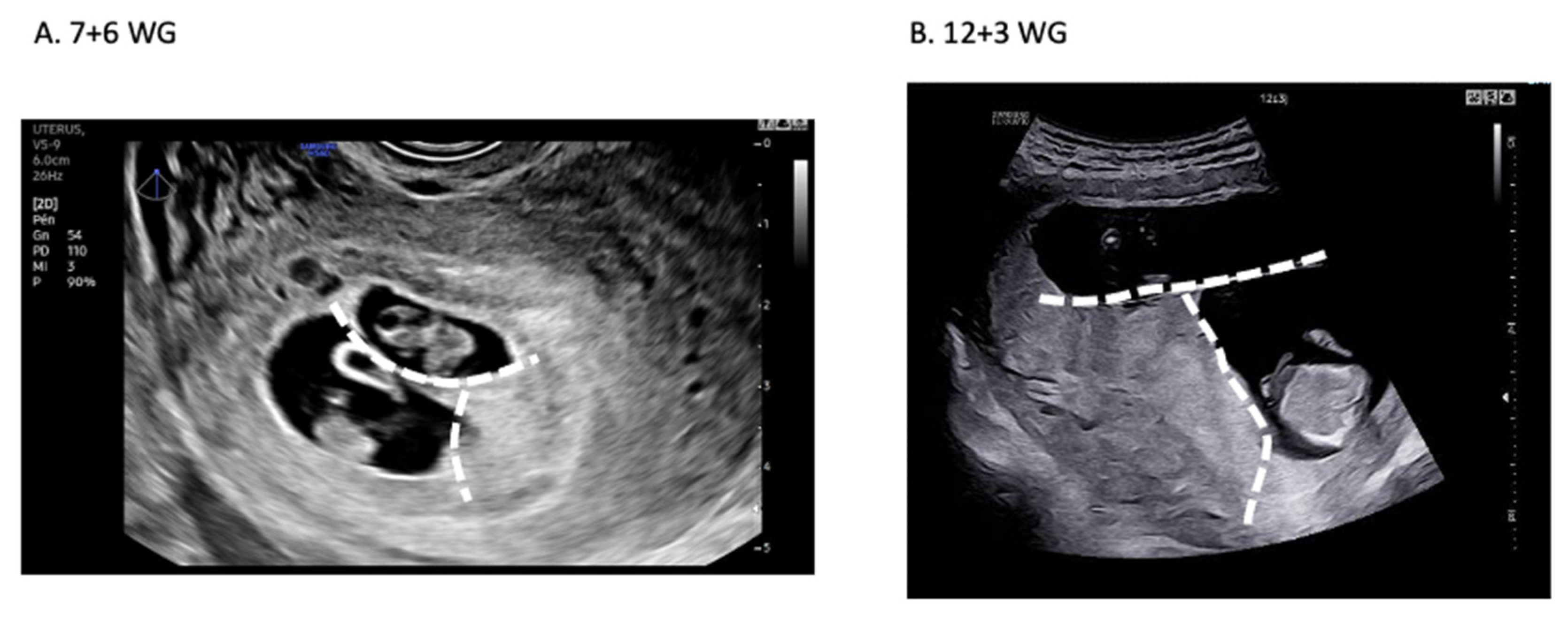
- Pregnancy
- Noninvasive prenatal diagnosis
- Genetic testing of twins
3. Discussion
- (1)
- Inform the couple about the possibility that the pregnancy results from the splitting of the transferred embryo (regardless of its day of transfer) or from the occurrence of a spontaneous concurrent pregnancy, which exposes them to have an affected child. After having informed them of this risk, ask them about unprotected sexual intercourse, which could be the origin of a spontaneous pregnancy;
- (2)
- Recommend a prenatal diagnosis to evaluate the genetic status of the fetus. This becomes strongly recommended if there has been potentially fertilizing sexual intercourse or if the fetuses are of different genders. Clearly inform the couple that same-sex twins do not guarantee monozygosity or genetic concordance with the diagnosed embryo. If possible, a noninvasive approach should be proposed to minimize the risk of miscarriage associated with choriocentesis or amniocentesis [37,38];
- (3)
- We propose the evaluation of twin zygosity using genetic testing. If there remains DNA from the biopsied embryo that was subsequently transferred (this may occur if a whole genome amplification technique has been performed before locus-specific amplification), systematically evaluate the genetic concordance between the transferred embryo and the twins. Indeed, the underlying mechanisms leading to blastocyst splitting are still unknown. The increase in the number of case reports will allow us to better understand the risk factors, assuring the safety of dichorionic pregnancy after single embryo transfer in PGT. Moreover, it will also contribute to the improvement of our knowledge of pre- and peri-implantation embryonic development.
4. Materials and Methods
4.1. Ethics
4.2. Patients
4.3. PGT Work-Up
4.4. IVF-PGT-M
4.4.1. Ovarian Stimulation
4.4.2. Ovarian Puncture and Gamete Fertilization
4.4.3. Embryo Biopsy and PGT-M
4.5. Pregnancy
4.6. Noninvasive Prenatal Diagnosis by Paternal Variant Exclusion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Duguet, A.-M.; Boyer-Beviere, B. Preimplantation Genetic Diagnosis: The Situation in France and in Other European Countries. Eur. J. Health Law 2017, 24, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Ginoza, M.; Isasi, R. Regulating Preimplantation Genetic Testing across the World: A Comparison of International Policy and Ethical Perspectives. Cold Spring Harb. Perspect. Med. 2019, 10, a036681. [Google Scholar] [CrossRef] [PubMed]
- Bayefsky, M.J. Comparative preimplantation genetic diagnosis policy in Europe and the USA and its implications for reproductive tourism. Reprod. Biomed. Soc. Online 2016, 3, 41–47. [Google Scholar] [CrossRef]
- The European IVF-Monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE); Wyns, C.; De Geyter, C.; Calhaz-Jorge, M.; Kupka, M.S.; Motrenko, T.; Smeenk, J.; Bergh, C.; Tandler-Schneider, A.; Rugescu, I.A.; et al. ART in Europe, 2017: Results generated from European registries by ESHRE. Hum. Reprod. Open 2021, 2021, hoab026. [Google Scholar] [PubMed]
- Hviid, K.V.R.; Malchau, S.S.; Pinborg, A.; Nielsen, H.S. Determinants of monozygotic twinning in ART: A systematic review and a meta-analysis. Hum. Reprod. Updat. 2018, 24, 468–483. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Tsutsumi, O.; Noda, Y.; Ibuki, Y.; Hiroi, M. Do assisted reproductive technologies have effects on the demography of monozygotic twinning? Fertil. Steril. 2000, 74, 178–179. [Google Scholar] [CrossRef]
- Parazzini, F.; Cipriani, S.; Bianchi, S.; Bulfoni, C.; Bortolus, R.; Somigliana, E. Risk of Monozygotic Twins After Assisted Reproduction: A Population-Based Approach. Twin Res. Hum. Genet. 2016, 19, 72–76. [Google Scholar] [CrossRef]
- Busnelli, A.; Dallagiovanna, C.; Reschini, M.; Paffoni, A.; Fedele, L.; Somigliana, E. Risk factors for monozygotic twinning after in vitro fertilization: A systematic review and meta-analysis. Fertil. Steril. 2019, 111, 302–317. [Google Scholar] [CrossRef]
- Ikemoto, Y.; Kuroda, K.; Ochiai, A.; Yamashita, S.; Ikuma, S.; Nojiri, S.; Itakura, A.; Takeda, S. Prevalence and risk factors of zygotic splitting after 937 848 single embryo transfer cycles. Hum. Reprod. 2018, 33, 1984–1991. [Google Scholar] [CrossRef]
- Li, H.; Shen, T.; Sun, X. Monozygotic dichorionic-diamniotic pregnancies following single frozen-thawed blastocyst transfer: A retrospective case series. BMC Pregnancy Childbirth 2020, 20, 768. [Google Scholar] [CrossRef]
- Corner, G.W. The observed embryology of human single-ovum twins and other multiple births. Am. J. Obstet. Gynecol. 1955, 70, 933–951. [Google Scholar] [CrossRef]
- Knopman, J.; Krey, L.C.; Lee, J.; Fino, M.E.; Novetsky, A.P.; Noyes, N. Monozygotic twinning: An eight-year experience at a large IVF center. Fertil. Steril. 2010, 94, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Kyono, K. The Precise Timing of Embryo Splitting for Monozygotic Dichorionic Diamniotic Twins: When Does Embryo Splitting for Monozygotic Dichorionic Diamniotic Twins Occur? Evidence for Splitting at the Morula/Blastocyst Stage from Studies of In Vitro Fertilization. Twin Res. Hum. Genet. 2013, 16, 827–832. [Google Scholar] [CrossRef]
- Sundaram, V.; Ribeiro, S.; Noel, M. Multi-chorionic pregnancies following single embryo transfer at the blastocyst stage: A case series and review of the literature. J. Assist. Reprod. Genet. 2018, 35, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.G. Twinning. Lancet 2003, 362, 735–743. [Google Scholar] [CrossRef]
- Tocino, A.; Blasco, V.; Prados, N.; Vargas, M.J.; Requena, A.; Pellicer, A.; Fernández-Sánchez, M. Monozygotic twinning after assisted reproductive technologies: A case report of asymmetric development and incidence during 19 years in an international group of in vitro fertilization clinics. Fertil. Steril. 2015, 103, 1185–1189. [Google Scholar] [CrossRef]
- Konno, H.; Murakoshi, T.; Miura, K.; Masuzaki, H. The Incidence of Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Embryo Transfer and Zygosity: 8 Years of Single-Center Experience. Twin Res. Hum. Genet. 2020, 23, 51–54. [Google Scholar] [CrossRef]
- Osianlis, T.; Rombauts, L.; Gabbe, M.; Motteram, C.; Vollenhoven, B. Incidence and zygosity of twin births following transfers using a single fresh or frozen embryo. Hum. Reprod. 2014, 29, 1438–1443. [Google Scholar] [CrossRef]
- Knopman, J.M.; Krey, L.C.; Oh, C.; Lee, J.; McCaffrey, C.; Noyes, N. Withdrawn: What makes them split? Identifying risk factors that lead to monozygotic twins after in vitro fertilization. Fertil. Steril. 2013. [Google Scholar] [CrossRef]
- Gardner, D.K.; Lane, M.; Stevens, J.; Schlenker, T.; Schoolcraft, W.B. Blastocyst score affects implantation and pregnancy outcome: Towards a single blastocyst transfer. Fertil. Steril. 2000, 73, 1155–1158. [Google Scholar] [CrossRef]
- Soothill, P.W.; Abdel-Fattah, S.A.; Porter, H.; Montague, I.; Kyle, P.M. Prediction of chorionicity in twin pregnancies at 10–14 weeks of gestation. BJOG 2002, 109, 182–186. [Google Scholar]
- Wang, C.-Y.; Tang, Y.-A.; Lee, I.-W.; Chang, F.-M.; Chien, C.-W.; Pan, H.-A.; Sun, H.S. Development and validation of an expanded targeted sequencing panel for non-invasive prenatal diagnosis of sporadic skeletal dysplasia. BMC Med Genom. 2021, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Pacault, M.; Verebi, C.; Lopez, M.; Vaucouleur, N.; Orhant, L.; Deburgrave, N.; Leturcq, F.; Vidaud, D.; Girodon, E.; Bienvenu, T.; et al. Non-invasive prenatal diagnosis of single gene disorders by paternal mutation exclusion: 3 years of clinical experience. BJOG Int. J. Obstet. Gynaecol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Vink, J.; Fuchs, K.; D’Alton, M.E. Amniocentesis in twin pregnancies: A systematic review of the literature. Prenat. Diagn. 2011, 32, 409–416. [Google Scholar] [CrossRef]
- Di Mascio, D.; Khalil, A.; Rizzo, G.; Buca, D.; Liberati, M.; Martellucci, C.A.; Flacco, M.E.; Manzoli, L.; D’Antonio, F. Risk of fetal loss following amniocentesis or chorionic villus sampling in twin pregnancy: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2020, 56, 647–655. [Google Scholar] [CrossRef]
- Yovich, J.L.; Stanger, J.D.; Grauaug, A.; A Barter, R.; Lunay, G.; Dawkins, R.L.; Mulcahy, M.T. Monozygotic twins from in vitro fertilization. Fertil. Steril. 1984, 41, 833–837. [Google Scholar] [CrossRef]
- Schlueter, R.; Arnett, C.; Huang, C.; Burlingame, J. Successful quintuplet pregnancy of monochorionic male quadruplets and single female after double embryo transfer: Case report and review of the literature. Fertil. Steril. 2018, 109, 284–288. [Google Scholar] [CrossRef]
- Tsunoda, Y.; McLaren, A. Effect of various procedures on the viability of mouse embryos containing half the normal number of blastomeres. Reproduction 1983, 69, 315–322. [Google Scholar] [CrossRef]
- Willadsen, S.M. A method for culture of micromanipulated sheep embryos and its use to produce monozygotic twins. Nat. 1979, 277, 298–300. [Google Scholar] [CrossRef]
- Willadsen, S.; Lehn-Jensen, H.; Fehilly, C.; Newcomb, R. The production of monozygotic twins of preselected parentage by micromanipulation of non-surgically collected cow embryos. Theriogenology 1981, 15, 23–29. [Google Scholar] [CrossRef]
- Van de Velde, H.; Cauffman, G.; Tournaye, H.; Devroey, P.; Liebaers, I. The four blastomeres of a 4-cell stage human embryo are able to develop individually into blastocysts with inner cell mass and trophectoderm. Hum. Reprod. 2008, 23, 1742–1747. [Google Scholar] [CrossRef] [PubMed]
- Herranz, G. The timing of monozygotic twinning: A criticism of the common model. Zygote 2013, 23, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Denker, H.W. Comment on G. Herranz: The timing of monozygotic twinning: A criticism of the common model. Zygote 2015, 23, 312–314. [Google Scholar] [CrossRef]
- Shibuya, Y.; Kyono, K. A successful birth of healthy monozygotic dichorionic diamniotic (DD) twins of the same gender following a single vitrified-warmed blastocyst transfer. J. Assist. Reprod. Genet. 2012, 29, 255–257. [Google Scholar] [CrossRef][Green Version]
- Van Langendonckt, A.; Wyns, C.; Godin, P.; Toussaint-Demylle, D.; Donnez, J. Atypical hatching of a human blastocyst leading to monozygotic twinning: A case report. Fertil. Steril. 2000, 74, 1047–1050. [Google Scholar] [CrossRef]
- Vega, M.; Zaghi, S.; Buyuk, E.; Jindal, S. Not all twins are monozygotic after elective single embryo transfer: Analysis of 32,600 elective single embryo transfer cycles as reported to the Society for Assisted Reproductive Technology. Fertil. Steril. 2018, 109, 118–122. [Google Scholar] [CrossRef]
- Gil, M.M.; Molina, F.S.; Rodríguez-Fernández, M.; Delgado, J.L.; Carrillo, M.P.; Jani, J.; Plasencia, W.; Stratieva, V.; Maíz, N.; Carretero, P.; et al. New approach for estimating risk of miscarriage after chorionic villus sampling. Ultrasound Obstet. Gynecol. 2020, 56, 656–663. [Google Scholar] [CrossRef]
- Salomon, L.J.; Sotiriadis, A.; Wulff, C.B.; Odibo, A.; Akolekar, R. Risk of miscarriage following amniocentesis or chorionic villus sampling: Systematic review of literature and updated meta-analysis. Ultrasound Obstet. Gynecol. 2019, 54, 442–451. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.; Wu, N.; Li, J.; Liu, H.; Wang, J. Integrated analysis of COL2A1 variant data and classification of type II collagenopathies. Clin. Genet. 2019, 97, 383–395. [Google Scholar] [CrossRef]
- Barat-Houari, M.; Dumont, B.; Fabre, A.; Them, F.T.; Alembik, Y.; Alessandri, J.-L.; Amiel, J.; Audebert, S.; Baumann-Morel, C.; Blanchet, P.; et al. The expanding spectrum of COL2A1 gene variants IN 136 patients with a skeletal dysplasia phenotype. Eur. J. Hum. Genet. 2015, 24, 992–1000. [Google Scholar] [CrossRef][Green Version]
- ESHRE PGT-M Working Group; Carvalho, F.; Moutou, C.; Dimitriadou, E.; Dreesen, J.; Giménez, C.; Goossens, V.; Kakourou, G.; Vermeulen, N.; Zuccarello, D.; et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum. Reprod. Open 2020, 2020, hoaa018. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Magli, M.; Podini, D.; Ferraretti, A.; Nuccitelli, A.; Vitale, N.; Baldi, M.; Gianaroli, L. The minisequencing method: An alternative strategy for preimplantation genetic diagnosis of single gene disorders. Mol. Hum. Reprod. 2003, 9, 399–410. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cui, X.F.; Li, H.H.; Goradia, T.M.; Lange, K.; Kazazian, H.H.; Galas, D.; Arnheim, N. Single-sperm typing: Determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc. Natl. Acad. Sci. USA 1989, 86, 9389–9393. [Google Scholar] [CrossRef]
- Gruber, A.; Pacault, M.; El Khattabi, L.A.; Vaucouleur, N.; Orhant, L.; Bienvenu, T.; Girodon, E.; Vidaud, D.; Leturcq, F.; Costa, C.; et al. Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: Detection of NF1 and CFTR mutations using droplet digital PCR. Clin. Chem. Lab. Med. 2018, 56, 728–738. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Value |
|---|---|
| Maternal age (years) | 34 |
| Maternal Body Mass Index (kg/m2) | 17.8 |
| Antral follicle count (number) | 26 |
| Anti-mullerian hormone (ng/mL) | 4.5 |
| Paternal age (years) | 35 |
| Paternal Body Mass Index (kg/m2) | 23.3 |
| Sperm parameters | Normal |
| Gravidity and Parity | 0/0 |
| Number of days of stimulation | 9 |
| Total gonadotrophin dose (IU) | 1012.5 |
| Estradiol peak (pg/mL) | 2189 |
| Number of retrieved oocytes (Metaphase II) | 17 |
| Embryo biopsy | Day 3 |
| Embryo transfer | Single blastocyst on Day 5 |
| Embryo stage | 4AA |
| Gestational weeks at delivery | 36 |
| Fetal weights (g) | 2400/2090 |
| Fetal gender | F/F |
| Zygosity | Monozygotic |
| Markers/Variation (Location on Chromosome 12/GRCh38) | Primer Sequences | Distance to the COL2A1 Pathogenic Variant (bp) | Amplicon Size (bp) |
|---|---|---|---|
| (TC)47.54 (47,541,504–47,542,343) | F: Hex-TGT GGT TTC TGT CTT GGG AGT R: TGC CAT ACT CCT TCT GTG TTC C | 440,963 | 170–184 |
| (TG)47.55 (47,557,867–47,558,698) | F: Hex-ATC TAT TTC AGG GCC CAG AGG R: ATC CTT GGA ACG ACA ATG GGT | 424,604 | 213–227 |
| D12S2175 (47,603,861–47,603,888) | F: Fam-AGC AAA TCA GTC TGT GTG CCT A R: TGC TTT GCA TAA TGC CTA TTT C | 379,012 | 108–110 |
| (GT)48.22 (47,833,117–47,833,147) | F: Fam-AAA AAC AGG CAG TGG AA R: GGA ACC CCA AAG CCT TAC TG | 149,754 | 272–282 |
| rs374243830 (47,982,664) | F: Hex-CCT GGG GAG GGA GGT AAG AG R: GGA AGG AAG AGG GGT TTG GG | 222 | 101–102 |
| c.2155C>T (g.47,982,886) | Multiplex PCR F: TGCTTCTCCCTGGACCTTCT R: GTTCATGGAGCCTGGGTAAC Mini sequencing F: TGCTTCTCCCTGGACCTTCT R: ACCCCTCTTCCTTCCCTTCC SnapShot MSF: TCTCCCGGTGCCCAGGGCCTCCAGGGTCCC | - | - |
| (TG)48.60 (48,215,576–48,215,618) | F: Hex-TTT TCT CTT CTG TGC CTT ATT GC R: TTC AGC TGT TCC ATG GCA TT | 232,711 | 246–258 |
| (TG)48.84 (48,447,213–48,447,251) | F: Rox-CTA CTG GCA CAG ATT CTA ACA GG R: TAT AGC CAC TTC CTG GCT TGT AG | 464,346 | 254–258 |
| (GT)49.25 (48,864,385–48,864,417) | F: Fam-GCT TTG CTC TGG GGA AGT AAA A R: GTG GGC AGT AAA ACA GGA CTT CT | 881,515 | 146–149 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brouillet, S.; Mereuze, S.; Ranisavljevic, N.; Chauveau, C.; Hamamah, S.; Cattin, J.; Verebi, C.; Cabrol, C.; Ishmukhametova, A.; Girardet, A.; et al. Molecular Characterization of a Rare Case of Monozygotic Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Transfer in Preimplantation Genetic Testing (PGT). Int. J. Mol. Sci. 2022, 23, 10835. https://doi.org/10.3390/ijms231810835
Brouillet S, Mereuze S, Ranisavljevic N, Chauveau C, Hamamah S, Cattin J, Verebi C, Cabrol C, Ishmukhametova A, Girardet A, et al. Molecular Characterization of a Rare Case of Monozygotic Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Transfer in Preimplantation Genetic Testing (PGT). International Journal of Molecular Sciences. 2022; 23(18):10835. https://doi.org/10.3390/ijms231810835
Chicago/Turabian StyleBrouillet, Sophie, Sandie Mereuze, Noémie Ranisavljevic, Claire Chauveau, Samir Hamamah, Julie Cattin, Camille Verebi, Christelle Cabrol, Aliya Ishmukhametova, Anne Girardet, and et al. 2022. "Molecular Characterization of a Rare Case of Monozygotic Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Transfer in Preimplantation Genetic Testing (PGT)" International Journal of Molecular Sciences 23, no. 18: 10835. https://doi.org/10.3390/ijms231810835
APA StyleBrouillet, S., Mereuze, S., Ranisavljevic, N., Chauveau, C., Hamamah, S., Cattin, J., Verebi, C., Cabrol, C., Ishmukhametova, A., Girardet, A., Anahory, T., & Willems, M. (2022). Molecular Characterization of a Rare Case of Monozygotic Dichorionic Diamniotic Twin Pregnancy after Single Blastocyst Transfer in Preimplantation Genetic Testing (PGT). International Journal of Molecular Sciences, 23(18), 10835. https://doi.org/10.3390/ijms231810835






