Targeting of the Mitochondrial TET1 Protein by Pyrrolo[3,2-b]pyrrole Chelators
Abstract
:1. Introduction
2. Results and Discussion
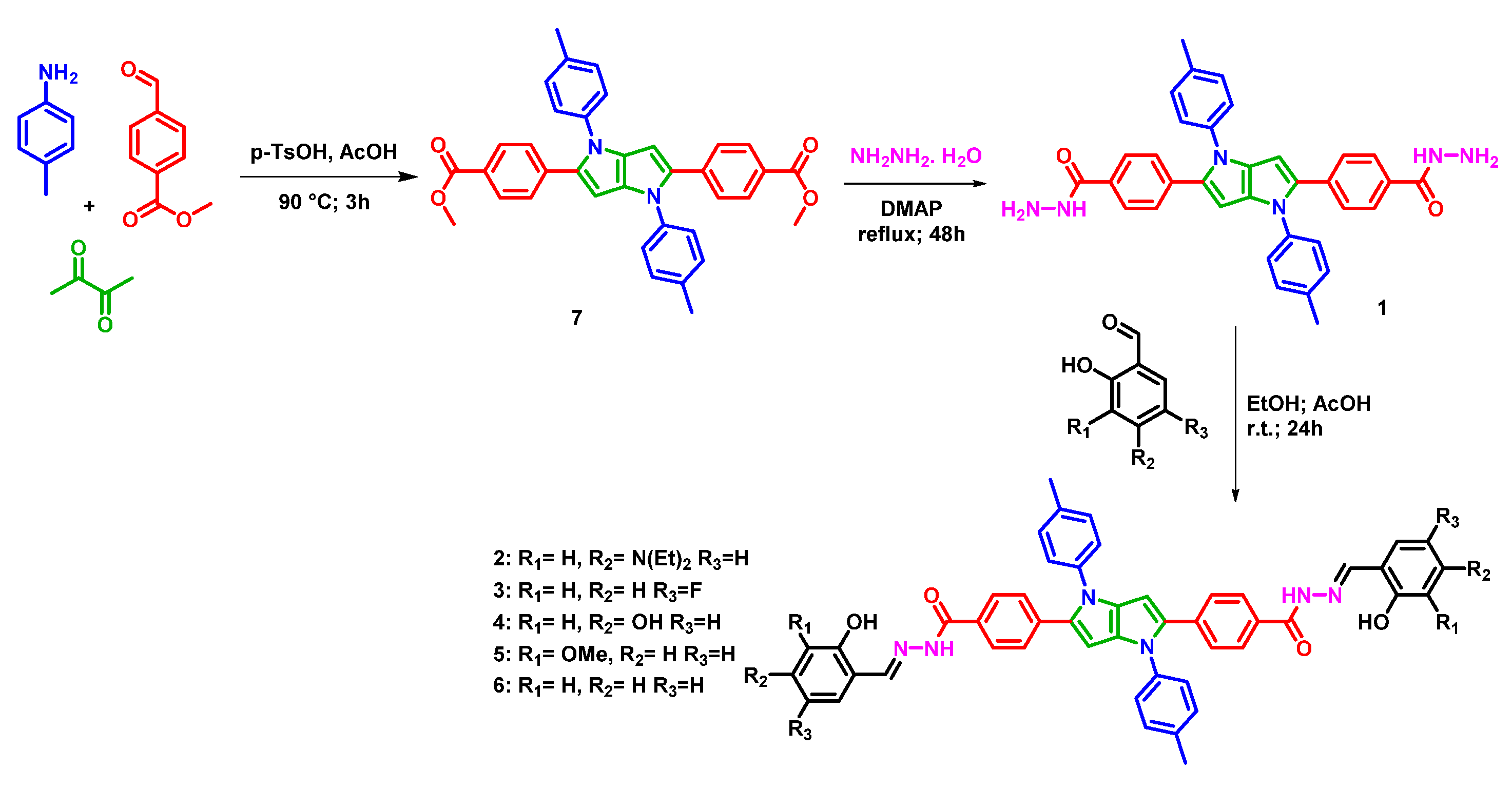
2.1. Synthesis of Compounds 1–7
2.2. FTIR and Raman Analysis of Compounds 1–7
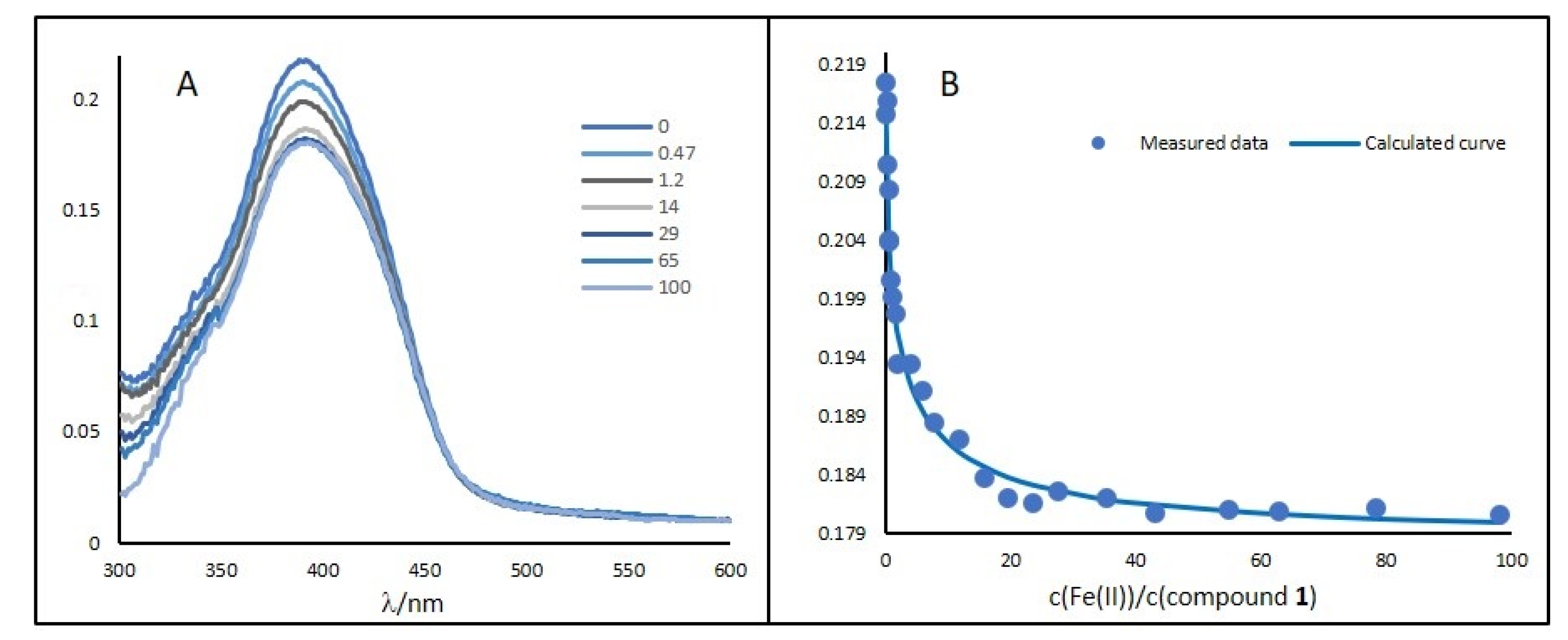
2.3. Binding Study of Compounds 1–6 with Fe(II) Ions
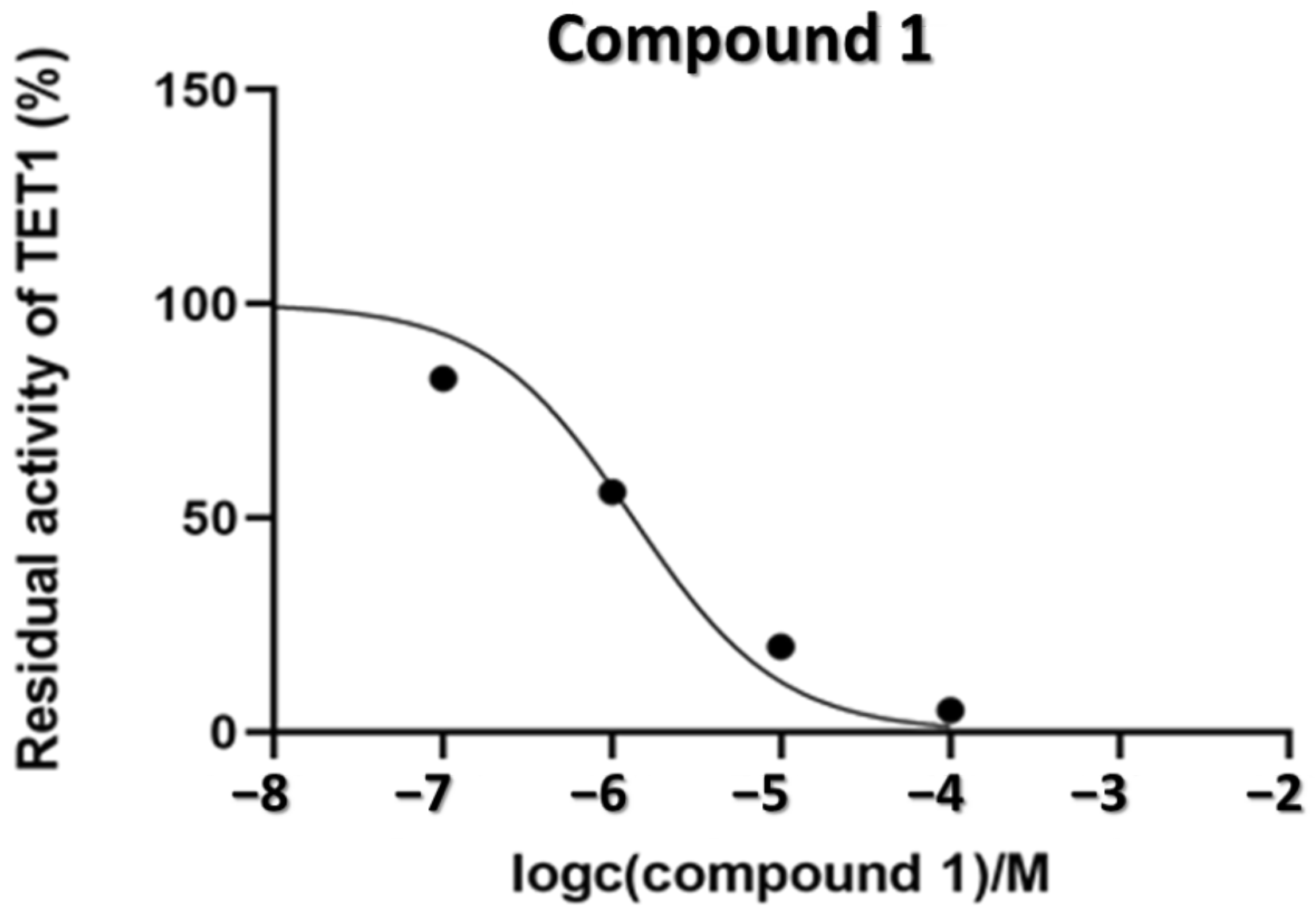
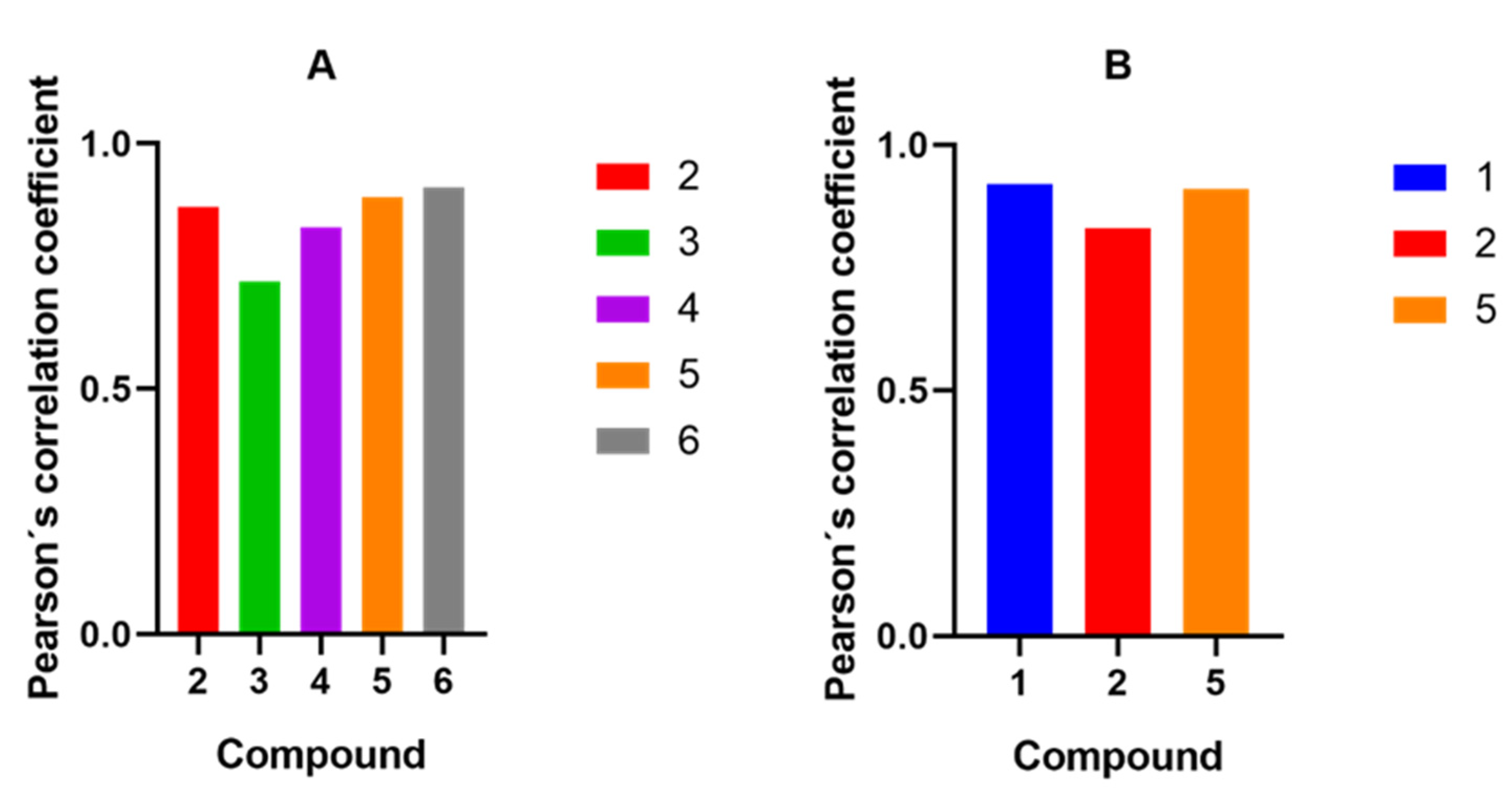
2.4. Inhibition of the TET1 Protein by Compounds 1–6
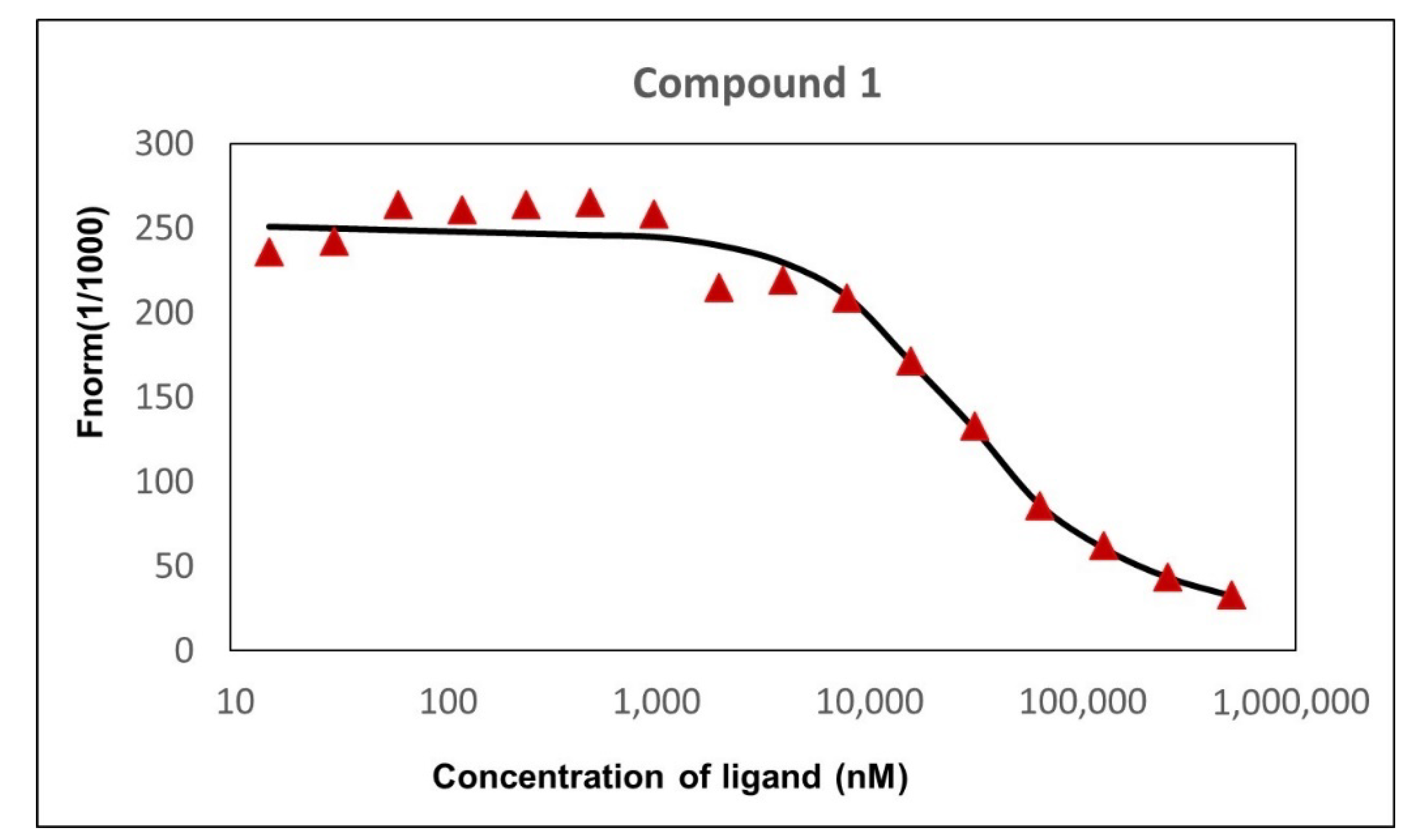
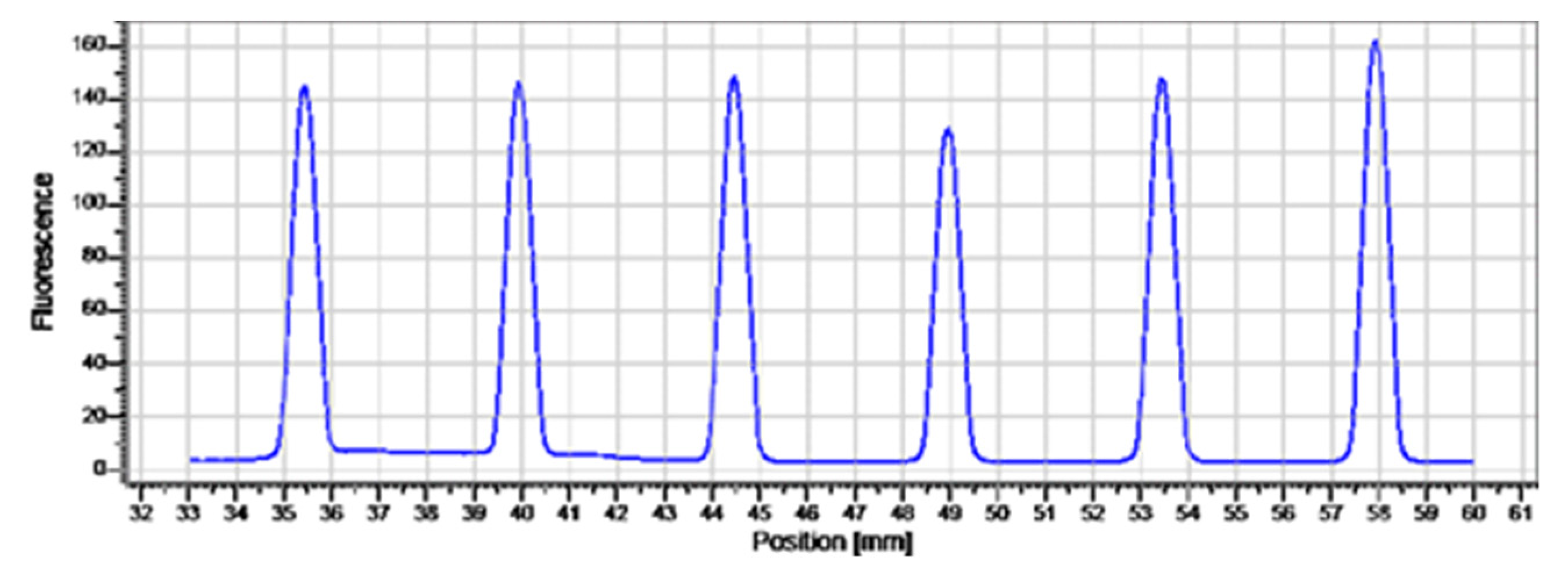
2.5. Affinity of Compound 1 to TET1 Protein 1
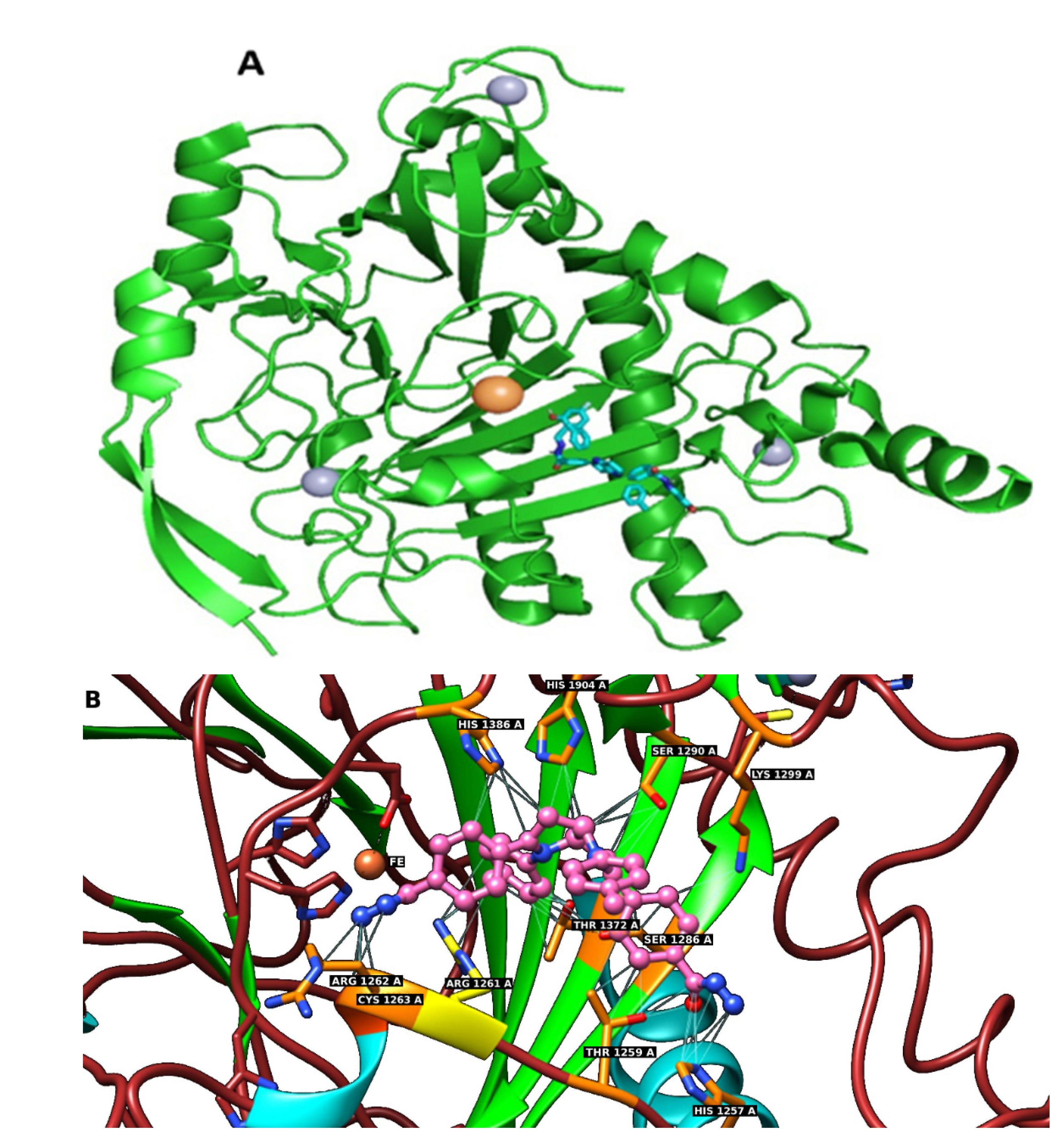
2.6. In Silico Docking of Compounds 1–7 to the TET1 Model
2.7. Influence of the Compounds 1–6 on the Cell Viability
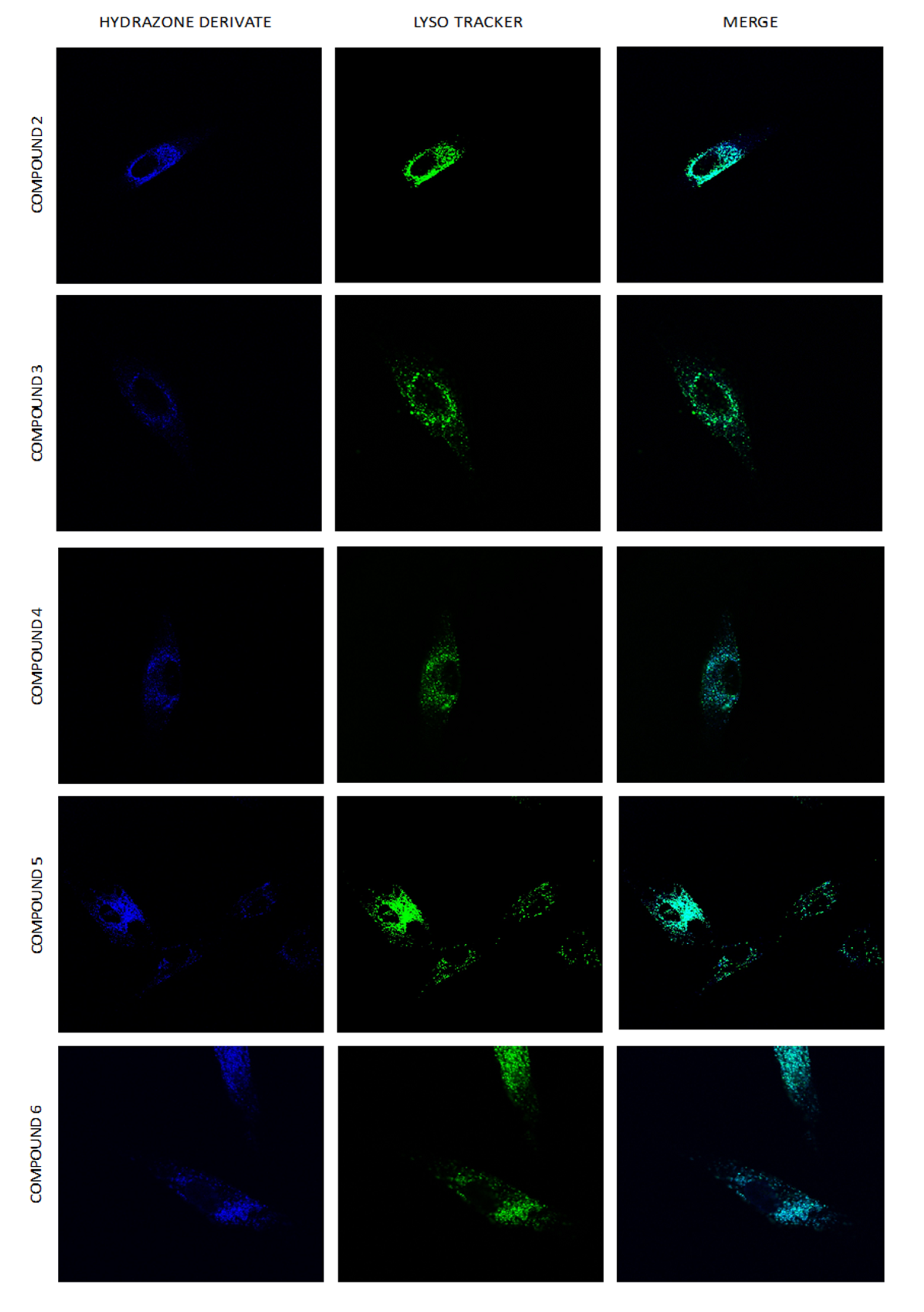
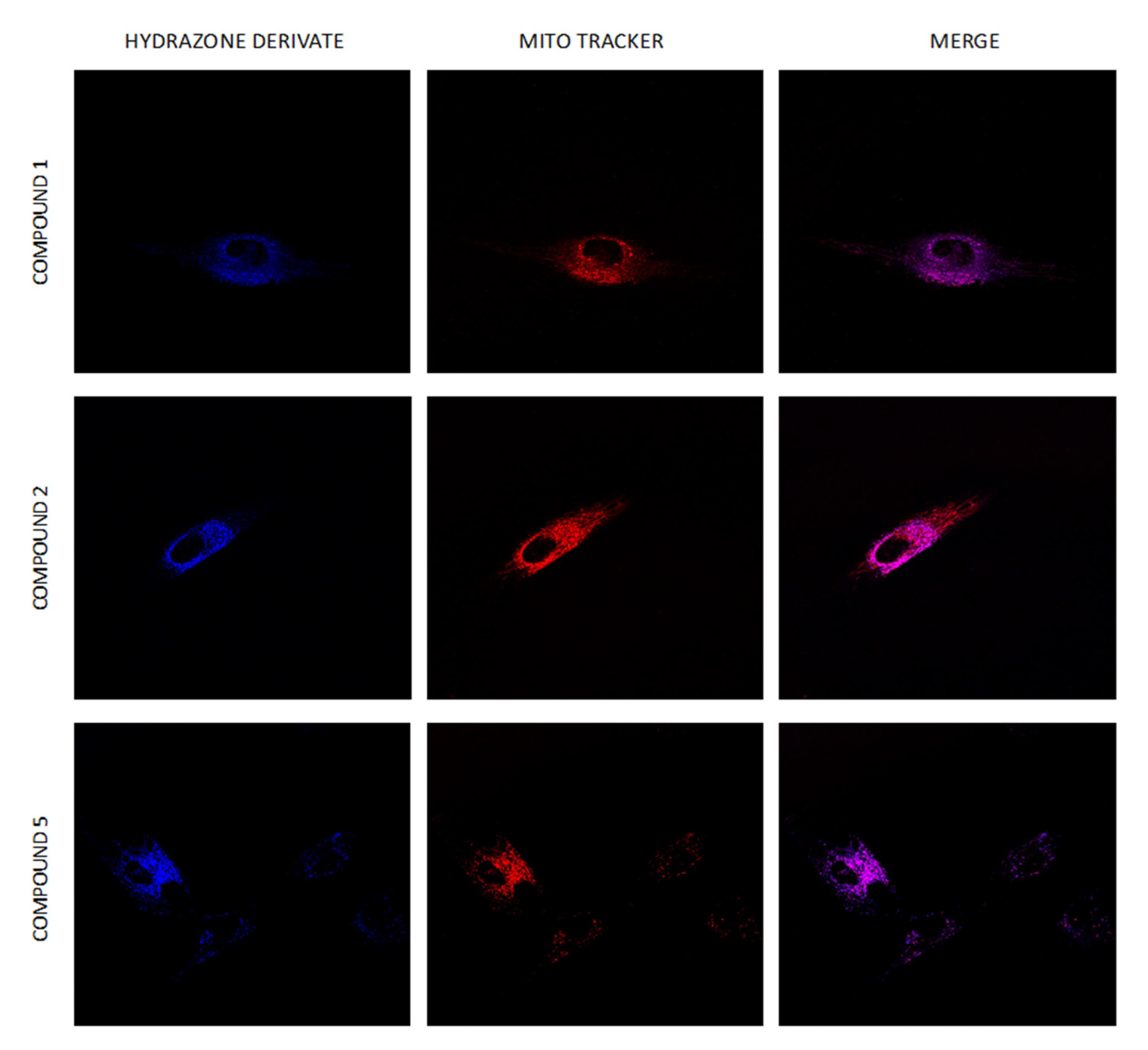
2.8. Intracellular Distribution of Compounds 1–6
3. Materials and Methods
3.1. Synthesis
3.1.1. Diester 7
3.1.2. Compound 1
3.1.3. General Procedure for the Synthesis of Compounds 2–6
3.1.4. Compound 2 (82% Yield)
3.1.5. Compound 3 (79% Yield)
3.1.6. Compound 4 (81% Yield)
3.1.7. Compound 5 (72% Yield)
3.1.8. Compound 6 (89% Yield)
3.2. Vibrational Spectroscopy
3.3. Determination of Conditional Binding Constants and Complex
3.4. Inhibition of TET1 Protein
3.5. Microscale Thermophoresis
3.6. Docking to the TET1 Protein
3.7. Cell Cultures
3.8. Determination of IC50 of Tested Compounds
3.9. Intracellular Localization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef] [PubMed]
- Mehler, M.F. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog. Neurobiol. 2008, 86, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Dean, W.; Santos, F.; Stojkovic, M.; Zakhartchenko, V.; Walter, J.; Wolf, E.; Reik, W. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA 2001, 98, 13734–13738. [Google Scholar] [CrossRef]
- Day, J.J.; Kennedy, A.J.; Sweatt, J.D. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 591–611. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome-biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.L.; Song, H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.; Christensen, J.; Pedersen, M.T.; Johansen, J.V.; Cloos, P.A.C.; Rappsilber, J.; Helin, K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011, 473, 343–348. [Google Scholar] [CrossRef]
- Jin, C.; Lu, Y.; Jelinek, J.; Liang, S.; Estecio, M.R.; Barton, M.C.; Issa, J.P. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014, 42, 6956–6971. [Google Scholar] [CrossRef]
- Wu, H.; D’Alessio, A.C.; Ito, S.; Xia, K.; Wang, Z.; Cui, K.; Zhao, K.; Sun, Y.E.; Zhang, Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 2011, 473, 389–393. [Google Scholar] [CrossRef]
- Iyer, L.M.; Tahiliani, M.; Rao, A.; Aravind, L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 2009, 8, 1698–1710. [Google Scholar] [CrossRef]
- Han, J.A.; An, J.; Ko, M. Functions of TET Proteins in Hematopoietic Transformation. Mol. Cells 2015, 38, 925–935. [Google Scholar] [CrossRef]
- Liu, W.; Wu, G.; Xiong, F.; Chen, Y. Advances in the DNA methylation hydroxylase TET1. Biomark. Res. 2021, 9, 76. [Google Scholar] [CrossRef]
- Bray, J.K.; Dawlaty, M.M.; Verma, A.; Maitra, A. Roles and Regulations of TET Enzymes in Solid Tumors. Trends Cancer 2021, 7, 635–646. [Google Scholar] [CrossRef]
- Huang, H.; Jiang, X.; Li, Z.; Li, Y.; Song, C.X.; He, C.; Sun, M.; Chen, P.; Gurbuxani, S.; Wang, J.; et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 11994–11999. [Google Scholar] [CrossRef]
- Good, C.R.; Panjarian, S.; Kelly, A.D.; Madzo, J.; Patel, B.; Jelinek, J.; Issa, J.J. TET1-Mediated Hypomethylation Activates Oncogenic Signaling in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 4126–4137. [Google Scholar] [CrossRef]
- Zhang, W.; Barger, C.J.; Link, P.A.; Mhawech-Fauceglia, P.; Miller, A.; Akers, S.N.; Odunsi, K.; Karpf, A.R. DNA hypomethylation-mediated activation of Cancer/Testis Antigen 45 (CT45) genes is associated with disease progression and reduced survival in epithelial ovarian cancer. Epigenetics 2015, 10, 736–748. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Y.; Huang, R.L.; Chan, M.W.; Yan, P.S.; Huang, T.S.; Wu, R.C.; Suryo Rahmanto, Y.; Su, P.H.; Weng, Y.C.; Chou, J.L.; et al. TET1 reprograms the epithelial ovarian cancer epigenome and reveals casein kinase 2α as a therapeutic target. J. Pathol. 2019, 248, 363–376. [Google Scholar] [CrossRef]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef]
- Filipczak, P.T.; Leng, S.; Tellez, C.S.; Do, K.C.; Grimes, M.J.; Thomas, C.L.; Walton-Filipczak, S.R.; Picchi, M.A.; Belinsky, S.A. p53-Suppressed Oncogene TET1 Prevents Cellular Aging in Lung Cancer. Cancer Res. 2019, 79, 1758–1768. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, D.; Yan, F.; Yang, L.; Huang, B. Circ_0007919 exerts an anti-tumor role in colorectal cancer through targeting miR-942-5p/TET1 axis. Pathol. Res. Pract. 2022, 229, 153704. [Google Scholar] [CrossRef]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ranawat, A.S.; Tolani, P.; Scaria, V. MitoepigenomeKB a comprehensive resource for human mitochondrial epigenetic data. Mitochondrion 2018, 42, 54–58. [Google Scholar] [CrossRef]
- Chen, H.; Dzitoyeva, S.; Manev, H. Effect of valproic acid on mitochondrial epigenetics. Eur. J. Pharmacol. 2012, 690, 51–59. [Google Scholar] [CrossRef]
- Dzitoyeva, S.; Chen, H.; Manev, H. Effect of aging on 5-hydroxymethylcytosine in brain mitochondria. Neurobiol. Aging 2012, 33, 2881–2891. [Google Scholar] [CrossRef]
- Bellizzi, D.; D’Aquila, P.; Scafone, T.; Giordano, M.; Riso, V.; Riccio, A.; Passarino, G. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013, 20, 537–547. [Google Scholar] [CrossRef]
- Brodie, S.A.; Brandes, J.C. Could valproic acid be an effective anticancer agent? The evidence so far. Expert Rev. Anticancer Ther. 2014, 14, 1097–1100. [Google Scholar] [CrossRef] [Green Version]
- Kejík, Z.; Jakubek, M.; Kaplánek, R.; Králová, J.; Mikula, I.; Martásek, P.; Král, V. Epigenetic agents in combined anticancer therapy. Future Med. Chem. 2018, 10, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Antonyová, V.; Kejík, Z.; Brogyanyi, T.; Kaplánek, R.; Veselá, K.; Abramenko, N.; Ocelka, T.; Masařík, M.; Matkowski, A.; Gburek, J.; et al. Non-psychotropic cannabinoids as inhibitors of TET1 protein. Bioorg. Chem. 2022, 124, 105793. [Google Scholar] [CrossRef] [PubMed]
- Jakubek, M.; Kejík, Z.; Kaplánek, R.; Antonyová, V.; Hromádka, R.; Šandriková, V.; Sýkora, D.; Martásek, P.; Král, V. Hydrazones as novel epigenetic modulators: Correlation between TET 1 protein inhibition activity and their iron(II) binding ability. Bioorg. Chem. 2019, 88, 102809. [Google Scholar] [CrossRef] [PubMed]
- Friese, D.H.; Mikhaylov, A.; Krzeszewski, M.; Poronik, Y.M.; Rebane, A.; Ruud, K.; Gryko, D.T. Pyrrolo[3,2-b]pyrroles—From Unprecedented Solvatofluorochromism to Two-Photon Absorption. Chem. A Eur. J. 2015, 21, 18364–18374. [Google Scholar] [CrossRef]
- Popiołek, Ł. Hydrazide–hydrazones as potential antimicrobial agents: Overview of the literature since 2010. Med. Chem. Res. 2017, 26, 287–301. [Google Scholar] [CrossRef]
- Kaplánek, R.; Jakubek, M.; Rak, J.; Kejík, Z.; Havlík, M.; Dolenský, B.; Frydrych, I.; Hajdúch, M.; Kolář, M.; Bogdanová, K.; et al. Caffeine–hydrazones as anticancer agents with pronounced selectivity toward T-lymphoblastic leukaemia cells. Bioorg. Chem. 2015, 60, 19–29. [Google Scholar] [CrossRef]
- Popiołek, Ł. Updated Information on Antimicrobial Activity of Hydrazide-Hydrazones. Int. J. Mol. Sci. 2021, 22, 9389. [Google Scholar] [CrossRef]
- Kaplánek, R.; Havlík, M.; Dolenský, B.; Rak, J.; Džubák, P.; Konečný, P.; Hajdúch, M.; Králová, J.; Král, V. Synthesis and biological activity evaluation of hydrazone derivatives based on a Tröger’s base skeleton. Bioorg. Med. Chem. 2015, 23, 1651–1659. [Google Scholar] [CrossRef]
- Çıkla-Süzgün, P.; Küçükgüzel, Ş.G. Recent Advances in Apoptosis: THE Role of Hydrazones. Mini Rev. Med. Chem. 2019, 19, 1427–1442. [Google Scholar] [CrossRef]
- Kumar, P.; Narasimhan, B. Hydrazides/Hydrazones as Antimicrobial and Anticancer Agents in the New Millennium. Mini Rev. Med. Chem. 2013, 13, 971–987. [Google Scholar] [CrossRef]
- Zhu, X.; Duan, Y.; Li, P.; Fan, H.; Han, T.; Huang, X. A highly selective and instantaneously responsive Schiff base fluorescent sensor for the “turn-off” detection of iron(III), iron(II), and copper(II) ions. Anal. Methods 2019, 11, 642–647. [Google Scholar] [CrossRef]
- Hawes, C.S.; Máille, G.M.Ó.; Byrne, K.; Schmitt, W.; Gunnlaugsson, T. Tetraarylpyrrolo[3,2-b]pyrroles as versatile and responsive fluorescent linkers in metal–organic frameworks. Dalton Trans. 2018, 47, 10080–10092. [Google Scholar] [CrossRef]
- Badal, S.; Her, Y.F.; Maher, L.J. Nonantibiotic Effects of Fluoroquinolones in Mammalian Cells. J. Biol. Chem. 2015, 290, 22287–22297. [Google Scholar] [CrossRef]
- Seydel, J.K.; Schaper, K.J.; Wempe, E.; Cordes, H.P. Mode of action and quantitative structure-activity correlations of tuberculostatic drugs of the isonicotinic acid hydrazide type. J. Med. Chem. 1976, 19, 483–492. [Google Scholar] [CrossRef]
- Chua, G.N.L.; Wassarman, K.L.; Sun, H.; Alp, J.A.; Jarczyk, E.I.; Kuzio, N.J.; Bennett, M.J.; Malachowsky, B.G.; Kruse, M.; Kennedy, A.J. Cytosine-Based TET Enzyme Inhibitors. ACS Med. Chem. Lett. 2019, 10, 180–185. [Google Scholar] [CrossRef]
- Dereka, B.; Rosspeintner, A.; Krzeszewski, M.; Gryko, D.T.; Vauthey, E. Symmetry-Breaking Charge Transfer and Hydrogen Bonding: Toward Asymmetrical Photochemistry. Angew. Chem. Int. Ed. 2016, 55, 15624–15628. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.; Qiao, X.; Wang, D.; Yang, X.; Li, H. Pyrrolo[3,2-b]pyrrole-Based Quinoidal Compounds For High Performance n-Channel Organic Field-Effect Transistor. Chem. Mater. 2018, 30, 6992–6997. [Google Scholar] [CrossRef]
- Liu, H.; Ye, J.; Zhou, Y.; Fu, L.; Lu, Q.; Zhang, C. New pyrrolo[3,2-b]pyrrole derivatives with multiple-acceptor substitution: Efficient fluorescent emission and near-infrared two-photon absorption. Tetrahedron Lett. 2017, 58, 4841–4844. [Google Scholar] [CrossRef]
- Gerecke, C.; Egea Rodrigues, C.; Homann, T.; Kleuser, B. The Role of Ten-Eleven Translocation Proteins in Inflammation. Front. Immunol. 2022, 13, 861351. [Google Scholar] [CrossRef]
- Barszcz, B.; Kędzierski, K.; Jeong, H.Y.; Kim, T.-D. Spectroscopic properties of diketopyrrolopyrrole derivatives with long alkyl chains. J. Lumin. 2017, 185, 219–227. [Google Scholar] [CrossRef]
- Socrates, G. Infrared and Raman Characteristic Group Frequencies, 3rd ed.; John Wiley & Sons, Inc.: Chichester, UK, 2001; p. 368. [Google Scholar]
- Narayana, L.; Suvarapu; Somala, A.R.; Bobbala, P.; Inseong, H.; Ammireddy, V.R. Simultaneous Spectrophotometric Determination of Chromium(VI) and Vanadium(V) by using 3,4-Dihydroxybenzaldehyde isonicotinoyl hydrazone (3,4-DHBINH). J. Chem. 2009, 6, 718738. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Petrat, F.; de Groot, H.; Sustmann, R.; Rauen, U. The chelatable iron pool in living cells: A methodically defined quantity. Biol. Chem. 2002, 383, 489–502. [Google Scholar] [CrossRef]
- Lv, H.; Shang, P. The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics 2018, 10, 899–916. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef]
- Liu, H.; Dai, C.; Fan, Y.; Guo, B.; Ren, K.; Sun, T.; Wang, W. From autophagy to mitophagy: The roles of P62 in neurodegenerative diseases. J. Bioenerg. Biomembr. 2017, 49, 413–422. [Google Scholar] [CrossRef]
- Corti, O.; Blomgren, K.; Poletti, A.; Beart, P.M. Autophagy in neurodegeneration: New insights underpinning therapy for neurological diseases. J. Neurochem. 2020, 154, 354–371. [Google Scholar] [CrossRef]
- Antonyová, V.; Kejík, Z.; Brogyányi, T.; Kaplánek, R.; Pajková, M.; Talianová, V.; Hromádka, R.; Masařík, M.; Sýkora, D.; Mikšátková, L.; et al. Role of mtDNA disturbances in the pathogenesis of Alzheimer’s and Parkinson’s disease. DNA Repair 2020, 92, 102871. [Google Scholar] [CrossRef]
- Xu, W.; Zeng, Z.; Jiang, J.-H.; Chang, Y.-T.; Yuan, L. Discerning the Chemistry in Individual Organelles with Small-Molecule Fluorescent Probes. Angew. Chem. Int. Ed. 2016, 55, 13658–13699. [Google Scholar] [CrossRef]
- Santo-Domingo, J.; Demaurex, N. Perspectives on: SGP symposium on mitochondrial physiology and medicine: The renaissance of mitochondrial pH. J. Gen. Physiol. 2012, 139, 415–423. [Google Scholar] [CrossRef]
- Geldon, S.; Fernández-Vizarra, E.; Tokatlidis, K. Redox-Mediated Regulation of Mitochondrial Biogenesis, Dynamics, and Respiratory Chain Assembly in Yeast and Human Cells. Front. Cell Dev. Biol. 2021, 9, 720656. [Google Scholar] [CrossRef]
- Manev, H.; Dzitoyeva, S. Progress in mitochondrial epigenetics. Biomol. Concepts 2013, 4, 381–389. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Frankowska, M.; Filip, M. Mitoepigenetics and drug addiction. Pharmacol. Ther. 2014, 144, 226–233. [Google Scholar] [CrossRef]
- Pirola, C.J.; Scian, R.; Gianotti, T.F.; Dopazo, H.; Rohr, C.; Martino, J.S.; Castaño, G.O.; Sookoian, S. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine 2015, 94, e1480. [Google Scholar] [CrossRef]
- Ghosh, S.; Sengupta, S.; Scaria, V. Hydroxymethyl cytosine marks in the human mitochondrial genome are dynamic in nature. Mitochondrion 2016, 27, 25–31. [Google Scholar] [CrossRef]
- Sun, X.; Vaghjiani, V.; Jayasekara, W.S.N.; Cain, J.E.; St. John, J.C. The degree of mitochondrial DNA methylation in tumor models of glioblastoma and osteosarcoma. Clin. Epigenetics 2018, 10, 157. [Google Scholar] [CrossRef]
- Upadhyay, M.; Agarwal, S. Ironing the mitochondria: Relevance to its dynamics. Mitochondrion 2020, 50, 82–87. [Google Scholar] [CrossRef]
- Bogenhagen, D.; Clayton, D.A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 1974, 249, 7991–7995. [Google Scholar] [CrossRef]
- Laukka, T.; Mariani, C.J.; Ihantola, T.; Cao, J.Z.; Hokkanen, J.; Kaelin, W.G.; Godley, L.A.; Koivunen, P. Fumarate and Succinate Regulate Expression of Hypoxia-inducible Genes via TET Enzymes. J. Biol. Chem. 2016, 291, 4256–4265. [Google Scholar] [CrossRef] [Green Version]
- Salminen, A.; Haapasalo, A.; Kauppinen, A.; Kaarniranta, K.; Soininen, H.; Hiltunen, M. Impaired mitochondrial energy metabolism in Alzheimer’s disease: Impact on pathogenesis via disturbed epigenetic regulation of chromatin landscape. Prog. Neurobiol. 2015, 131, 1–20. [Google Scholar] [CrossRef]
- Kaas, G.A.; Zhong, C.; Eason, D.E.; Ross, D.L.; Vachhani, R.V.; Ming, G.-l.; King, J.R.; Song, H.; Sweatt, J.D. TET1 Controls CNS 5-Methylcytosine Hydroxylation, Active DNA Demethylation, Gene Transcription, and Memory Formation. Neuron 2013, 79, 1086–1093. [Google Scholar] [CrossRef]
- Stoccoro, A.; Coppedè, F. Mitochondrial DNA Methylation and Human Diseases. Int. J. Mol. Sci. 2021, 22, 4594. [Google Scholar] [CrossRef]
- Melamed, P.; Yosefzon, Y.; David, C.; Tsukerman, A.; Pnueli, L. Tet Enzymes, Variants, and Differential Effects on Function. Front. Cell Dev. Biol. 2018, 6, 22. [Google Scholar] [CrossRef]
- Nedoluzhko, A.; Mjelle, R.; Renström, M.; Skjærven, K.H.; Piferrer, F.; Fernandes, J.M.O. The first mitochondrial 5-methylcytosine map in a non-model teleost (Oreochromis niloticus) reveals extensive strand-specific and non-CpG methylation. Genomics 2021, 113, 3050–3057. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, C.; Wang, X.; Sun, Y.; Zhang, J.; Chen, J.; Shi, Y. An Epigenetic Role of Mitochondria in Cancer. Cells 2022, 11, 2518. [Google Scholar] [CrossRef] [PubMed]
- Sillen, L.G. High-speed computers as supplement to graphical methods.III. twist matrix methods for minimizing error-square sum in problems with many unknown constants. Acta Chem. Scand. 1964, 18, 1085–1098. [Google Scholar] [CrossRef]
- Jakubek, M.; Kejík, Z.; Antonyová, V.; Kaplánek, R.; Sýkora, D.; Hromádka, R.; Vyhlídalová, K.; Martásek, P.; Král, V. Benzoisothiazole-1,1-dioxide-based synthetic receptor for zinc ion recognition in aqueous medium and its interaction with nucleic acids. Supramol. Chem. 2019, 31, 19–27. [Google Scholar] [CrossRef]
- Jakubek, M.; Kejík, Z.; Kaplánek, R.; Veselá, H.; Sýkora, D.; Martásek, P.; Král, V. Perimidine-based synthetic receptors for determination of copper(II) in water solution. Supramol. Chem. 2018, 30, 218–226. [Google Scholar] [CrossRef]
- Jakubek, M.; Kejík, Z.; Parchaňský, V.; Kaplánek, R.; Vasina, L.; Martásek, P.; Král, V. Water soluble chromone Schiff base derivatives as fluorescence receptor for aluminium(III). Supramol. Chem. 2017, 29, 1–7. [Google Scholar] [CrossRef]
- Abramenko, N.; Kejík, Z.; Kaplánek, R.; Tatar, A.; Brogyányi, T.; Pajková, M.; Sýkora, D.; Veselá, K.; Antonyová, V.; Dytrych, P.; et al. Spectroscopic study of in situ-formed metallocomplexes of proton pump inhibitors in water. Chem. Biol. Drug Des. 2021, 97, 305–314. [Google Scholar] [CrossRef]
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100. [Google Scholar] [CrossRef]
- Hu, L.; Li, Z.; Cheng, J.; Rao, Q.; Gong, W.; Liu, M.; Shi, Y.G.; Zhu, J.; Wang, P.; Xu, Y. Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell 2013, 155, 1545–1555. [Google Scholar] [CrossRef]
- Hu, L.; Lu, J.; Cheng, J.; Rao, Q.; Li, Z.; Hou, H.; Lou, Z.; Zhang, L.; Li, W.; Gong, W.; et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 2015, 527, 118–122. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., 3rd; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Szabó, P.; Kolář, M.; Dvořánková, B.; Lacina, L.; Štork, J.; Vlček, Č.; Strnad, H.; Tvrdek, M.; Smetana, K., Jr. Mouse 3T3 fibroblasts under the influence of fibroblasts isolated from stroma of human basal cell carcinoma acquire properties of multipotent stem cells. Biol. Cell 2011, 103, 233–248. [Google Scholar] [CrossRef]
- Dvořánková, B.; Lacina, L.; Smetana, K., Jr. Isolation of Normal Fibroblasts and Their Cancer-Associated Counterparts (CAFs) for Biomedical Research. Methods Mol. Biol. 2019, 1879, 393–406. [Google Scholar] [CrossRef]




| Compound | Log(K) | Fe(II) Ions: Compound | Absorption Maximum | pKa |
|---|---|---|---|---|
| 1 | 8.4/M | 1:1 | 391 | N/A |
| 17.2/M2 | 1:2 | |||
| 2 | 7.2/M | 1:1 | 411 | 9.293 |
| 17.2/M2 | 1:2 | |||
| 3 | - | - | 415 | 8.772 |
| 4 | 6.9/M | 1:1 | 414 | |
| 17.4/M2 | 1:2 | 9.011 and 12.851 | ||
| 10.9/M2 | 2:1 | |||
| 5 | - | - | 418 | 9.000 |
| 6 | - | - | 419 | 9.213 |
| Compound | Binding Energy |
|---|---|
| 1 | −11.9 kcal/mol |
| 2 | −3.97 kcal/mol |
| 3 | −7.87 kcal/mol |
| 4 | −8.24 kcal/mol |
| 5 | −6.33 kcal/mol |
| 6 | −7.56 kcal/mol |
| 7 | −5.24 kcal/mol |
| Compound | HF-P4 (μM) | H-1299 (μM) | Caov-3 (μM) | U-118 MG (μM) | BT-20 (μM) |
|---|---|---|---|---|---|
| 1 | >20 | 6.89 | 6.48 | >20 | 5.94 |
| 2 | >20 | 8.94 | 13.34 | >20 | 11.90 |
| 3 | >20 | 10.6 | 17.78 | 19.50 | 13.68 |
| 4 | 13.14 | 5.75 | 9.13 | 6.81 | 6.22 |
| 5 | >20 | 10.13 | 9.48 | 6.85 | 11.15 |
| 6 | 16.39 | 14.62 | 15.4 | 8.12 | 9.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonyová, V.; Tatar, A.; Brogyányi, T.; Kejík, Z.; Kaplánek, R.; Vellieux, F.; Abramenko, N.; Sinica, A.; Hajduch, J.; Novotný, P.; et al. Targeting of the Mitochondrial TET1 Protein by Pyrrolo[3,2-b]pyrrole Chelators. Int. J. Mol. Sci. 2022, 23, 10850. https://doi.org/10.3390/ijms231810850
Antonyová V, Tatar A, Brogyányi T, Kejík Z, Kaplánek R, Vellieux F, Abramenko N, Sinica A, Hajduch J, Novotný P, et al. Targeting of the Mitochondrial TET1 Protein by Pyrrolo[3,2-b]pyrrole Chelators. International Journal of Molecular Sciences. 2022; 23(18):10850. https://doi.org/10.3390/ijms231810850
Chicago/Turabian StyleAntonyová, Veronika, Ameneh Tatar, Tereza Brogyányi, Zdeněk Kejík, Robert Kaplánek, Fréderic Vellieux, Nikita Abramenko, Alla Sinica, Jan Hajduch, Petr Novotný, and et al. 2022. "Targeting of the Mitochondrial TET1 Protein by Pyrrolo[3,2-b]pyrrole Chelators" International Journal of Molecular Sciences 23, no. 18: 10850. https://doi.org/10.3390/ijms231810850






