Transcriptome Reveals the Effects of Early Weaning on Lipid Metabolism and Liver Health of Yangtze Sturgeon (Acipenser dabryanus)
Abstract
:1. Introduction
2. Results
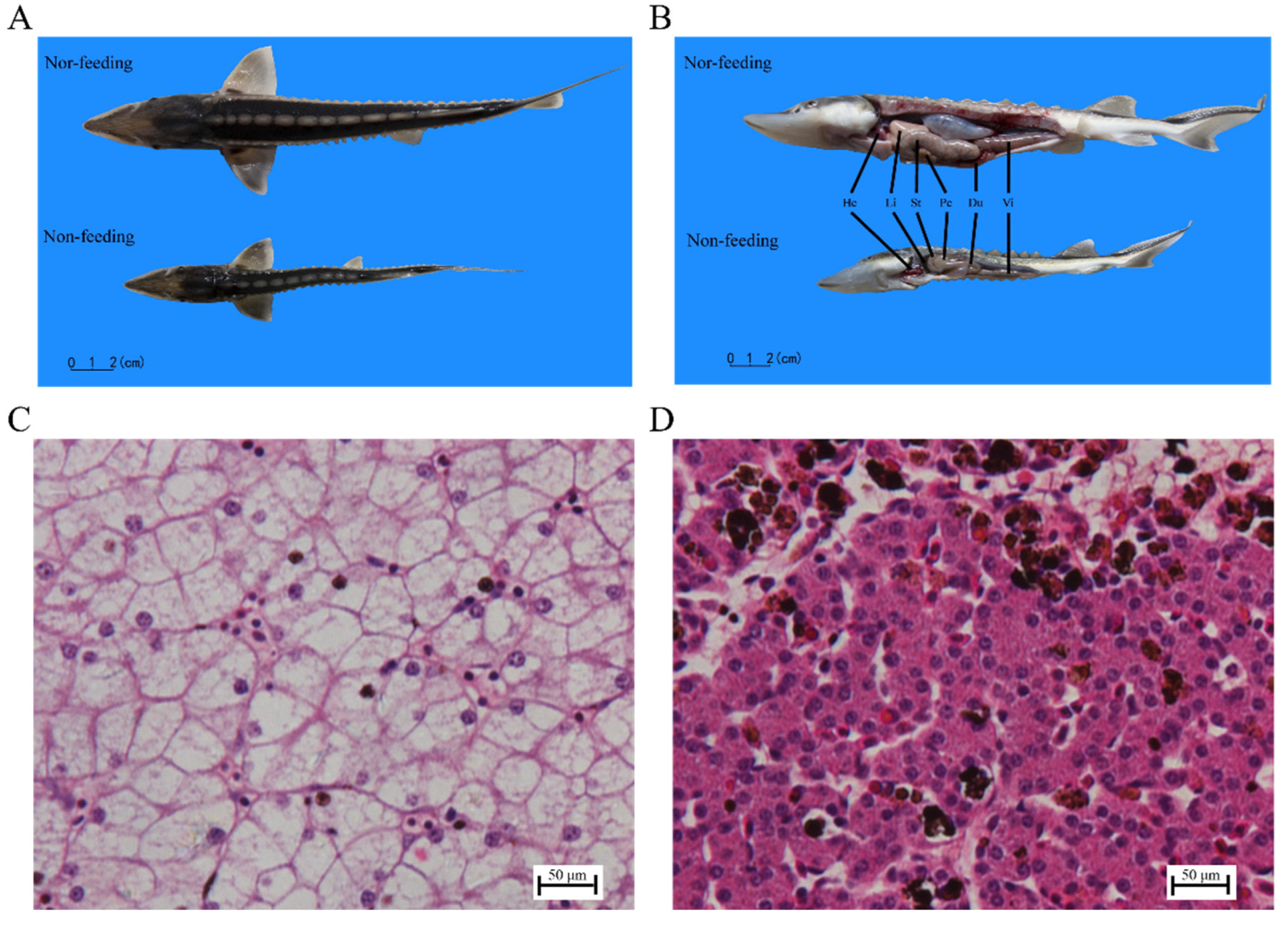
2.1. Histological Observation and Transaminase Activity of Liver
2.2. Transcriptome-Data Analysis
2.3. Gene Cloning and Sequence Analysis
2.4. Effects of Weaning on the Expressions of Genes Related to Liver Lipid Metabolism
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Experimental Design and Sample Collection
4.3. Histological Analysis of Liver
4.4. Transaminase Activity in Liver
4.5. RNA Extraction and Transcriptome Sequencing
4.6. Gene Cloning
4.7. Quantitative Real-Time PCR (qRT-PCR)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. FAO Yearbook. Fishery and Aquaculture Statistics 2019; FAO: Rome, Italy, 2021. [Google Scholar]
- Zhao, L.; He, K.; Luo, J.; Sun, J.; Liao, L.; Tang, X.; Liu, Q.; Yang, S. Co-modulation of Liver Genes and Intestinal Microbiome of Largemouth Bass Larvae (Micropterus salmoides) During Weaning. Front. Microbiol. 2020, 11, 1332. [Google Scholar] [CrossRef]
- Herath, S.S.; Atapaththu, K.S.S. Sudden weaning of angel fish pterophyllum scalare (Lichtenstein) (Pisces; Cichlidae) larvae from brine shrimp (Artemia sp) nauplii to formulated larval feed. SpringerPlus 2013, 2, 102. [Google Scholar] [CrossRef]
- Molnár, T.; Csuvár, A.; Benedek, I.; Molnár, M.; Kabai, P. Domestication affects exploratory behaviour of pikeperch (Sander lucioperca L.) during the transition to pelleted food. PLoS ONE 2018, 13, e0196118. [Google Scholar] [CrossRef]
- Žák, J.; Dyková, I.; Reichard, M. Good performance of turquoise killifish (Nothobranchius furzeri) on pelleted diet as a step towards husbandry standardization. Sci. Rep. 2020, 10, 8986. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, T.N.; Yanong, R.P.; Ramee, S.W.; DiMaggio, M.A. Histological, histochemical and biochemical characterization of larval digestive system ontogeny in black tetra Gymnocorymbus ternetzi to inform aquaculture weaning protocols. Aquaculture 2020, 520, 734957. [Google Scholar] [CrossRef]
- Campoverde, C.; Milne, D.J.; Secombes, C.J.; Estévez, A.; Gisbert, E.; Andree, K.B. Gene expression analysis of the innate immune system during early rearing and weaning of meagre (Argyrosomus regius). Fish Shellfish Immunol. 2019, 94, 819–832. [Google Scholar] [CrossRef] [PubMed]
- He, S.; You, J.-J.; Liang, X.-F.; Zhang, Z.-L.; Zhang, Y.-P. Transcriptome sequencing and metabolome analysis of food habits domestication from live prey fish to artificial diets in mandarin fish (Siniperca chuatsi). BMC Genom. 2021, 22, 129. [Google Scholar] [CrossRef]
- Moguel-Hernández, I.; Peña, R.; Andree, K.B.; Tovar-Ramirez, D.; Bonacic, K.; Dumas, S.; Gisbert, E. Ontogeny changes and weaning effects in gene expression patterns of digestive enzymes and regulatory digestive factors in spotted rose snapper (Lutjanus guttatus) larvae. Fish Physiol. Biochem. 2016, 42, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, E.; Mozanzadeh, M.T. Weaning European glass eels (Anguilla anguilla) with plant protein-based diets and its effects on intestinal maturation. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 228, 43–50. [Google Scholar] [CrossRef]
- Peng, J.; Dou, Y.-Q.; Liang, H.; He, S.; Liang, X.-F.; Shi, L.-J. Social Learning of Acquiring Novel Feeding Habit in Mandarin Fish (Siniperca chuatsi). Int. J. Mol. Sci. 2019, 20, 4399. [Google Scholar] [CrossRef] [Green Version]
- Betancor, M.B.; Ortega, A.; de la Gándara, F.; Tocher, D.R.; Mourente, G. Lipid metabolism-related gene expression pattern of Atlantic bluefin tuna (Thunnus thynnus L.) larvae fed on live prey. Fish Physiol. Biochem. 2017, 43, 493–516. [Google Scholar] [CrossRef] [PubMed]
- Betancor, M.B.; Ortega, A.; de la Gándara, F.; Tocher, D.R.; Mourente, G. Performance, feed utilization, and hepatic metabolic response of weaned juvenile Atlantic bluefin tuna (Thunnus thynnus L.): Effects of dietary lipid level and source. Fish Physiol. Biochem. 2019, 45, 697–718. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, Z.; Liang, X.; Matulic, D.; Hussein, M.; Gao, J. Growth performance, fatty-acid composition, lipid deposition and hepatic-lipid metabolism-related gene expression in juvenile pond loach Misgurnus anguillicaudatus fed diets with different dietary soybean oil levels. J. Fish Biol. 2018, 92, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liao, K.; Wang, T.; Mai, K.; Xu, W.; Ai, Q. Dietary Lipid Levels Influence Lipid Deposition in the Liver of Large Yellow Croaker (Larimichthys crocea) by Regulating Lipoprotein Receptors, Fatty Acid Uptake and Triacylglycerol Synthesis and Catabolism at the Transcriptional Level. PLoS ONE 2015, 10, e0129937. [Google Scholar] [CrossRef] [PubMed]
- Chandra, G.; Fopp-Bayat, D. Trends in aquaculture and conservation of sturgeons: A review of molecular and cytogenetic tools: Genetic tools in sturgeon aquaculture. Rev. Aquac. 2020, 13, 119–137. [Google Scholar] [CrossRef]
- Gisbert, E.; Solovyev, M.; Bonpunt, E.; Mauduit, C. Weaning in Siberian Sturgeon Larvae. In The Siberian Sturgeon (Acipenser baerii, Brandt, 1869) Volume 2–Farming; Williot, P., Nonnotte, G., Chebanov, M., Eds.; Springer: Cham, Switzerland, 2018; pp. 59–72. [Google Scholar]
- Shakourian, M.; Pourkazemi, M.; Sadati, M.A.Y.; Hassani, M.H.S.; Pourali, H.R.; Arshad, U. Effects of replacing live food with formulated diets on growth and survival rates in Persian sturgeon (Acipenser persicus) larvae. J. Appl. Ichthyol. 2011, 27, 771–774. [Google Scholar] [CrossRef]
- Laczynska, B.; Siddique, M.A.M.; Ziomek, E.; Shelton, W.L.; Fopp-Bayat, D. Early Weaning Effects on Survival, Growth, and Histopathology of Larval Sterlet Acipenser ruthenus. N. Am. J. Aquac. 2020, 82, 181–189. [Google Scholar] [CrossRef]
- Rónyai, A.; Feledi, T. Co-feeding as a weaning procedure in sterlet (Acipenser ruthenus L.) larvae. Aquac. Res. 2012, 44, 1489–1491. [Google Scholar] [CrossRef]
- Palmegiano, G.B.; Gai, F.; Daprà, F.; Gasco, L.; Pazzaglia, M.; Peiretti, P.G. Effects of Spirulina and plant oil on the growth and lipid traits of white sturgeon (Acipenser transmontanus) fingerlings. Aquac. Res. 2008, 39, 587–595. [Google Scholar] [CrossRef]
- Markin, E.L.; Lazur, A.; Hengst, A. Assessment of Prerelease Diet Regimes on Growth of Juvenile Atlantic Sturgeon. N. Am. J. Aquac. 2010, 72, 172–176. [Google Scholar] [CrossRef]
- Yang, H.; Leng, X.; Du, H.; Luo, J.; Wu, J.; Wei, Q. Adjusting the Prerelease Gut Microbial Community by Diet Training to Improve the Postrelease Fitness of Captive-Bred Acipenser dabryanus. Front. Microbiol. 2020, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Ping, Z.; Ke, F.; Wei, Q.; He, X.; Cen, Y. Biology and life history of Dabry’s sturgeon, Acipenser dabryanus, in the Yangtze River. Environ. Biol. Fish 1997, 48, 257–264. [Google Scholar]
- Zhang, H.; Wei, Q.W.; Du, H.; Li, L.X. Present status and risk for extinction of the Dabry’s sturgeon (Acipenser dabryanus) in the Yangtze River watershed: A concern for intensified rehabilitation needs. J. Appl. Ichthyol. 2011, 27, 181–185. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–22. [Google Scholar] [CrossRef]
- Wang, B.; Chen, S.H.; Zhang, X.; Tang, N.; Li, Y.; Liu, Y.L.; Hu, Q.; Tang, P.; Chen, D.F.; Li, Z.Q.; et al. The Exploration of Appetite Regulation of Yangtze Sturgeon (Acipenser dabryanu) during Weaning. Aquaculture, 2022; submitted. [Google Scholar]
- Bennett, H.; Troutman, T.D.; Sakai, M.; Glass, C.K. Epigenetic Regulation of Kupffer Cell Function in Health and Disease. Front. Immunol. 2021, 11, 3600. [Google Scholar] [CrossRef]
- Deng, L.; He, K.; Pan, Y.; Wang, H.; Luo, Y.; Xia, Q. The role of tumor-associated macrophages in primary hepatocellular carcinoma and its related targeting therapy. Int. J. Med. Sci. 2021, 18, 2109–2116. [Google Scholar] [CrossRef]
- Feng, W.; Qiao, J.; Jiang, J.; Sun, B.; Li, Z.; Qi, L. Development of alanine aminotransferase reactor based on polymer@Fe3O4 nanoparticles for enzyme inhibitors screening by chiral ligand exchange capillary electrophoresis. Talanta 2018, 182, 600–605. [Google Scholar] [CrossRef]
- Karatas, T.; Onalan, S.; Yildirim, S. Effects of prolonged fasting on levels of metabolites, oxidative stress, immune-related gene expression, histopathology, and DNA damage in the liver and muscle tissues of rainbow trout (Oncorhynchus mykiss). Fish Physiol. Biochem. 2021, 47, 1119–1132. [Google Scholar] [CrossRef]
- Zheng, J.-L.; Luo, Z.; Zhu, Q.-L.; Tan, X.-Y.; Chen, Q.-L.; Sun, L.-D.; Hu, W. Molecular cloning and expression pattern of 11 genes involved in lipid metabolism in yellow catfish Pelteobagrus fulvidraco. Gene 2013, 531, 53–63. [Google Scholar] [CrossRef]
- Tang, Z.; Sun, C.; Yan, A.; Wu, S.; Qin, C.; Zhang, Y.; Li, W. Genes involved in fatty acid metabolism: Molecular characterization and hypothalamic mRNA response to energy status and neuropeptide Y treatment in the orange-spotted grouper Epinephelus coioides. Mol. Cell. Endocrinol. 2013, 376, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; He, L.; Wang, Y.; Li, D.; Chen, W.; Ye, J. Growth performance, fatty acid composition, and lipid metabolism are altered in groupers (Epinephelus coioides) by dietary fish oil replacement with palm oil. Anim. Nutr. 2021, 8, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Wakil, S.J.; Stoops, J.K.; Joshi, V.C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983, 52, 537–579. [Google Scholar] [CrossRef]
- Ohlrogge, J.B.; Jaworski, J.G. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 109–136. [Google Scholar] [CrossRef]
- Peng, M.; Xu, W.; Mai, K.; Zhou, H.; Zhang, Y.; Liufu, Z.; Zhang, K.; Ai, Q. Growth performance, lipid deposition and hepatic lipid metabolism related gene expression in juvenile turbot (Scophthalmus maximus L.) fed diets with various fish oil substitution levels by soybean oil. Aquaculture 2014, 433, 442–449. [Google Scholar] [CrossRef]
- Zimmermann, R.; Strauss, J.G.; Haemmerle, G.; Schoiswohl, G.; Birner-Gruenberger, R.; Riederer, M.; Lass, A.; Neuberger, G.; Eisenhaber, F.; Hermetter, A.; et al. Fat Mobilization in Adipose Tissue Is Promoted by Adipose Triglyceride Lipase. Science 2004, 306, 1383–1386. [Google Scholar] [CrossRef]
- McGarry, J.D.; Woeltje, K.F.; Kuwajima, M.; Foster, D.W. Regulation of ketogenesis and the renaissance of carnitine palmitoyltransferase. Diabetes/Metab. Rev. 1989, 5, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, M.; Li, Y.; Tang, N.; Zhang, X.; Chen, H.; Zhang, S.; Liu, Y.; Wang, J.; Chen, D.; et al. Cloning and expression of kiss genes and regulation of feeding in Siberian sturgeon (Acipenser baerii). Fish Physiol. Biochem. 2022, 48, 419–436. [Google Scholar] [CrossRef]
- Chen, H.; Wang, M.; Li, Y.; Liu, Y.; Zhou, B.; Tang, N.; Zhang, S.; Xu, S.; Yu, N.; Long, Q.; et al. Using the transcriptome to evaluate the best reference genes for studying nutrition of the critically endangered Yangtze sturgeon (Acipenser dabryanus). Aquaculture 2021, 543, 736894. [Google Scholar] [CrossRef]







| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error Rate (%) | Q20 | Sample | Raw Reads |
|---|---|---|---|---|---|---|---|---|
| F_L | 54,101,748 | 8,169,363,948 | 53,678,894 | 8,008,681,153 | 0.0244 | 98.33 | 94.72 | 48.04 |
| S_L | 64,480,944 | 9,736,622,544 | 63,958,528 | 9,527,555,685 | 0.0242 | 98.38 | 94.86 | 47.77 |
| Type | Unigene | Transcript |
|---|---|---|
| Total number | 52,559 | 78,286 |
| Total base | 46,907,651 | 77,903,931 |
| Largest length (bp) | 15,546 | 15,546 |
| Smallest length (bp) | 201 | 201 |
| Average length (bp) | 892.48 | 995.12 |
| N50 length (bp) | 1613 | 1740 |
| E90N50 length (bp) | 2675 | 2399 |
| Fragment-mapped percent (%) | 67.268 | 79.695 |
| GC percent (%) | 44.37 | 44.4 |
| TransRate score | 0.37572 | 0.43717 |
| BUSCO score | C:95.3% [S:93.3%; D:2.0%] | C:95.3% [S:93.3%; D:2.0%] |
| Ingredients | Nutrient Composition (%) |
|---|---|
| Casein | 49 |
| Gelatin | 5 |
| α-starch | 25 |
| Fish oil | 8 |
| Choline chloride | 0.25 |
| Vitamin and mineral premix | 1 |
| CMC-Na | 4 |
| Microcrystalline cellulose | 2.75 |
| Potassium sorbate | 0.1 |
| Moisture | 6.35 |
| Crud protein | 42.55 |
| Crud lipid | 10.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, Y.; Chen, S.; Wang, B.; Wu, H.; Tang, N.; Zhao, L.; Yang, S.; Liu, Q.; Zhou, B.; et al. Transcriptome Reveals the Effects of Early Weaning on Lipid Metabolism and Liver Health of Yangtze Sturgeon (Acipenser dabryanus). Int. J. Mol. Sci. 2022, 23, 10866. https://doi.org/10.3390/ijms231810866
Zhang X, Liu Y, Chen S, Wang B, Wu H, Tang N, Zhao L, Yang S, Liu Q, Zhou B, et al. Transcriptome Reveals the Effects of Early Weaning on Lipid Metabolism and Liver Health of Yangtze Sturgeon (Acipenser dabryanus). International Journal of Molecular Sciences. 2022; 23(18):10866. https://doi.org/10.3390/ijms231810866
Chicago/Turabian StyleZhang, Xin, Youlian Liu, Shuhuang Chen, Bin Wang, Hongwei Wu, Ni Tang, Liulan Zhao, Song Yang, Qiao Liu, Bo Zhou, and et al. 2022. "Transcriptome Reveals the Effects of Early Weaning on Lipid Metabolism and Liver Health of Yangtze Sturgeon (Acipenser dabryanus)" International Journal of Molecular Sciences 23, no. 18: 10866. https://doi.org/10.3390/ijms231810866
APA StyleZhang, X., Liu, Y., Chen, S., Wang, B., Wu, H., Tang, N., Zhao, L., Yang, S., Liu, Q., Zhou, B., Chen, D., & Li, Z. (2022). Transcriptome Reveals the Effects of Early Weaning on Lipid Metabolism and Liver Health of Yangtze Sturgeon (Acipenser dabryanus). International Journal of Molecular Sciences, 23(18), 10866. https://doi.org/10.3390/ijms231810866







