Plasmids as Key Players in Acinetobacter Adaptation
Abstract
:1. Introduction
2. Results
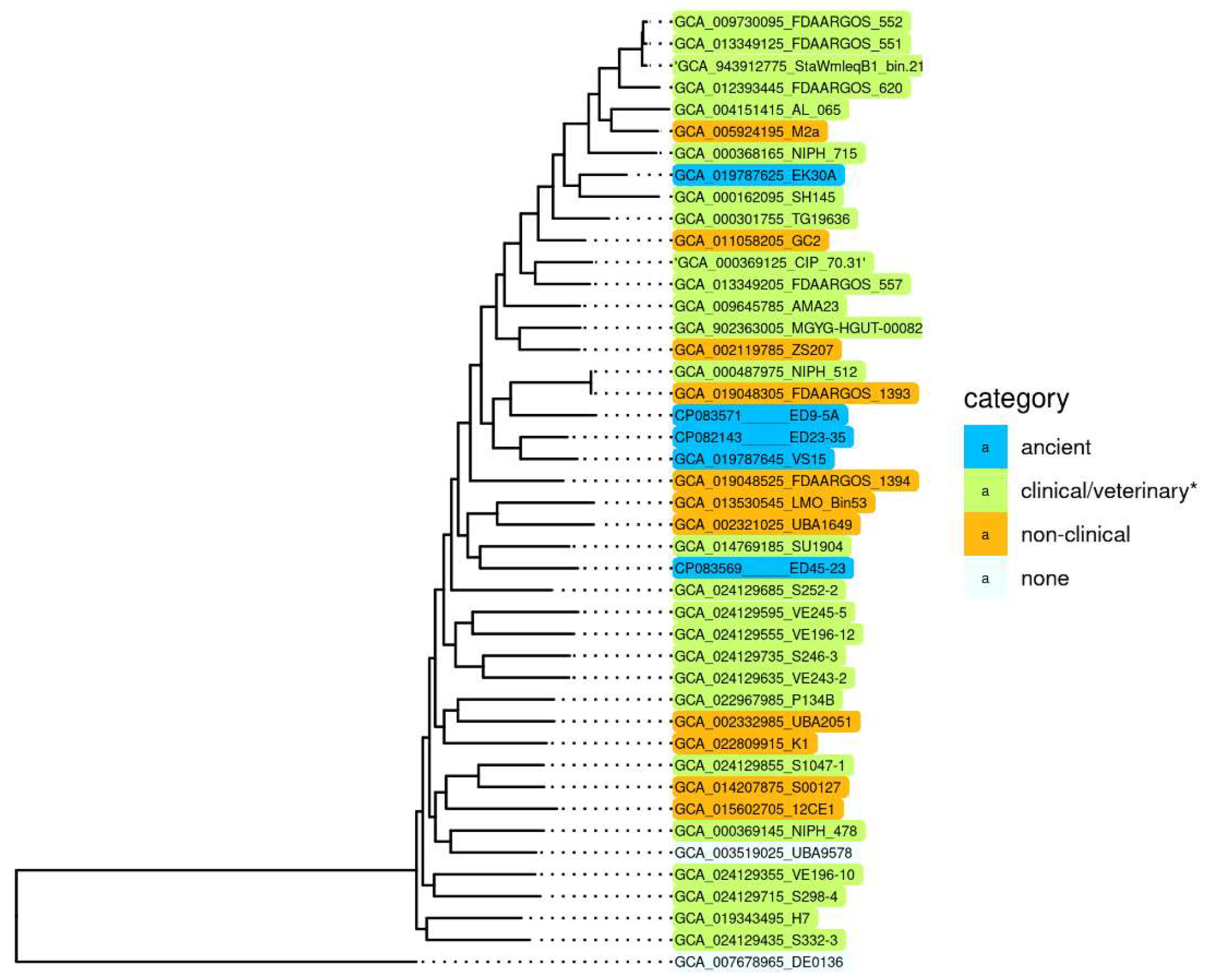
2.1. Comparison of the Chromosomes of A. lwoffii
2.2. Comparison of the Plasmids of A. lwoffii
2.3. Comparative Structure of A. lwoffii and A. baumannii Genomes
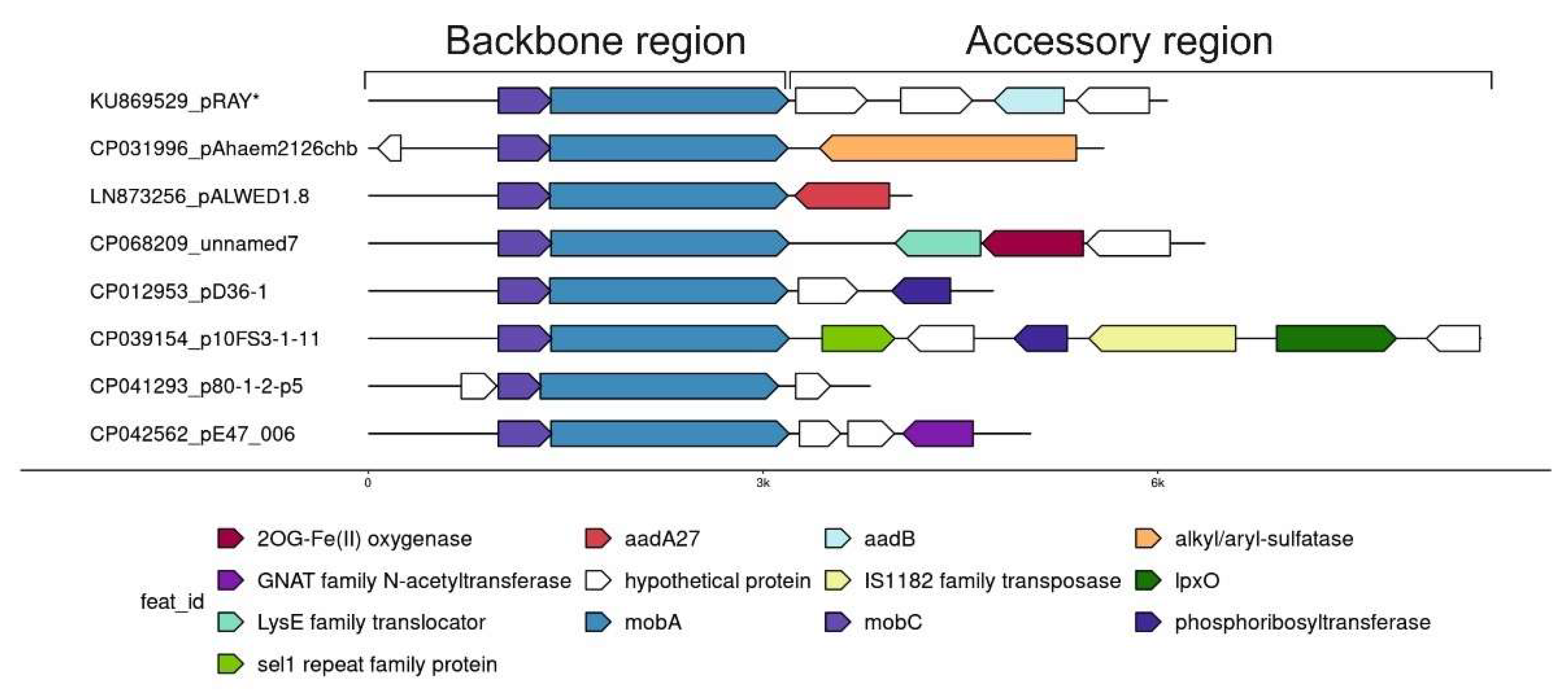
2.4. Acquisition of New Genes in Small Plasmids of Acinetobacter
2.5. Acquisition of New Genes by Acinetobacter Mega-Plasmids
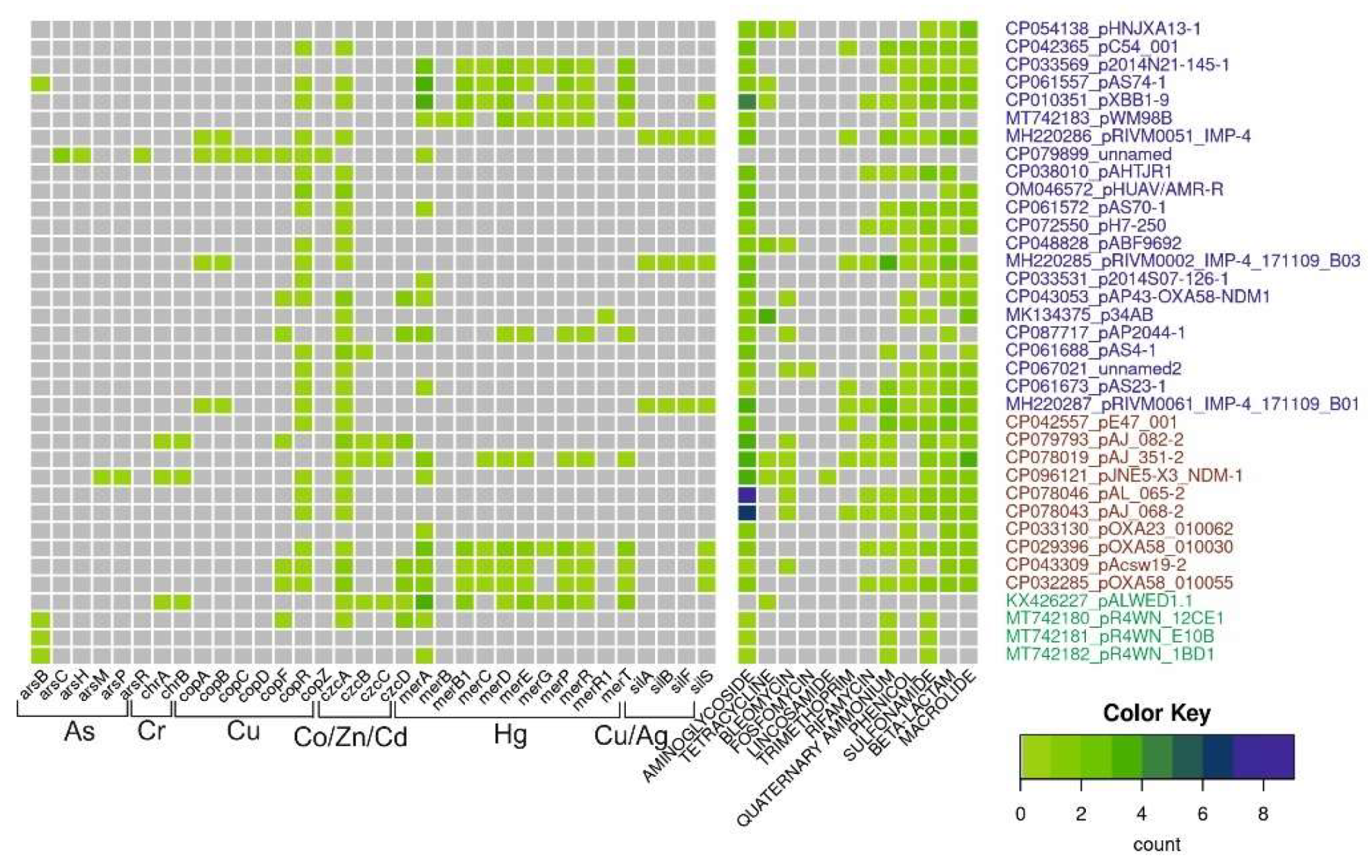
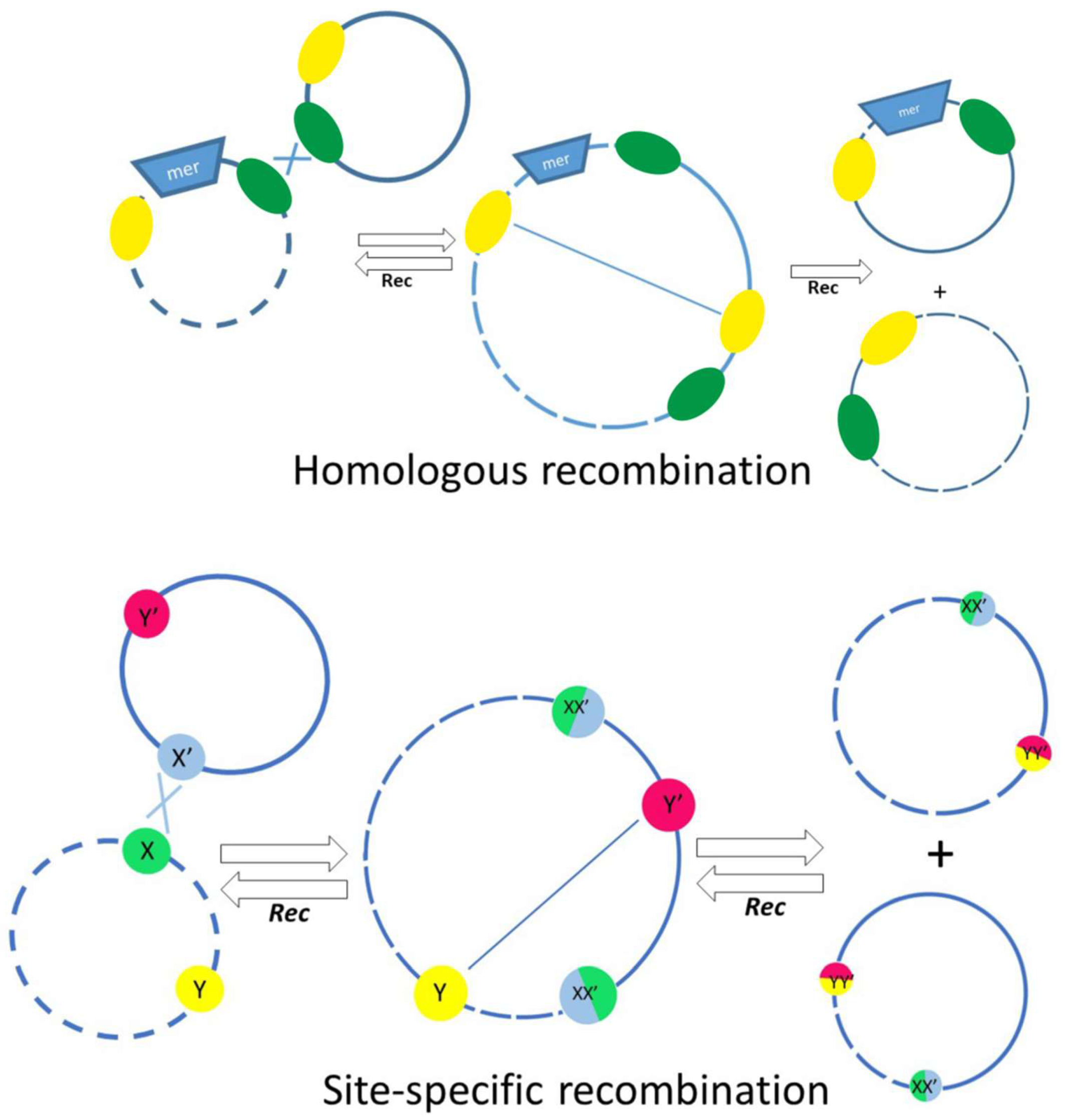
2.6. Mobile Genetic Elements and Mechanisms of Rearrangement of Acinetobacter Plasmids
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baumann, P.; Doudoroff, M.; Stanier, R.Y. A study of the Moraxella group. II. Oxidative-negative species (genus Acinetobacter). J. Bacteriol. 1968, 95, 1520–1541. [Google Scholar] [CrossRef] [PubMed]
- Juni, E. Genetics and physiology of Acinetobacter. Ann. Rev. Microbiol. 1978, 32, 349–371. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Jee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The Ecology, Biology and Pathogenesis of Acinetobacter spp.: An Overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; de Breij, A.; Adams, M.D.; Cerqueira, G.M.; Mocali, S.; Galardini, M.; Nibbering, P.H.; Earl, A.M.; Ward, D.V.; Paterson, D.L.; et al. The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 2012, 7, e46984. [Google Scholar] [CrossRef]
- Fondi, M.; Bacci, G.; Brilli, M.; Papaleo, M.C.; Mengoni, A.; Vaneechoutte, M.; Dijkshoorn, L.; Fani, R. Exploring the evolutionary dynamics of plasmids: The Acinetobacter pan-plasmidome. BMC Evol. Biol. 2010, 10, 59. [Google Scholar] [CrossRef]
- Imperi, F.; Antunes, L.C.; Blom, J.; Villa, L.; Iacono, M.; Visca, P.; Carattoli, A. The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 2011, 63, 1068–1074. [Google Scholar] [CrossRef]
- Jia, J.; Liu, M.; Feng, L.; Wang, Z. Comparative genomic analysis reveals the evolution and environmental adaptation of Acinetobacter johnsonii. Gene 2022, 808, 145985. [Google Scholar] [CrossRef]
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative analysis of Acinetobacters: Three genomes for three lifestyles. PLoS ONE 2008, 3, e1805. [Google Scholar] [CrossRef]
- Santala, S.; Santala, V. Acinetobacter baylyi ADP1-naturally competent for synthetic biology. Essays Biochem. 2021, 65, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Salto, I.P.; Torres Tejerizo, G.; Wibberg, D.; Pühler, A.; Schlüter, A.; Pistorio, M. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 2018, 8, 7783. [Google Scholar] [CrossRef]
- Figueiredo, S.; Poirel, L.; Seifert, H.; Mugnier, P.; Benhamou, D.; Nordmann, P. OXA-134, a naturally occurring carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 2010, 54, 5372–5375. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.C.; Hsueh, P.R.; Yang, P.C.; Luh, K.T. Clinical and microbiological characteristics of bacteremia caused by Acinetobacter lwoffii. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.Z.; Petrova, M.A.; Gorlenko, Z.M.; Soina, V.S.; Khachikian, N.A.; Karaevskaya, E.A. Multidrug-resistant bacteria in permafrost: Isolation, biodiversity, phenotypic and genotypic analysis. In New Permafrost and Glacier Research, 1st ed.; Krugger, M.I., Stern, H.P., Eds.; Nova Science: Hauppauge, NY, USA, 2009; pp. 89–105. [Google Scholar]
- Mindlin, S.; Petrenko, A.; Kurakov, A.; Beletsky, A.; Mardanov, A.; Petrova, M. Resistance of Permafrost and Modern Acinetobacter lwoffii Strains to Heavy Metals and Arsenic Revealed by Genome Analysis. Biomed. Res. Int. 2016, 16, 3970831. [Google Scholar] [CrossRef]
- Rakitin, A.L.; Ermakova, A.Y.; Beletsky, A.V.; Petrova, M.A.; Mardanov, A.V.; Ravin, N.V. Genome Analysis of Acinetobacter lwoffii Strains Isolated from Permafrost Soils Aged from 15 Thousand to 1.8 Million Years Revealed Their Close Relationships with Present-Day Environmental and Clinical Isolates. Biology 2021, 10, 871. [Google Scholar] [CrossRef]
- Walter, T.; Klim, J.; Jurkowski, M.; Gawor, J.; Köhling, I.; Słodownik, M.; Zielenkiewicz, U. Plasmidome of an environmental Acinetobacter lwoffii strain originating from a former gold and arsenic mine. Plasmid 2020, 110, 102505. [Google Scholar] [CrossRef]
- Veress, A.; Nagy, T.; Wilk, T.; Kömüves, J.; Olasz, F.; Kiss, J. Abundance of mobile genetic elements in an Acinetobacter lwoffii strain isolated from Transylvanian honey sample. Sci. Rep. 2020, 10, 2969. [Google Scholar] [CrossRef]
- Acer, O.; Güven, K.; Poli, A.; Donato, P.D.; Leone, L.; Buono, L.; Güven, R.G.; Nicolaus, B.; Finore, I. Acinetobacter mesopotamicus sp. nov., Petroleum-degrading Bacterium, Isolated from Petroleum-contaminated Soil in Diyarbakir, in the Southeast of Turkey. Curr. Microbiol. 2020, 77, 3192–3200. [Google Scholar] [CrossRef]
- Nemec, A. Strain “Acinetobacter mesopotamicus” GC2 Does Not Represent a Novel Species, but Belongs to the Species Acinetobacter lwoffii as Revealed by Whole-Genome Sequence-Based Analysis. Curr. Microbiol. 2021, 78, 369–370. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 2011, 63, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Hall, R.M. AbGR14, a novel antibiotic resistance island in multiply antibiotic-resistant Acinetobacter baumannii clinical isolates. J. Antimicrob. Chemother. 2017, 72, 2944–2947. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.L.N.; Morais-Rodrigues, F.; Hurtado, R.; Dos Santos, R.G.; Costa, D.C.; Barh, D.; Ghosh, P.; Alzahrani, K.J.; Soares, S.C.; Ramos, R.; et al. Pan-Resistome Insights into the Multidrug Resistance of Acinetobacter baumannii. Antibiotics 2021, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Fondi, M.; Rizzi, E.; Emilian, I.G.; Orlandini, V.; Berna, L.; Papaleo, M.C.; Perrin, E.; Maida, I.; Corti, G.; de Bellis, G.; et al. The genome sequence of the hydrocarbon-degrading Acinetobacter venetianus VE-C3. Res. Microbiol. 2013, 164, 439–449. [Google Scholar] [CrossRef]
- Brovedan, M.A.; Cameranesi, M.M.; Limansky, A.S.; Morán-Barrio, J.; Marchiaro, P.; Repizo, G.D.; World, J. What do we know about plasmids carried by members of the Acinetobacter genus? Microbiol. Biotechnol. 2020, 36, 109. [Google Scholar] [CrossRef]
- Feng, Y.; Yang, P.; Wang, X.; Zong, Z. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J. Antimicrob. Chemother. 2016, 71, 71–75. [Google Scholar] [CrossRef]
- Brovedan, M.; Repizo, G.D.; Marchiaro, P.; Viale, A.M.; Limansky, A. Characterization of the diverse plasmid pool harbored by the blaNDM-1- containing Acinetobacter bereziniae HPC229 clinical strain. PLoS ONE 2019, 14, e0220584. [Google Scholar] [CrossRef]
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.; Guo, W.; Jia, Q.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef]
- Kurakov, A.; Mindlin, S.; Beletsky, A.; Shcherbatova, N.; Rakitin, A.; Ermakova, A.; Mardanov, A.; Petrova, M. The ancient small mobilizable plasmid pALWED1.8 harboring a new variant of the non-cassette streptomycin/spectinomycin resistance gene aadA27. Plasmid 2016, 84–85, 36–43. [Google Scholar] [CrossRef]
- Hamidian, M.; Nigro, S.J.; Hall, R.M. Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 2012, 67, 2833–2836. [Google Scholar] [CrossRef] [Green Version]
- Ghaly, T.M.; Paulsen, I.T.; Sajjad, A.; Tetu, S.G.; Gillings, M.R. A novel family of Acinetobacter mega-plasmids are disseminating multi-drug resistance across the globe while acquiring location-specific accessory genes. Front. Microbiol. 2020, 11, 605952. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.; Beletsky, A.; Rakitin, A.; Mardanov, A.; Petrova, M. Acinetobacter Plasmids: Diversity and Development of Classification Strategies. Front. Microbiol. 2020, 11, 588410. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.; Maslova, O.; Beletsky, A.; Nurmukanova, V.; Zong, Z.; Mardanov, A.; Petrova, M. Ubiquitous Conjugative Mega-Plasmids of Acinetobacter Species and Their Role in Horizontal Transfer of Multi-Drug Resistance. Front. Microbiol. 2021, 12, 728644. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R. Class 1 integrons as invasive species. Curr. Opin. Microbiol. 2017, 38, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, A.D.; Yin, X.L.; Zhang, T. The Prevalence of Integrons as the Carrier Resistance Genes in Natural and Man-Made Environments. Environ. Sci. Technol. 2017, 51, 5721–5728. [Google Scholar] [CrossRef]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The Natural History of Integrons. Microorganisms 2021, 9, 2212. [Google Scholar] [CrossRef]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Nigro, S.J.; Hall, R.M. Structure and Context of acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 2016, 71, 1135–1147. [Google Scholar] [CrossRef]
- Repizo, G.D.; Viale, A.M.; Borges, V.; Cameranesi, M.M.; Taib, N.; Espariz, M.; Brochier-Armanet, C.; Gomes, J.P.; Salcedo, S.P. The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol. Evol. 2017, 9, 2292–2307. [Google Scholar] [CrossRef]
- Ross, K.; Varani, A.M.; Snesrud, E.; Huang, H.; Alvarenga, D.O.; Zhang, J.; Wu, C.; McGann, P.; Chandler, M. TnCentral: A Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis. mBio 2021, 12, e0206021. [Google Scholar] [CrossRef]
- Kholodii, G.; Mindlin, S.; Gorlenko Zh Petrova, M.; Hobman, J.; Nikirofov, V. Translocation of transposition-deficient (TndPKLH2-like) transposons in the natural environment: Mechanistic insights from the study of adjacent DNA sequences. Microbiology 2004, 150, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Kholodii, G. The shuffling function of resolvases. Gene 2001, 269, 121–130. [Google Scholar] [CrossRef]
- Moon, H.S.; Abercrombie, L.L.; Eda, S.; Blanvillain, R.; Thomson, J.G.; Ow, D.W.; Stewart, C.N., Jr. Transgene excision in pollen using a codon optimized serine resolvase CinH-RS2 site-specific recombination system. Plant Mol. Biol. 2011, 5, 621–631. [Google Scholar] [CrossRef]
- Thomson, J.G.; Ow, D.W. Site-specific recombination systems for the genetic manipulation of eukaryotic genomes. Genesis 2006, 44, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Balalovski, P.; Grainge, I. Mobilization of pdif modules in Acinetobacter: A novel mechanism for antibiotic resistance gene shuffling? Mol. Microbiol. 2020, 114, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Blakely, G.W.; Sherratt, D.J. Interactions of the site-specific recombinases XerC and XerD with the recombination site dif. Nucleic Acids Res. 1994, 22, 5613–5620. [Google Scholar] [CrossRef]
- Hayes, F.; Sherratt, D.J. Recombinase binding specificity at the chromosome dimer resolution site dif of Escherichia coli. J. Mol. Biol. 1997, 266, 525–537. [Google Scholar] [CrossRef]
- Merino, M.; Acosta, J.; Poza, M.; Sanz, F.; Beceiro, A.; Chaves, F.; Bou, G. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 2010, 54, 2724–2727. [Google Scholar] [CrossRef]
- Blackwell, G.A.; Hall, R.M. The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 2017, 61, e780-17. [Google Scholar] [CrossRef]
- Mindlin, S.; Beletsky, A.; Mardanov, A.; Petrova, M. Adaptive dif Modules in Permafrost Strains of Acinetobacter lwoffii and Their Distribution and Abundance Among Present Day Acinetobacter Strains. Front. Microbiol. 2019, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Mindlin, S.; Petrenko, A.; Petrova, M. Chromium resistance genetic element flanked by XerC/XerD recombination sites and its distribution in environmental and clinical Acinetobacter strains. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, M.M.; Giani, T.; D’Arezzo, S.; Capone, A.; Petrosillo, N.; Visca, P.; Luzzaro, F.; Rossolini, G.M. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 3528–3533. [Google Scholar] [CrossRef] [PubMed]
- Grosso, F.; Quinteira, S.; Poirel, L.; Novais, A.; Peixe, L. Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 2012, 56, 3969–3972. [Google Scholar] [CrossRef] [PubMed]
- Povilonis, J.; Seputiene, V.; Krasauskas, R.; Juskaite, R.; Miskinyte, M.; Suziedelis, K.; Suziedeliene, E. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 2013, 68, 1000–1006. [Google Scholar] [CrossRef]
- Liu, H.; Moran, R.A.; Chen, Y.; Doughty, E.L.; Hua, X.; Jiang, Y.; Xu, Q.; Zhang, L.; Blair, J.M.A.; McNally, A.; et al. Transferable Acinetobacter baumannii plasmid pDETAB2 encodes OXA-58 and NDM-1 and represents a new class of antibiotic resistance plasmids. J. Antimicrob. Chemother. 2021, 76, 1130–1134. [Google Scholar] [CrossRef]
- Ibrahim, S.; Al-Saryi, N.; Al-Kadmy, I.M.S.; Aziz, S.N. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 2021, 48, 6987–6998. [Google Scholar] [CrossRef]
- Hamidian, M.; Ambrose, S.J.; Blackwell, G.A.; Nigro, S.J.; Hall, R.M. An outbreak of multiply antibiotic-resistant ST49:ST128:KL11:OCL8 Acinetobacter baumannii isolates at a Sydney hospital. J. Antimicrob. Chemother. 2021, 7, 893–900. [Google Scholar] [CrossRef]
- Wang, Z.; Li, H.; Zhang, J.; Wang, X.; Zhang, Y.; Wang, H. Identification of a novel plasmid-mediated tigecycline resistance-related gene, tet(Y), in Acinetobacter baumannii. J. Antimicrob. Chemother. 2021, 77, 58–68. [Google Scholar] [CrossRef]
- Hernández-González, I.L.; Mateo-Estrada, V.; Castillo-Ramirez, S. The promiscuous and highly mobile resistome of Acinetobacter baumannii. Microb. Genom. 2022, 8, 000762. [Google Scholar] [CrossRef]
| Strain | Isolation Depth (m) | Age of Permafrost (Thousand Years) | Resistance to Antibiotics | Resistance to Heavy Metals | Accession Number |
|---|---|---|---|---|---|
| ED25-35 | 4.5 | 20–40 | Sm, Sp | Hg, Cr, Co, Cd, Zn, Ni | CP082143.1 |
| ED45-23 | 2.9 | 20–40 | - | Hg, As, Cu | CP083569.1 |
| ED9-5A | 6.5 | 15–30 | - | Hg, As, Cr, Cd, Zn, Cu | CP083571.1 |
| VS15 | 34.0 | 20–40 | Ap, Cm, Sm, Sp | Co, Cd, Zn, Cu | CP080576.1 |
| EK30A | 47.9 | 1600–1800 | Ap, Sm, Sp | Cr, Co, Cd, Cu | CP080636.1 |
| Group of Plasmids | Plasmid Number | Number of Genes (Determinants) of | |||
|---|---|---|---|---|---|
| Heavy Metal Resistance | Antibiotic Resistance | ||||
| Total | Per Plasmid | Total | Per Plasmid | ||
| From permafrost strains | 41 | 30 | 30/41 = 0.7 | 3 | 3/41 = 0.07 |
| From modern strains | 75 | 33 | 33/75 = 0.44 | 41 | 41/75 = 0.55 |
| Species | Genome Size | Plasmid Number | Content of Plasmid DNA, % of the Genome | Location of Heavy Metal Resistance Genes |
|---|---|---|---|---|
| A. lwoffii | 3,408,464 | 5–15 | 7.5% | mainly in plasmids |
| A. baumannii | 4,018,426 | 0–4 | 2.0% | mainly in the chromosome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maslova, O.; Mindlin, S.; Beletsky, A.; Mardanov, A.; Petrova, M. Plasmids as Key Players in Acinetobacter Adaptation. Int. J. Mol. Sci. 2022, 23, 10893. https://doi.org/10.3390/ijms231810893
Maslova O, Mindlin S, Beletsky A, Mardanov A, Petrova M. Plasmids as Key Players in Acinetobacter Adaptation. International Journal of Molecular Sciences. 2022; 23(18):10893. https://doi.org/10.3390/ijms231810893
Chicago/Turabian StyleMaslova, Olga, Sofia Mindlin, Alexey Beletsky, Andrey Mardanov, and Mayya Petrova. 2022. "Plasmids as Key Players in Acinetobacter Adaptation" International Journal of Molecular Sciences 23, no. 18: 10893. https://doi.org/10.3390/ijms231810893
APA StyleMaslova, O., Mindlin, S., Beletsky, A., Mardanov, A., & Petrova, M. (2022). Plasmids as Key Players in Acinetobacter Adaptation. International Journal of Molecular Sciences, 23(18), 10893. https://doi.org/10.3390/ijms231810893