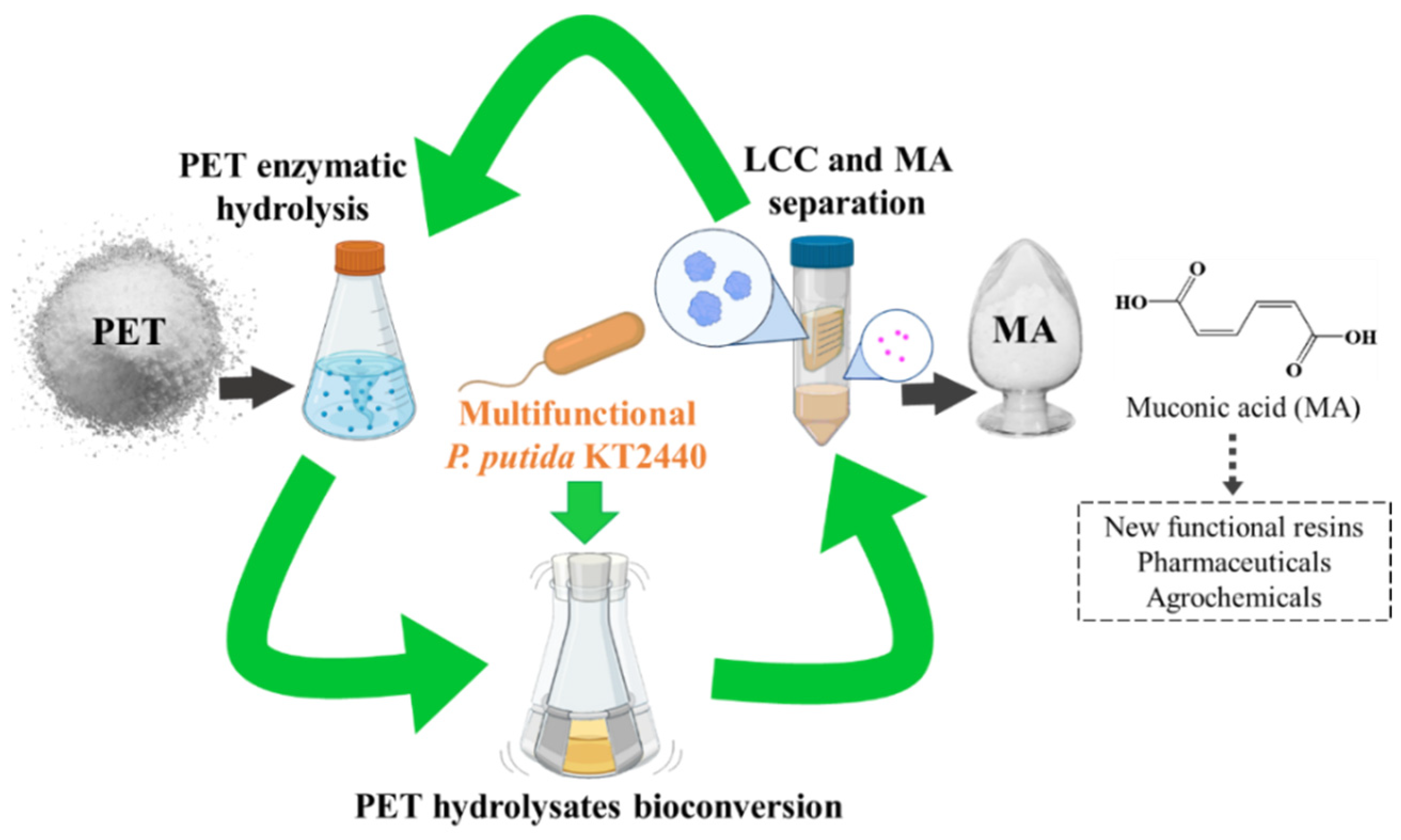
Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida
Abstract
:1. Introduction
2. Results and Discussion
2.1. Construction of a Multifunctional P. Putida KT2440
2.2. Enzymatic Hydrolysis of PET Using LCC Crude Enzyme
2.3. Bioconversion of the PET Hydrolysates into MA by the Engineered Multifunctional Strain
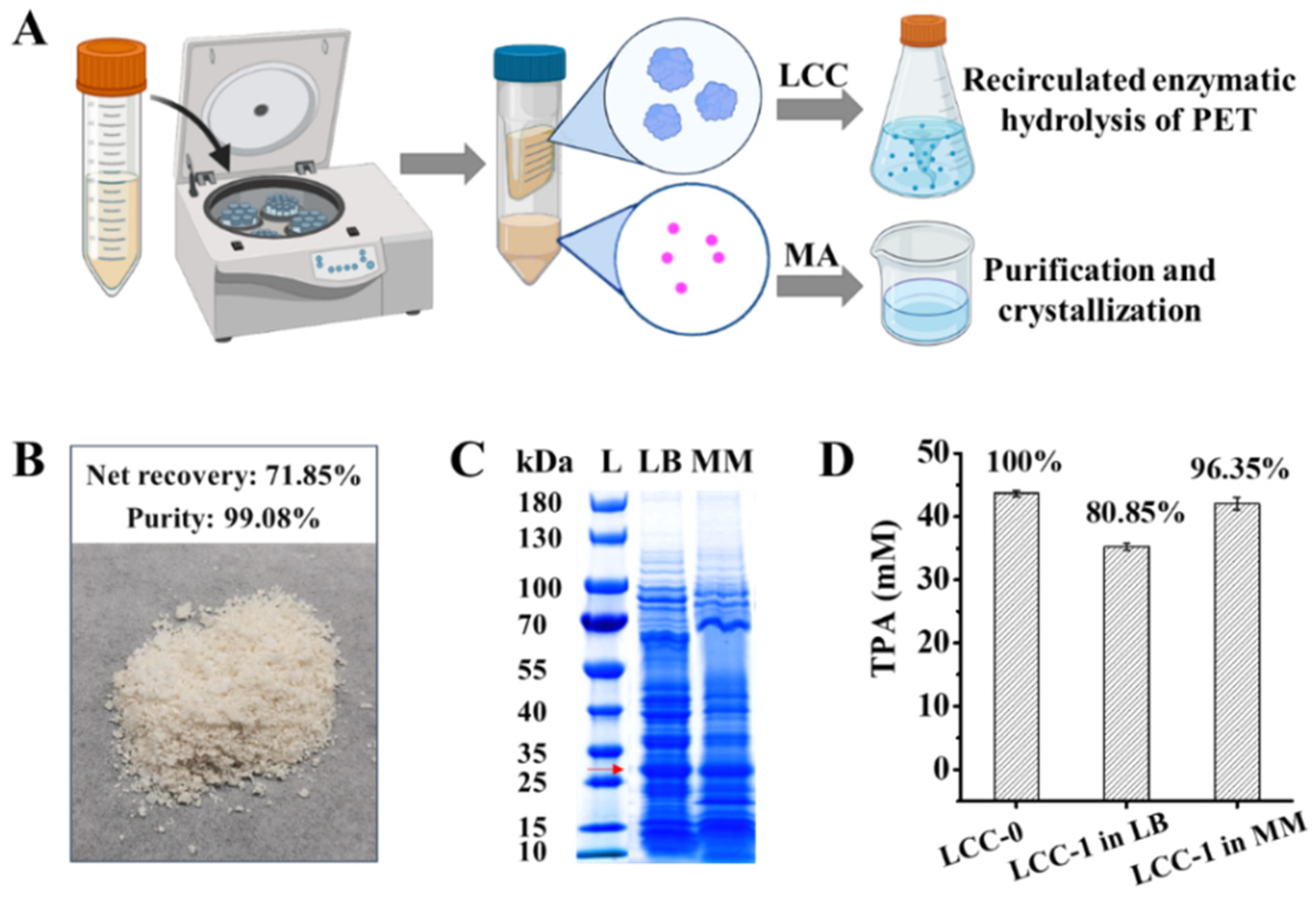
2.4. Separation of LCC from MA in Culture Supernatant for Continuous PET Valorization
3. Materials and Methods
3.1. Strains and Cultivation
3.2. Plasmid Construction and Strain Engineering
3.3. Preparatin of LCC Crude Enzyme
3.4. Enzyme Assays on Bis(2-Hydroxyethyl) Terephthalate (BHET)
3.5. Enzymatic Hydrolysis of PET
3.6. Well Plate Cultivation for the Bioconversion of TPA
3.7. Shake Flask Cultivation on the PET Hydrolysates
3.8. Separation of LCC and MA
3.9. Substrate and Product Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsironi, T.N.; Chatzidakis, S.M.; Stoforos, N.G. The future of polyethylene terephthalate bottles: Challenges and sustainability. Packag. Technol. Sci. 2022, 35, 317–325. [Google Scholar] [CrossRef]
- Kubowicz, S.; Booth, A.M. Biodegradability of Plastics: Challenges and Misconceptions. Environ. Sci. Technol. 2017, 51, 12058–12060. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Pantelic, B.; Azeem, M.; Taxeidis, G.; Babu, R.; Topakas, E.; Brennan Fournet, M.; Nikodinovic-Runic, J. Progressing Plastics Circularity: A Review of Mechano-Biocatalytic Approaches for Waste Plastic (Re)valorization. Front. Bioeng. Biotechnol. 2021, 9, 696040. [Google Scholar] [CrossRef] [PubMed]
- Barnard, E.; Rubio Arias, J.J.; Thielemans, W. Chemolytic depolymerisation of PET: A review. Green Chem. 2021, 23, 3765–3789. [Google Scholar] [CrossRef]
- Qi, X.; Yan, W.; Cao, Z.; Ding, M.; Yuan, Y. Current Advances in the Biodegradation and Bioconversion of Polyethylene Terephthalate. Microorganisms 2021, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Kenny, S.T.; Runic, J.N.; Kaminsky, W.; Woods, T.; Babu, R.P.; Keely, C.M.; Blau, W.; O’Connor, K.E. Up-cycling of PET (polyethylene terephthalate) to the biodegradable plastic PHA (polyhydroxyalkanoate). Environ. Sci. Technol. 2008, 42, 7696–7701. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.-H.; Song, B.K.; Park, S.J.; et al. Biological Valorization of Poly(ethylene terephthalate) Monomers for Upcycling Waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Cao, G.; Ma, X.; Zhou, J.; Xu, W. Bacterial cellulose production from terylene ammonia hydrolysate by Taonella mepensis WT-6. Int. J. Biol. Macromol. 2021, 166, 251–258. [Google Scholar] [CrossRef]
- Kim, D.H.; Han, D.O.; In Shim, K.; Kim, J.K.; Pelton, J.G.; Ryu, M.H.; Joo, J.C.; Han, J.W.; Kim, H.T.; Kim, K.H. One-Pot Chemo-bioprocess of PET Depolymerization and Recycling Enabled by a Biocompatible Catalyst, Betaine. ACS Catal. 2021, 11, 3996–4008. [Google Scholar] [CrossRef]
- Werner, A.Z.; Clare, R.; Mand, T.D.; Pardo, I.; Ramirez, K.J.; Haugen, S.J.; Bratti, F.; Dexter, G.N.; Elmore, J.R.; Huenemann, J.D.; et al. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to beta-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261. [Google Scholar] [CrossRef]
- Kawai, F. The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts 2021, 11, 206. [Google Scholar] [CrossRef]
- Then, J.; Wei, R.; Oeser, T.; Gerdts, A.; Schmidt, J.; Barth, M.; Zimmermann, W. A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 2016, 6, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zheng, Y.; Su, T.; Wang, Q.; Liang, Q.; Zhang, Z.; Qi, Q.; Tian, J. Computational design of a cutinase for plastic biodegradation by mining molecular dynamics simulations trajectories. Comput. Struct. Biotechnol. J. 2022, 20, 459–470. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.e.; Qiao, Y.; Mitra, R.; Han, J.; Li, C.; Han, X.; et al. Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catal. 2021, 11, 1340–1350. [Google Scholar] [CrossRef]
- Sadler, J.C.; Wallace, S. Microbial synthesis of vanillin from waste poly(ethylene terephthalate). Green Chem. 2021, 23, 4665–4672. [Google Scholar] [CrossRef]
- Tiso, T.; Narancic, T.; Wei, R.; Pollet, E.; Beagan, N.; Schroder, K.; Honak, A.; Jiang, M.; Kenny, S.T.; Wierckx, N.; et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021, 66, 167–178. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, T.; Zheng, Y.; Li, Q.; Su, T.; Qi, Q. Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Eng. Microbiol. 2021, 1, 100003. [Google Scholar] [CrossRef]
- Rorrer, N.A.; Dorgan, J.R.; Vardon, D.R.; Martinez, C.R.; Yang, Y.; Beckham, G.T. Renewable Unsaturated Polyesters from Muconic Acid. ACS Sustain. Chem. Eng. 2016, 4, 6867–6876. [Google Scholar] [CrossRef]
- Yang, M.; Liu, D.; Baral, N.R.; Lin, C.Y.; Simmons, B.A.; Gladden, J.M.; Eudes, A.; Scown, C.D. Comparing in planta accumulation with microbial routes to set targets for a cost-competitive bioeconomy. Proc. Natl. Acad. Sci. USA 2022, 119, e2122309119. [Google Scholar] [CrossRef]
- Khalil, I.; Quintens, G.; Junkers, T.; Dusselier, M. Muconic acid isomers as platform chemicals and monomers in the biobased economy. Green Chem. 2020, 22, 1517–1541. [Google Scholar] [CrossRef]
- Xie, N.Z.; Liang, H.; Huang, R.B.; Xu, P. Biotechnological production of muconic acid: Current status and future prospects. Biotechnol. Adv. 2014, 32, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.W.; Salvachua, D.; Khanna, P.; Smith, H.; Peterson, D.J.; Beckham, G.T. Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 2016, 3, 111–119. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, T.; Zheng, Y.; Li, Q.B.; Liang, Q.F.; Qi, Q.S. Screening and genome analysis of a Pseudomonas stutzeri that degrades PET monomer terephthalate. Acta Microbiol. Sin. 2021, 62, 200–212. [Google Scholar] [CrossRef]
- White, M.D.; Payne, K.A.; Fisher, K.; Marshall, S.A.; Parker, D.; Rattray, N.J.; Trivedi, D.K.; Goodacre, R.; Rigby, S.E.; Scrutton, N.S.; et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 2015, 522, 502–506. [Google Scholar] [CrossRef]
- Salvachúa, D.; Johnson, C.W.; Singer, C.A.; Rohrer, H.; Peterson, D.J.; Black, B.A.; Knapp, A.; Beckham, G.T. Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 2018, 20, 5007–5019. [Google Scholar] [CrossRef]
- Vardon, D.R.; Franden, M.A.; Johnson, C.W.; Karp, E.M.; Guarnieri, M.T.; Linger, J.G.; Salm, M.J.; Strathmann, T.J.; Beckham, G.T. Adipic acid production from lignin. Energy Environ. Sci. 2015, 8, 617–628. [Google Scholar] [CrossRef]
- Weber, C.; Bruckner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef]
- Curran, K.A.; Leavitt, J.M.; Karim, A.S.; Alper, H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013, 15, 55–66. [Google Scholar] [CrossRef]
- Sonoki, T.; Morooka, M.; Sakamoto, K.; Otsuka, Y.; Nakamura, M.; Jellison, J.; Goodell, B. Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J. Biotechnol. 2014, 192, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Franden, M.A.; Jayakody, L.N.; Li, W.J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J.; et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Li, W.J.; Jayakody, L.N.; Franden, M.A.; Wehrmann, M.; Daun, T.; Hauer, B.; Blank, L.M.; Beckham, G.T.; Klebensberger, J.; Wierckx, N. Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environ. Microbiol. 2019, 21, 3669–3682. [Google Scholar] [CrossRef]
- Su, L.; Woodard, R.W.; Chen, J.; Wu, J. Extracellular location of Thermobifida fusca cutinase expressed in Escherichia coli BL21(DE3) without mediation of a signal peptide. Appl. Environ. Microbiol. 2013, 79, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Shirke, A.N.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: Mechanism and effect on PET hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Alonso Villela, S.M.; Kraiem, H.; Bouhaouala-Zahar, B.; Bideaux, C.; Aceves Lara, C.A.; Fillaudeau, L. A protocol for recombinant protein quantification by densitometry. Microbiologyopen 2020, 9, 1175–1182. [Google Scholar] [CrossRef]
- Johnson, C.W.; Salvachúa, D.; Rorrer, N.A.; Black, B.A.; Vardon, D.R.; St. John, P.C.; Cleveland, N.S.; Dominick, G.; Elmore, J.R.; Grundl, N.; et al. Innovative Chemicals and Materials from Bacterial Aromatic Catabolic Pathways. Joule 2019, 3, 1523–1537. [Google Scholar] [CrossRef]
- Yu, X.; Jin, H.; Liu, W.; Wang, Q.; Qi, Q. Engineering Corynebacterium glutamicum to produce 5-aminolevulinic acid from glucose. Microb. Cell Fact. 2015, 14, 183. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., 2nd; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Wang, Y.; Horlamus, F.; Henkel, M.; Kovacic, F.; Schläfle, S.; Hausmann, R.; Wittgens, A.; Rosenau, F. Growth of engineered Pseudomonas putida KT2440 on glucose, xylose, and arabinose: Hemicellulose hydrolysates and their major sugars as sustainable carbon sources. GCB Bioenergy 2019, 11, 249–259. [Google Scholar] [CrossRef]
- Cui, Z.; Zheng, H.; Jiang, Z.; Wang, Z.; Hou, J.; Wang, Q.; Liang, Q.; Qi, Q. Identification and Characterization of the Mitochondrial Replication Origin for Stable and Episomal Expression in Yarrowia lipolytica. ACS Synth. Biol. 2021, 10, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wei, R.; Cui, Q.; Bornscheuer, U.T.; Liu, Y.J. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2021, 14, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Vardon, D.R.; Rorrer, N.A.; Salvachúa, D.; Settle, A.E.; Johnson, C.W.; Menart, M.J.; Cleveland, N.S.; Ciesielski, P.N.; Steirer, K.X.; Dorgan, J.R.; et al. cis,cis-Muconic acid: Separation and catalysis to bio-adipic acid for nylon-6,6 polymerization. Green Chem. 2016, 18, 3397–3413. [Google Scholar] [CrossRef]
- Lu, H.; Diaz, D.J.; Czarnecki, N.J.; Zhu, C.; Kim, W.; Shroff, R.; Acosta, D.J.; Alexander, B.R.; Cole, H.O.; Zhang, Y.; et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 2022, 604, 662–667. [Google Scholar] [CrossRef]



| Strains and Plasmids | Relevant Properties | Sources |
|---|---|---|
| pK18mobsacB | Allelic exchange vector, oriColE1 Mob+, sacB, Kmr | Lab Stock [39] |
| pBBR1MCS-2 | Protein expression vector, pBBR1 replicon, Mob+, Kmr | Lab Stock [40] |
| pBBR-LCC | LCC expression driven by lac promoter on pBBR1MCS-2 | This study |
| E. coli DH5α | F– φ80lacZ∆M15 ∆(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1, used for plasmids construction and clone. | Lab Stock |
| P. putida KT2440 | Wild-type | Lab Stock |
| P. putida KT2440-t | P. putida KT2440 (ΔpcaHG::tph), genomic replacement of pcaHG with tph cluster (tphRA2A3A1BK). | This study |
| P. putida KT2440-ta | P. putida KT2440 (ΔpcaHG::tph::aroY:ecdB), additional insertion of codon-optimized aroY and ecdB follow tph cluster. | This study |
| P. putida KT2440-tac | P. putida KT2440 (ΔpcaHG::tph::aroY:ecdB ΔcatRBC::Ptac:catA), additional genomic replacement of catRBC with tac promoter, which enabled constitutive expression of catA | This study |
| P. putida KT2440-tacR | P. putida KT2440 (ΔpcaHG::tph::aroY:ecdB ΔcatRBC::Ptac:catA ΔgclR), additional gclR deletion. | This study |
| P. putida KT2440-tacRD | P. putida KT2440 (ΔpcaHG::tph::aroY:ecdB ΔcatRBC::Ptac:catA ΔgclR::Ptac:glcDEF), additional promoter replacement of glcDEF wih tac promoter. | This study |
| P. putida KT2440-tacRDL | P. putida KT2440 (ΔpcaHG::tph::aroY:ecdB ΔcatRBC::Ptac:catA ΔgclR::Ptac:glcDEF) (pBBR-LCC), additional LCC expression on pBBR1MCS-2 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zheng, Y.; Yuan, Y.; Zhang, T.; Li, Q.; Liang, Q.; Su, T.; Qi, Q. Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida. Int. J. Mol. Sci. 2022, 23, 10997. https://doi.org/10.3390/ijms231910997
Liu P, Zheng Y, Yuan Y, Zhang T, Li Q, Liang Q, Su T, Qi Q. Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida. International Journal of Molecular Sciences. 2022; 23(19):10997. https://doi.org/10.3390/ijms231910997
Chicago/Turabian StyleLiu, Pan, Yi Zheng, Yingbo Yuan, Tong Zhang, Qingbin Li, Quanfeng Liang, Tianyuan Su, and Qingsheng Qi. 2022. "Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida" International Journal of Molecular Sciences 23, no. 19: 10997. https://doi.org/10.3390/ijms231910997
APA StyleLiu, P., Zheng, Y., Yuan, Y., Zhang, T., Li, Q., Liang, Q., Su, T., & Qi, Q. (2022). Valorization of Polyethylene Terephthalate to Muconic Acid by Engineering Pseudomonas Putida. International Journal of Molecular Sciences, 23(19), 10997. https://doi.org/10.3390/ijms231910997





