Mutation of OsSAC3, Encoding the Xanthine Dehydrogenase, Caused Early Senescence in Rice
Abstract
:1. Introduction
2. Results
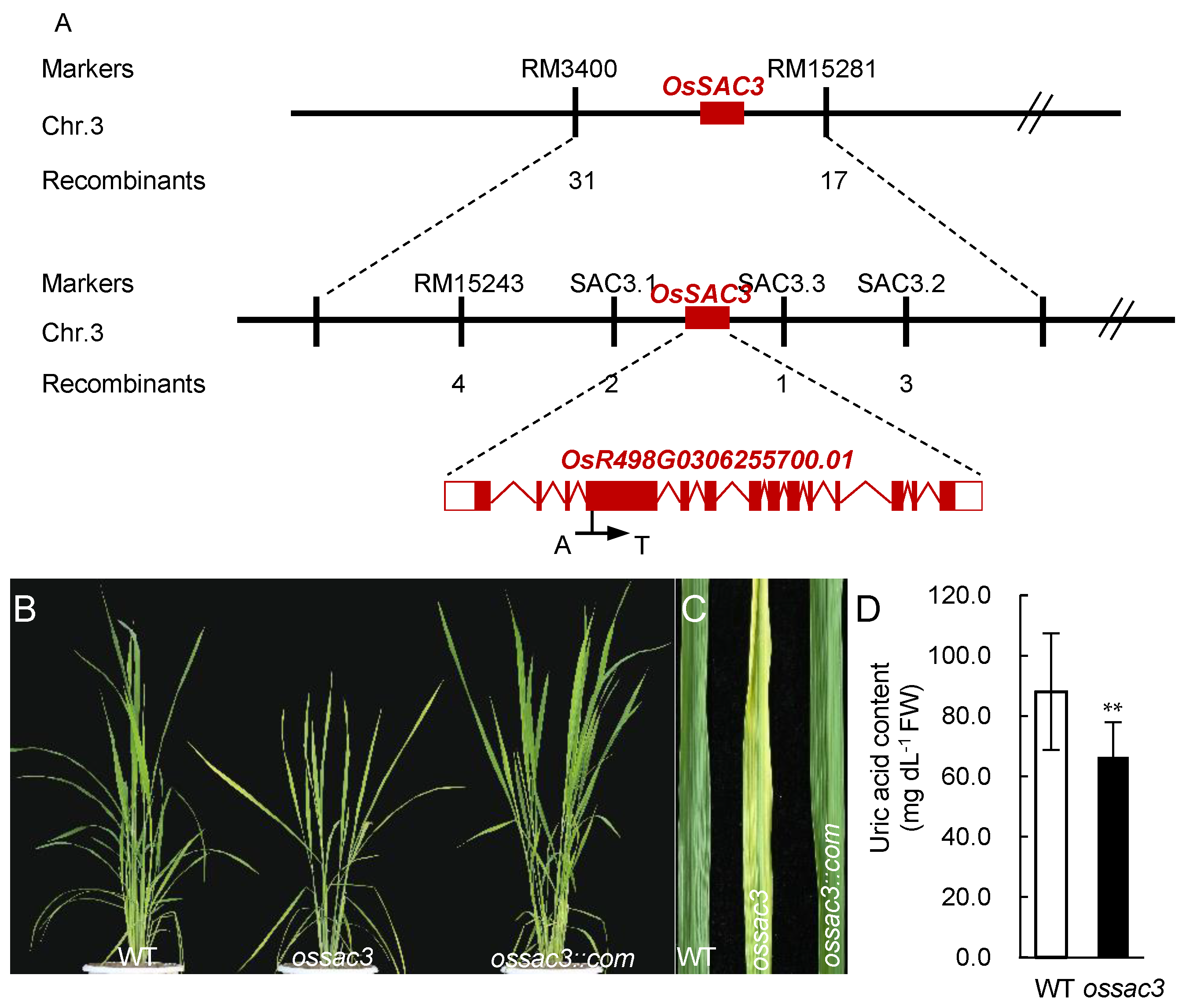
2.1. Map-Based Clone and Identification of OsSAC3
2.2. Bioinformatic Analysis of OsSAC3
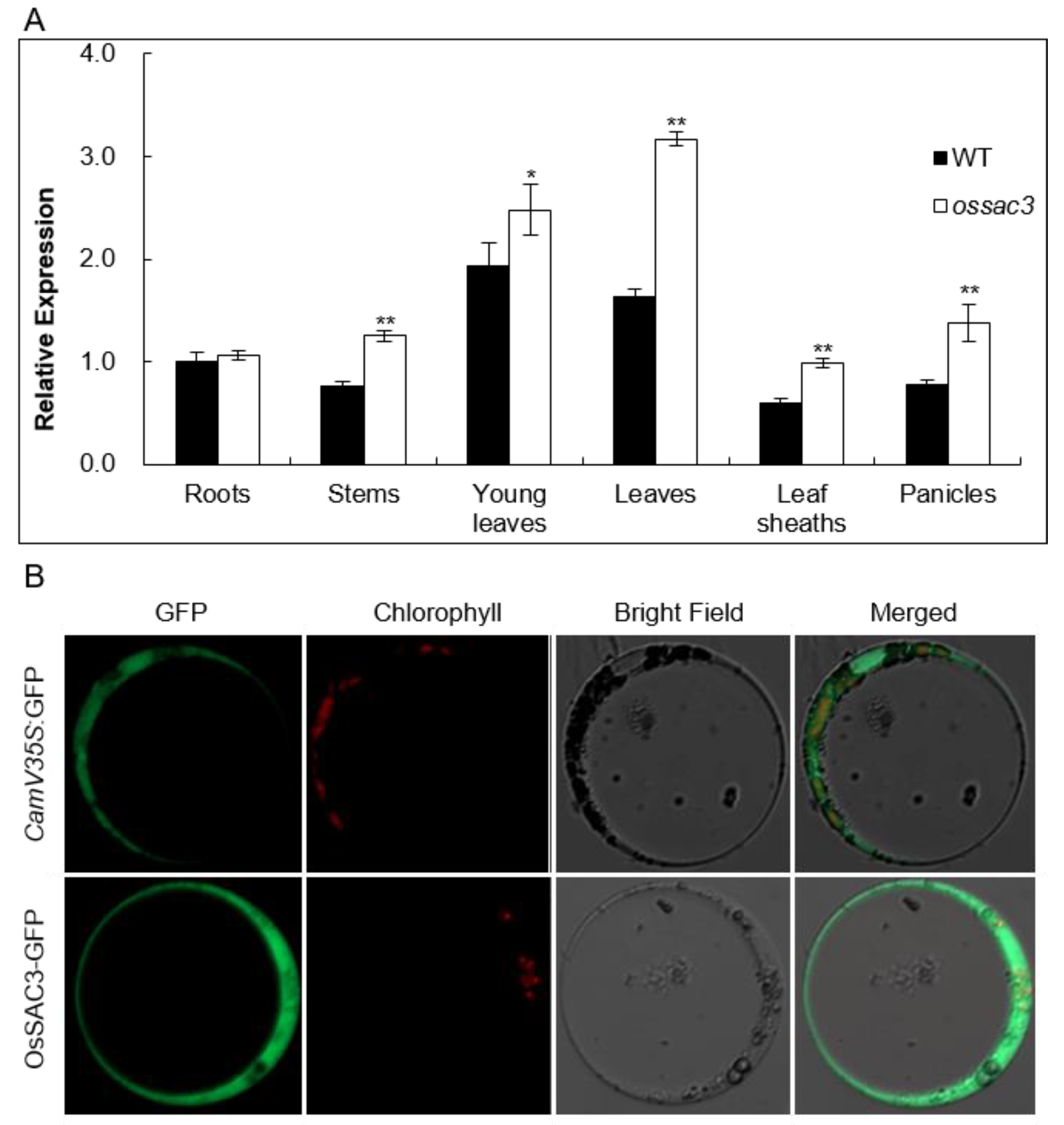
2.3. OsSAC3 Encodes a Cytoplasmic Xanthine Dehydrogenase with Constitutively Expression Pattern
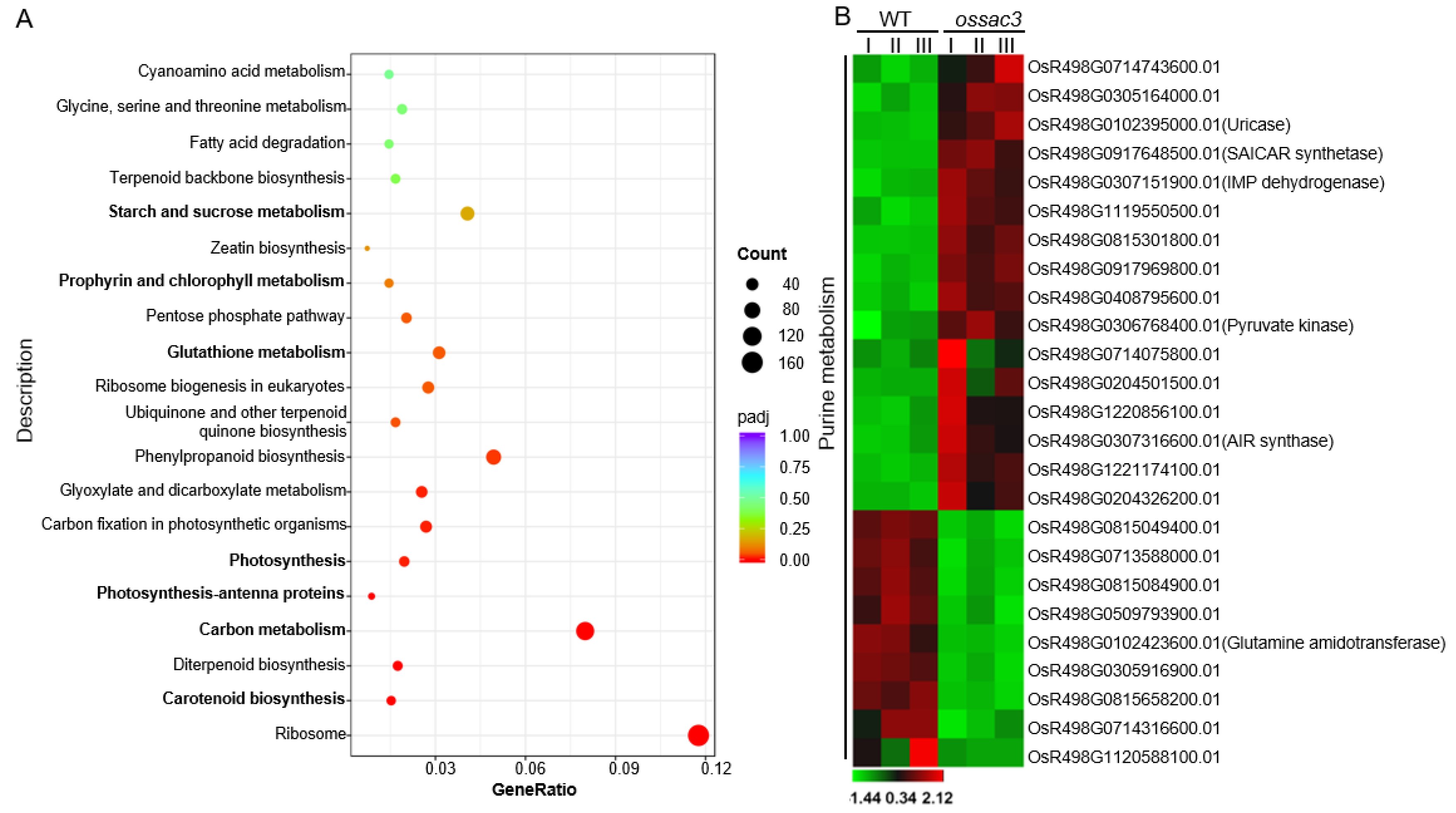
2.4. Transcriptome and Gene Ontology Enrichment Analysis
2.5. Gene Expression Related to Purine Metabolism Was Altered in ossac3
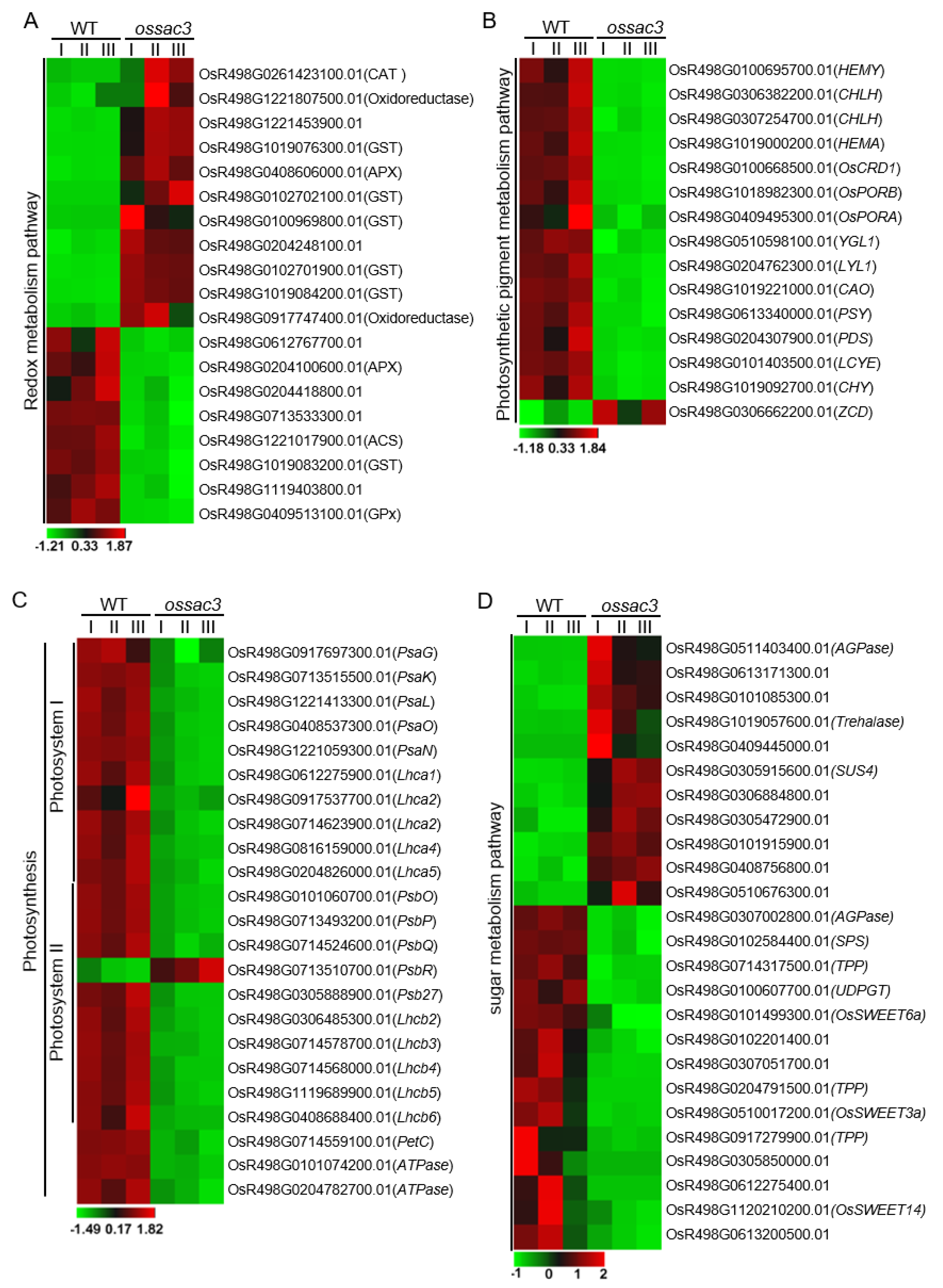
2.6. Redox Process Was Altered in ossac3
2.7. Photosynthesis Was Damaged in ossac3
2.8. Mutation of OsSAC3 Alters Expression of Sugar Metabolism/Distribution Related Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Fine Mapping and Isolation of the OsSAC3 Gene
4.3. Physical and Chemical Analysis
4.4. Multiple Sequence Alignment and Evolutionary Analysis
4.5. Vector Construction, Transformation and Identification
4.6. Gene Expression Analysis
4.7. Transcriptome and KEGG Pathway Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leng, Y.; Ye, G.; Zeng, D. Genetic dissection of leaf senescence in rice. Int. J. Molecualr Sci. 2017, 18, 2686. [Google Scholar] [CrossRef]
- Dai, N.; Schaffer, A.; Petreikov, M.; Shahak, Y.; Giller, Y.; Ratner, K.; Levine, A.; Granot, D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 1999, 11, 1253–1266. [Google Scholar] [CrossRef]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.-H.; Liu, Y.-X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Wingler, A.; Delatte, T.L.; O′Hara, L.; Primavesi, L.; Jhurreea, D.; Paul, M.; Schluepmann, H. Trehalose 6-Phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 2012, 158, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, W.; Huang, J.; Zhang, T.; Zhang, X.; Cui, Y.; Sang, X.; Ling, Y.; Li, Y.; Wang, N.; et al. Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa). Plant Cell Physiol. 2018, 59, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Alikulov, Z.; Fluhr, R.; Sagi, M. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 2008, 54, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, H.K.; Glassman, E.; Hadorn, E. Hypoxanthine in Rosy and Maroon-like Mutants of Drosophila melanogaster. Science 1959, 129, 268–269. [Google Scholar] [CrossRef]
- Glatigny, A.; Scazzocchio, C. Cloning and Molecular Characterization of hxA, the Gene Coding for the Xanthine Dehydrogenase (Purine Hydroxylase I) of Aspergillus nidulans. J. Biol. Chem. 1995, 270, 3534–3550. [Google Scholar] [CrossRef]
- Leimkühler, S.; Hodson, R.; George, G.; Rajagopalan, K.V. Recombinant Rhodobacter capsulatus Xanthine Dehydrogenase, a Useful Model System for the Characterization of Protein Variants Leading to Xanthinuria I in Humans. J. Biol. Chem. 2003, 278, 20802–20811. [Google Scholar] [CrossRef]
- Nakagawa, A.; Sakamoto, S.; Takahashi, M.; Morikawa, H.; Sakamoto, A. The RNAi-mediated silencing of xanthine dehydrogenase impairs growth and fertility and accelerates leaf senescence in transgenic Arabidopsis plants. Plant Cell Physiol. 2007, 48, 1484–1495. [Google Scholar] [CrossRef]
- Huang, J.; Yan, M.; Zhu, X.; Zhang, T.; Shen, W.; Yu, P.; Wang, Y.; Sang, X.; Yu, G.; Zhao, B.; et al. Gene mapping of starch accumulation and premature leaf senescence in the ossac3 mutant of rice. Euphytica 2018, 214, 177. [Google Scholar] [CrossRef]
- Nishino, T.; Okamoto, K.; Eger, B.T.; Pai, E.F.; Nishino, T. Mammalian xanthine oxidoreductase—mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 2008, 275, 3278–3289. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Okamoto, K.; Kawaguchi, Y.; Hori, H.; Matsumura, T.; Eger, B.T.; Pai, E.F.; Nishino, T. Mechanism of the conversion of xanthine dehydrogenase to xanthine oxidase: Identification of the two cysteine disulfide bonds and crystal structure of a non-convertible rat liver xanthine dehydrogenase mutant. J. Biol. Chem. 2005, 280, 24888–24894. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Okamoto, K. The role of the [2Fe–2S] cluster centers in xanthine oxidoreductase. J. Inorg. Biochem. 2000, 82, 43–49. [Google Scholar] [CrossRef]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine piosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Saed-Moucheshi, A.; Shekoofa, A.; Pessarakli, M. Reactive Oxygen Species (ROS) generation and detoxifying in plants. J. Plant Nutr. 2014, 37, 1573–1585. [Google Scholar] [CrossRef]
- Hilaire, E.; Young, S.A.; Willard, L.H.; McGee, J.D.; Sweat, T.; Chittoor, J.M.; Guikema, J.A.; Leach, J.E. Vascular Defense Responses in Rice: Peroxidase Accumulation in Xylem Parenchyma Cells and Xylem Wall Thickening. Mol. Plant-Microbe Interact. 2001, 14, 1411–1419. [Google Scholar] [CrossRef]
- Passardi, F.; Longet, D.; Penel, C.; Dunand, C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 2004, 65, 1879–1893. [Google Scholar] [CrossRef]
- Dixon, D.P.; Cummins, I.; Cole, D.J.; Edwards, R. Glutathione-mediated detoxification systems in plants. Curr. Opin. Plant Biol. 1998, 1, 258–266. [Google Scholar] [CrossRef]
- Estévez, I.H.; Hernández, M.R. Plant Glutathione S-transferases: An overview. Plant Gene. 2020, 3, 100233–100238. [Google Scholar] [CrossRef]
- Moons, A. Osgstu3 and osgtu4, encoding tau class glutathioneS-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots1. FEBS Lett. 2003, 553, 427–432. [Google Scholar] [CrossRef]
- Jing, X.-Q.; Zhou, M.-R.; Nie, X.-M.; Zhang, L.; Shi, P.-T.; Shalmani, A.; Miao, H.; Li, W.-Q.; Liu, W.-T.; Chen, K.-M. OsGSTU6 Contributes to Cadmium Stress Tolerance in Rice by Involving in Intracellular ROS Homeostasis. J. Plant Growth Regul. 2020, 40, 945–961. [Google Scholar] [CrossRef]
- Cho, H.-Y.; Lee, H.J.; Kong, K.-H. A phi class glutathione S-transferase from Oryza sativa (OsGSTF5): Molecular cloning, expression and biochemical characteristics. J. Biochem. Mol. Biol. 2007, 40, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef]
- Quian-Ulloa, R.; Stange, C. Carotenoid biosynthesis and plastid development in plants: The role of light. Int. J. Mol. Sci. 2021, 22, 1184. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Su, X.; Pan, X.; Liu, Z.; Chang, W.; Li, M. Structure, assembly and energy transfer of plant photosystem II supercomplex. Biochim. Biophys. Acta 2018, 1859, 633–644. [Google Scholar] [CrossRef]
- Schöttler, M.A.; Albus, C.A.; Bock, R. Photosystem I: Its biogenesis and function in higher plants. J. Plant Physiol. 2011, 168, 1452–1461. [Google Scholar] [CrossRef]
- Kotting, O.; Kossmann, J.; Zeeman, S.C.; Lloyd, J.R. Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 2010, 13, 320–328. [Google Scholar] [CrossRef]
- Ohdan, T.; Francisco, P.B.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Tang, Z.; Zhang, Y.; Niu, L.; Yang, F.; Zhang, D.; Hu, Y. Rice SUT and SWEET transporters. Int. J. Mol. Sci. 2021, 22, 11198. [Google Scholar] [CrossRef] [PubMed]
- Fei, H.; Yang, Z.; Lu, Q.; Wen, X.; Zhang, Y.; Zhang, A.; Lu, C. OsSWEET14 cooperates with OsSWEET11 to contribute to grain filling in rice. Plant Sci. 2021, 306, 110851. [Google Scholar] [CrossRef] [PubMed]
- Morii, M.; Sugihara, A.; Takehara, S.; Kanno, Y.; Kawai, K.; Hobo, T.; Hattori, M.; Yoshimura, H.; Seo, M.; Ueguchi-Tanaka, M. The dual function of OsSWEET3a as a gibberellin and glucose transporter is important for young shoot development in rice. Plant Cell Physiol. 2020, 61, 1935–1945. [Google Scholar] [CrossRef]
- Hesberg, C. Tandem Orientation of Duplicated Xanthine Dehydrogenase Genes from Arabidopsis thaliana differential gene expression and enzyme activities. J. Biol. Chem. 2004, 279, 13547–13554. [Google Scholar] [CrossRef] [PubMed]
- Enroth, C.; Eger, B.T.; Okamoto, K.; Nishino, T.; Pai, E.F. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc. Natl. Acad. Sci. USA 2000, 97, 10723–10728. [Google Scholar] [CrossRef]
- Kameda, H.; Hirabayashi, K.; Wada, K.; Fukuyama, K. Mapping of Protein-Protein Interaction Sites in the Plant-Type [2Fe-2S] Ferredoxin. PLoS ONE 2011, 6, e21947. [Google Scholar] [CrossRef]
- Ichida, K.; Amaya, Y.; Kamatani, N.; Nishino, T.; Hosoya, T.; Sakai, O. Identification of two mutations in human xanthine dehydrogenase gene responsible for classical type I xanthinuria. J. Clin. Investig. 1997, 99, 2391–2397. [Google Scholar] [CrossRef]
- Lu, J.; Hou, X.; Yuan, X.; Cui, L.; Liu, Z.; Li, X.; Ma, L.; Cheng, X.; Xin, Y.; Wang, C.; et al. Knockout of the urate oxidase gene provides a stable mouse model of hyperuricemia associated with metabolic disorders. Kidney Int. 2018, 93, 69–80. [Google Scholar] [CrossRef]
- Tasaki, E.; Sakurai, H.; Nitao, M.; Matsuura, K.; Iuchi, Y. Uric acid, an important antioxidant contributing to survival in termites. PLoS ONE 2017, 12, e0179426. [Google Scholar] [CrossRef]
- Hauck, O.K.; Scharnberg, J.; Escobar, N.M.; Wanner, G.; Giavalisco, P.; Witte, C.-P. Uric Acid Accumulation in an Arabidopsis Urate Oxidase Mutant Impairs Seedling Establishment by Blocking Peroxisome Maintenance. Plant Cell 2014, 26, 3090–3100. [Google Scholar] [CrossRef] [Green Version]
- Stinefelt, B.; Leonard, S.S.; Blemings, K.P.; Shi, X.; Klandorf, H. Free radical scavenging, DNA protection, and inhibition of lipid peroxidation mediated by uric acid. Ann. Clin. Lab. Sci. 2005, 35, 37–45. [Google Scholar]
- Dietz, K.-J.; Pfannschmidt, T. Novel Regulators in Photosynthetic Redox Control of Plant Metabolism and Gene Expression. Plant Physiol. 2011, 155, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Katsiaryna, S.; Libero, G.; Francesca, S.; Paolo, T.; Andreas, B. Redox regulation of starch metabolism. Front. Plant Sci. 2018, 9, 1344–1352. [Google Scholar]
- Mikkelsen, R.; Mutenda, K.E.; Mant, A.; Schürmann, P.; Blennow, A. α-Glucan, water dikinase (GWD): A plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. USA 2005, 102, 1785–1790. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.M.; Silva, L.P.; Issakidis-Bourguet, E.; Glaring, M.A.; Schriemer, D.C.; Moorhead, G.B.G. Insight into the redox regulation of the phosphoglucan phosphatase SEX4 involved in starch degradation. FEBS J. 2013, 280, 538–548. [Google Scholar] [CrossRef]
- Valerio, C.; Costa, A.; Marri, L.; Issakidis-Bourguet, E.; Pupillo, P.; Trost, P.; Sparla, F. Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 2010, 62, 545–555. [Google Scholar] [CrossRef]
- Seung, D.; Thalmann, M.; Sparla, F.; Hachem, M.A.; Lee, S.K.; Issakidis-Bourguet, E.; Svensson, B.; Zeeman, S.; Santelia, D. Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic α-amylase. J. Biol. Chem. 2013, 288, 33620–33633. [Google Scholar] [CrossRef]
- Akihiro, T.; Mizuno, K.; Fujimura, T. Gene Expression of ADP-glucose Pyrophosphorylase and Starch Contents in Rice Cultured Cells are Cooperatively Regulated by Sucrose and ABA. Plant Cell Physiol. 2005, 46, 937–946. [Google Scholar] [CrossRef]
- You, S.; Zhu, B.; Wang, F.; Han, H.; Sun, M.; Zhu, H.; Peng, R.; Yao, Q. A Vitis vinifera xanthine dehydrogenase gene, VvXDH, enhances salinity tolerance in transgenic Arabidopsis. Plant Biotechnol. Rep. 2017, 11, 147–160. [Google Scholar] [CrossRef]
- Watanabe, S.; Nakagawa, A.; Izumi, S.; Shimada, H.; Sakamoto, A. RNA interference-mediated suppression of xanthine dehydrogenase reveals the role of purine metabolism in drought tolerance in Arabidopsis. FEBS Lett. 2010, 584, 1181–1186. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Z.; Zhao, B.; Zhang, M.; Sang, X.; Zhao, F.; Feng, P.; He, G.; Zhu, X. Mutation of OsSAC3, Encoding the Xanthine Dehydrogenase, Caused Early Senescence in Rice. Int. J. Mol. Sci. 2022, 23, 11053. https://doi.org/10.3390/ijms231911053
Xie Z, Zhao B, Zhang M, Sang X, Zhao F, Feng P, He G, Zhu X. Mutation of OsSAC3, Encoding the Xanthine Dehydrogenase, Caused Early Senescence in Rice. International Journal of Molecular Sciences. 2022; 23(19):11053. https://doi.org/10.3390/ijms231911053
Chicago/Turabian StyleXie, Ziyu, Bingbing Zhao, Mengxue Zhang, Xianchun Sang, Fangming Zhao, Ping Feng, Guanghua He, and Xiaoyan Zhu. 2022. "Mutation of OsSAC3, Encoding the Xanthine Dehydrogenase, Caused Early Senescence in Rice" International Journal of Molecular Sciences 23, no. 19: 11053. https://doi.org/10.3390/ijms231911053



